Submitted:
26 December 2023
Posted:
27 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Yersinia enterocolitica and Yersinia pseudotuberculosis strains
2.2. Phenotypic characterization of Yersinia strains
2.3. Genotypic characterization of Yersinia strains
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-Based Phylogeny and Taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales Ord. Nov. Divided into the Families Enterobacteriaceae, Erwiniaceae Fam. Nov., Pectobacteriaceae Fam. Nov., Yersiniaceae Fam. Nov., Hafniaceae Fam. Nov., Morganellaceae Fam. Nov., and Budviciaceae Fam. Nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson-Ahomaa, M. Enteropathogenic Yersinia Spp. In Zoonoses: Infections Affecting Humans and Animals; Sing, A., Ed.; Springer International Publishing: Cham, 2023; pp. 329–353. ISBN 978-3-031-27164-9. [Google Scholar]
- EFSA; ECDC The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, e8442. [CrossRef]
- Bonke, R.; Wacheck, S.; Stüber, E.; Meyer, C.; Märtlbauer, E.; Fredriksson-Ahomaa, M. Antimicrobial Susceptibility and Distribution of β-Lactamase A (BlaA) and β-Lactamase B (BlaB) Genes in Enteropathogenic Yersinia Species. Microb. Drug Resist. 2011, 17, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Koskinen, J.; Ortiz-Martínez, P.; Keto-Timonen, R.; Joutsen, S.; Fredriksson-Ahomaa, M.; Korkeala, H. Prudent Antimicrobial Use Is Essential to Prevent the Emergence of Antimicrobial Resistance in Yersinia Enterocolitica 4/O: 3 Strains in Pigs. Front. Microbiol. 2022, 13, 841841. [Google Scholar] [CrossRef] [PubMed]
- Guillier, L.; Fravalo, P.; Leclercq, A.; Thébaut, A.; Kooh, P.; Cadavez, V.; Gonzales-Barron, U. Risk Factors for Sporadic Yersinia Enterocolitica Infections: A Systematic Review and Meta-Analysis. Microb. Risk Anal. 2020, 100141. [Google Scholar] [CrossRef]
- Rosner, B.; Stark, K.; Höhle, M.; Werber, D. Risk Factors for Sporadic Yersinia Enterocolitica Infections, Germany 2009–2010. Epidemiol. Infect. 2012, 140, 1738–1747. [Google Scholar] [CrossRef]
- Boqvist, S.; Pettersson, H.; Svensson, Å.; Andersson, Y. Sources of Sporadic Yersinia Enterocolitica Infection in Children in Sweden, 2004: A Case-Control Study. Epidemiol. Infect. 2009, 137, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Fredriksson-Ahomaa, M.; Korte, T.; Korkeala, H. Transmission of Yersinia Enterocolitica 4/O:3 to Pets via Contaminated Pork. Lett. Appl. Microbiol. 2001, 32, 375–378. [Google Scholar] [CrossRef]
- Stamm, I.; Hailer, M.; Depner, B.; Kopp, P.A.; Rau, J. Yersinia Enterocolitica in Diagnostic Fecal Samples from European Dogs and Cats: Identification by Fourier Transform Infrared Spectroscopy and Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2013, 51, 887–893. [Google Scholar] [CrossRef]
- Laukkanen-Ninios, R.; Didelot, X.; Jolley, K.A.; Morelli, G.; Sangal, V.; Kristo, P.; Brehony, C.; Imori, P.F.; Fukushima, H.; Siitonen, A. Population Structure of the Yersinia Pseudotuberculosis Complex According to Multilocus Sequence Typing. Environ. Microbiol. 2011, 13, 3114–3127. [Google Scholar] [CrossRef]
- Sen, K. Rapid Identification of Yersinia Enterocolitica in Blood by the 5′ Nuclease PCR Assay. J. Clin. Microbiol. 2000, 38, 1953–1958. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, H.; Hensel, A.; Aleksic, S.; Meyer, H. Identification of Yersinia Enterocolitica within the Genus Yersinia. Syst. Appl. Microbiol. 2000, 23, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, H.N.; Rasmussen, O.F.; Andersen, J.K.; Olsen, J.E. Specific Detection of Pathogenic Yersinia Enterocolitica by Two-Step PCR Using Hot-Start and DMSO. Mol. Cell. Probes 1994, 8, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, H.; Inoue, M.; Mori, T.; Itoh, K.; Arakawa, E.; Watanabe, H. Detection and Identification of Yersinia Pseudotuberculosis and Pathogenic Yersinia Enterocolitica by an Improved Polymerase Chain Reaction Method. J. Clin. Microbiol. 1992, 30, 2484–2486. [Google Scholar] [CrossRef] [PubMed]
- Lambertz, S.T.; Nilsson, C.; Hallanvuo, S.; Lindblad, M. Real-Time PCR Method for Detection of Pathogenic Yersinia Enterocolitica in Food. Appl. Environ. Microbiol. 2008, 74, 6060–6067. [Google Scholar] [CrossRef] [PubMed]
- Lambertz, S.T.; Nilsson, C.; Hallanvuo, S. TaqMan-Based Real-Time PCR Method for Detection of Yersinia Pseudotuberculosis in Food. Appl. Environ. Microbiol. 2008, 74, 6465–6469. [Google Scholar] [CrossRef]
- Thoerner, P.; Bin Kingombe, C.; Bogli-Stuber, K.; Bissig-Choisat, B.; Wassenaar, T.; Frey, J.; Jemmi, T. PCR Detection of Virulence Genes in Yersinia Enterocolitica and Yersinia Pseudotuberculosis and Investigation of Virulence Gene Distribution. Appl. Environ. Microbiol. 2003, 69, 1810–1816. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, H.; Tsunomori, Y.; Seki, R. Duplex Real-Time SYBR Green PCR Assays for Detection of 17 Species of Food-or Waterborne Pathogens in Stools. J. Clin. Microbiol. 2003, 41, 5134–5146. [Google Scholar] [CrossRef] [PubMed]
- Wren, B.; Tabaqchali, S. Detection of Pathogenic Yersinia Eriterocolitica by the Polymerase Chain Reaction. The Lancet 1990, 336, 693. [Google Scholar] [CrossRef]
- Weynants, V.; Jadot, V.; Denoel, P.A.; Tibor, A.; Letesson, J.-J. Detection of Yersinia Enterocolitica Serogroup O: 3 by a PCR Method. J. Clin. Microbiol. 1996, 34, 1224–1227. [Google Scholar] [CrossRef]
- Jacobsen, N.; Bogdanovich, T.; Skurnik, M.; Lübeck, P.; Ahrens, P.; Hoorfar, J. A Real-Time PCR Assay for the Specific Identification of Serotype O: 9 of Yersinia Enterocolitica. J. Microbiol. Methods 2005, 63, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Fenwick, S.G.; Madie, P.; Wilks, C.R. Duration of Carriage and Transmission of Yersinia Enterocolitica Biotype 4, Serotype 0:3 in Dogs. Epidemiol. Infect. 1994, 113, 471–477. [Google Scholar] [CrossRef]
- Joutsen, S.; Laukkanen-Ninios, R.; Henttonen, H.; Niemimaa, J.; Voutilainen, L.; Kallio, E.R.; Helle, H.; Korkeala, H.; Fredriksson-Ahomaa, M. Yersinia Spp. in Wild Rodents and Shrews in Finland. Vector-Borne Zoonotic Dis. 2017, 17, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Joutsen, S.; Eklund, K.-M.; Laukkanen-Ninios, R.; Stephan, R.; Fredriksson-Ahomaa, M. Sheep Carrying Pathogenic Yersinia Enterocolitica Bioserotypes 2/O:9 and 5/O:3 in the Feces at Slaughter. Vet. Microbiol. 2016, 197, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Sihvonen, L.M.; Jalkanen, K.; Huovinen, E.; Toivonen, S.; Corander, J.; Kuusi, M.; Skurnik, M.; Siitonen, A.; Haukka, K. Clinical Isolates of Yersinia Enterocolitica Biotype 1A Represent Two Phylogenetic Lineages with Differing Pathogenicity-Related Properties. BMC Microbiol. 2012, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Blomvall, L.; Pelkola, K.; Lienemann, T.; Lehtoniemi, S.; Pohjola, L.; Fredriksson-Ahomaa, M. Osteomyelitis in a Slaughter Turkey Flock Caused by Yersinia Pseudotuberculosis Sequence Type ST42. Vet. Microbiol. 2022, 269, 109424. [Google Scholar] [CrossRef] [PubMed]
- Castro, H.; Jaakkonen, A.; Hakakorpi, A.; Hakkinen, M.; Isidro, J.; Korkeala, H.; Lindström, M.; Hallanvuo, S. Genomic Epidemiology and Phenotyping Reveal On-Farm Persistence and Cold Adaptation of Raw Milk Outbreak-Associated Yersinia Pseudotuberculosis. Front. Microbiol. 2019, 10, 1049. [Google Scholar] [CrossRef] [PubMed]
- Cabanel, N.; Galimand, M.; Bouchier, C.; Chesnokova, M.; Klimov, V.; Carniel, E. Molecular Bases for Multidrug Resistance in Yersinia Pseudotuberculosis. Int. J. Med. Microbiol. 2017, 307, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, P.A.; Tano, E.; Jernberg, C.; Hickman, R.A.; Guy, L.; Järhult, J.D.; Wang, H. Molecular Characterization of Multidrug-Resistant Yersinia Enterocolitica from Foodborne Outbreaks in Sweden. Front. Microbiol. 2021, 12, 664665. [Google Scholar] [CrossRef]
- Fredriksson-Ahomaa, M.; Heikkilä, T.; Pernu, N.; Kovanen, S.; Hielm-Björkman, A.; Kivistö, R. Raw Meat-Based Diets in Dogs and Cats. Vet. Sci. 2017, 4, 33. [Google Scholar] [CrossRef]
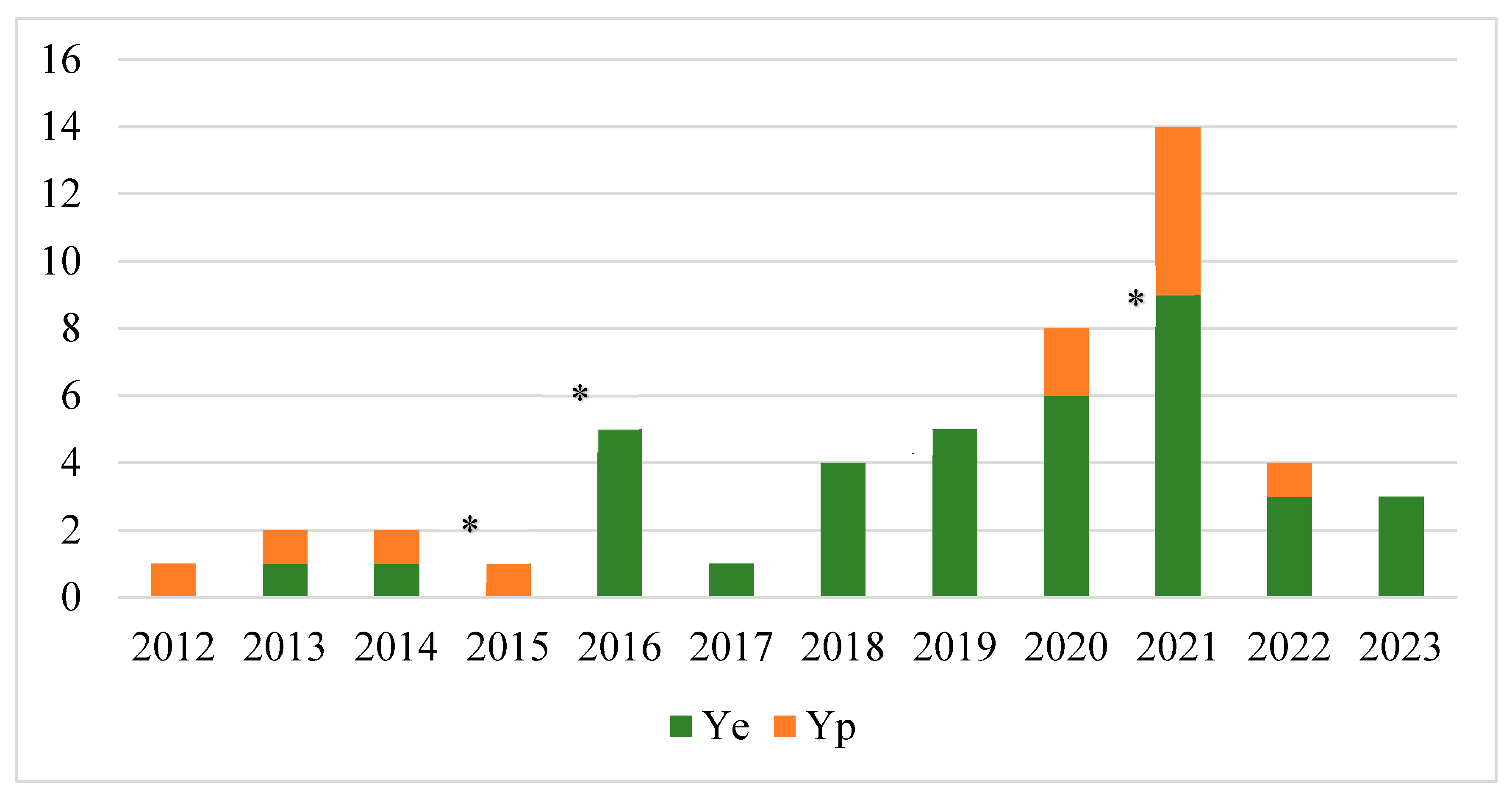
| Identification | Target | Gene | Ye | Yp | Reference |
|---|---|---|---|---|---|
| Species | 16SrRNA | rrn | x | x | [12] |
| rrn | x | [13] | |||
| Pathogenicity | Chromosome | inv | x | [14] | |
| inv | x | [15] | |||
| ail | x | [16] | |||
| ail | x | [17] | |||
| ystA | x | [18] | |||
| ystB | x | [18] | |||
| Plasmid | yadA | x | x | [19] | |
| virF | [20] | ||||
| Serotype | O:3 | rfbC | [21] | ||
| O:9 | per | [22] |
| Animal | Numberof strains | Sampletype | Ye | Yp | |
| Biotype | Serotype | Serotype | |||
| Dog | 6 | Faeces (5), abscess (1) | 1A | NTa | |
| 4 | Faeces | 2 | O:9 | ||
| 25 | Faeces | 4 | O:3 | ||
| 11 | Faeces (7), urine (2),abscess (1), blood (1) | O:1 | |||
| Cat | 2 | Faeces | 1A | NT | |
| 1 | Faeces | 4 | O:3 | ||
| 1 | Wound | O:1 | |||
| Sp. | Type | No. ofstrains | Virulence gene | Sequencetypea | ||||||
| ail | inv | irp | psaA | ystA | ystB | virF/yadA | ||||
| Ye | 1A | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 388 |
| 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 138 | ||
| 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 290, 738 | ||
| 4 | 0 | 4 | 0 | 0 | 0 | 4 | 0 | 148, 179, 365,479/551 | ||
| 2/O:9 | 4 | 4 | 4 | 0 | 4 | 4 | 0 | 4 | 139 | |
| 4/O:3 | 26 | 26 | 26 | 0 | 26 | 26 | 0 | 25 | 135 | |
| Yp | O:1 | 3 | 3 | 3 | 3 | 3 | 0 | 0 | 2 | 9 |
| 9 | 9 | 9 | 9 | 9 | 0 | 0 | 7 | 42 | ||
| Antimicrobial agent | Numberof strains | MIC (µg/mL) | AMR genepresent | |
| Breakpoint S ≤ | Observedvalue | |||
| Y. enterocolitica (n=38) | ||||
| Ampicillin | 38 | 8 | ≥32a | blaA |
| Chloramphenicol | 2 | 8 | >64 | catA1 |
| Streptomycin | 2 | NTb | aadA12 | |
| 1 | NT | aph(3)-lb, aph(6)-ld | ||
| Streptogramin | 38 | NT | vat(F) | |
| Sulfamethoxazole | 2 | NAc | >512 | sul1 |
| 1 | NA | >512 | sul2 | |
| Tetracycline | 1 | 4 | >32 | tet(A) |
| Y. pseudotuberculosis (n=12) | ||||
| Colistin | 12 | 2 | >16a | 0 |
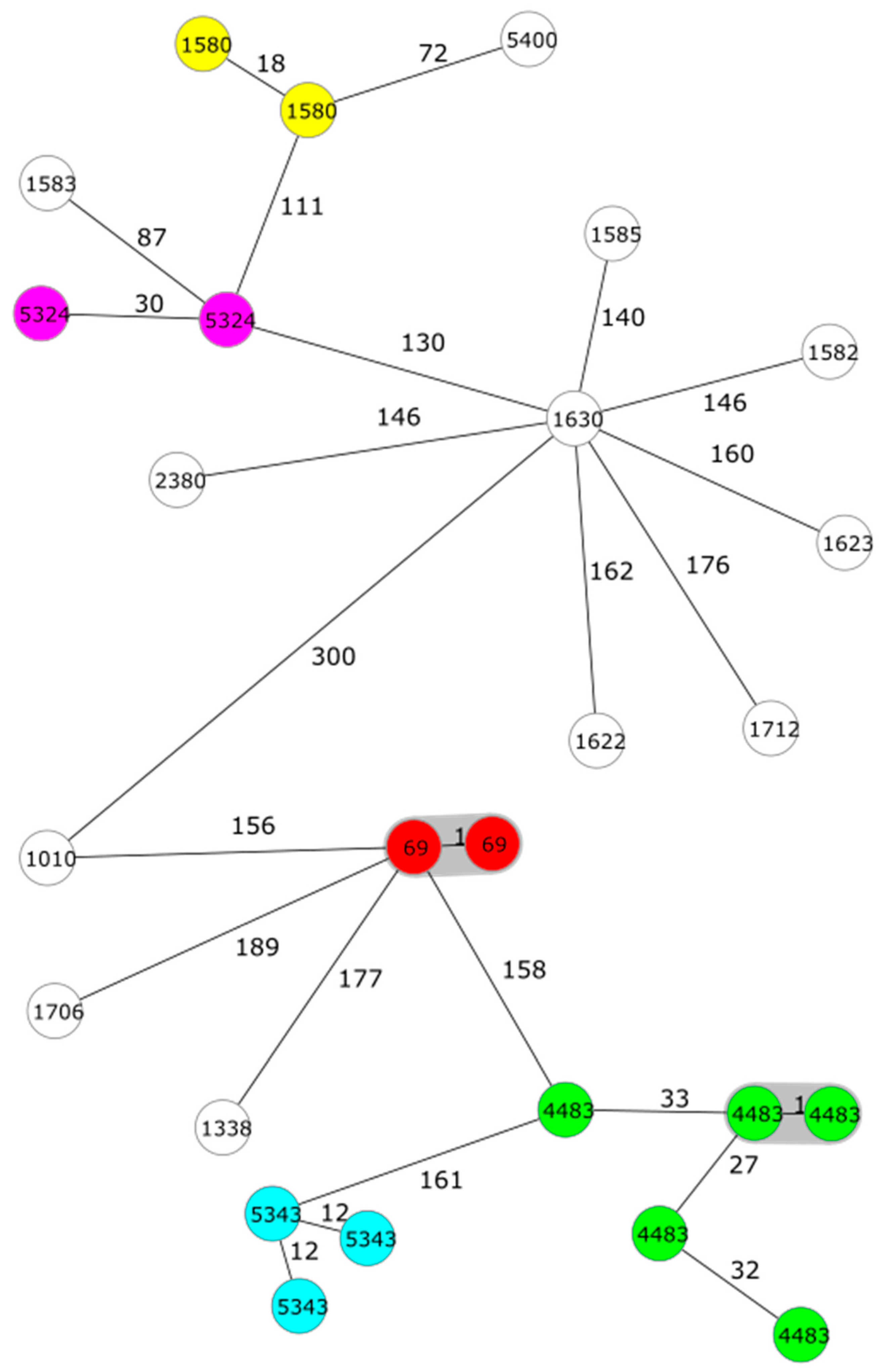
| Species | Type | ST | CTs (number of strains) | ADs |
|---|---|---|---|---|
| Ye | 4/O:3 | 135 | 69 (2), 1580 (2), 4483 (5), 5324 (2), 5343 (3) | 1–50 |
| 1583, 5400 | 51–100 | |||
| 1010, 1338, 1582, 1585, 1622, 1623, 1630, 1706, 1712, 2380 | 101–189 | |||
| 2/O:9 | 139 | 510 (2), 904, 5351 | 2-37 | |
| 1A | 138 | 5575 | 2025 | |
| 148 | 1401 | 1661 | ||
| 179 | 1295 | 1661 | ||
| 290 | 1379 | 2030 | ||
| 365 | 2282 | 2067 | ||
| 388 | 1710 | 2164 | ||
| 479/551 | 5505 | 2650 | ||
| 738 | 5332 | 2025 | ||
| Yp | O:1 | 9 | 782 (2), 806 | 20–93 |
| 42 | 816, 1728, 3900 (2), 5560 (3), 5610 (2) | 1–78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





