1. Introduction
Biomass is a renewable, carbon-neutral, abundantly available, and sustainable energy source that holds the potential to reduce or replace reliance on fossil fuels, contributing to energy savings and environmental preservation [
1]. Studies indicate that global lignocellulosic biomass, including crop residues, wood, and grass, generates approximately 181.5 billion tonnes each year [
2]. This highlights the substantial potential of utilizing agricultural residues and wood from fast−growing trees for bio-conversion. A primary concern is that biomass generally has a lower energy density compared to fossil fuels. Nevertheless, the application of diverse biomass energy technologies rooted in thermochemical treatments-namely, pyrolysis (300 to 1000 °C), gasification (500 to 1300 °C), liquefaction (250 to 550 °C), torrefaction (200 to 300 °C), and combustion (700 to 1000 °C)− displays the potential to effectively transform biomass into valuable energy resources [
3,
4].
The wide range of biomass sources, the diverse assortment of agricultural and tree waste materials, and the continuous variations in their properties during processing, storage, and transportation collectively pose challenges in effectively characterizing and understanding biomass as an efficient fuel. The characterization of biomass is crucial for reliably predicting its behavior as a fuel [
5]. In the context of the thermal conversion of biomass, proximate analysis emerges as a pivotal method for characterization [
4]. Proximate analysis is used to determine the mass percentages on a wet basis of MC, VM, FC, and ash content, all of which significantly influence both combustion behavior and plant design.
During the biomass combustion process, the rate of heat release is primarily influence by proximate analysis parameters, including MC, VM, FC, and ash content [
6]. Adjustments in these proximate analysis values can yield variations in crucial combustion performance indicators such as ignition time and temperature, burnout time and temperature, and the overall heat release. Higher moisture levels decrease the higher heating value of biomass [
7] and lead to reduced combustion efficiency, whereas higher ratios of VM to FC correspond to increased fuel reactivity [
8]. VM refers to the components in biomass that are converted into a gaseous form through thermal decomposition, whereas FC represents the non-volatile segment of the biomass. A high VM content does not necessarily ensure a high calorific value because some VM constituents are derived from non−combustible gases like CO
2 and H
2O. Lower MC and VM contents contribute to the improvement of the FC content in biomass samples [
9]. A high FC signifies a higher carbon content in the biomass, resulting in higher energy content and generating maximum heat during combustion [
10]. Therefore, the FC content of biomass positively influences its energy potential and calorific value. Conversely, a higher presence of VM in the biomass results in lower FC, consequently impacting the energy potential of the biomass negatively.
Ash is an inorganic residue remained after ignition of combustible substance [
11,
12]. The energy required for the thermal breakdown and phase transition of ash-forming inorganics is extracted from the biomass burning energy, resulting in a reduction of calorific value [
13]. The presence of high MC and elevated ash content in biomass lead to ignition and combustions problems [
5]. Also, the presence of ash influences corrosion and slag formation [
8].
Various standard procedures exist to measure these proximate analysis parameters. ASTM E−871−82 (2019) addresses moisture, ASTM E−872−82 (2019) handles volatile matter, and ASTM E1755−01 (2020) tackles ash in biomass. FC is usually determined by calculating the difference between 100% and the sum of MC, VM, and ash content [
12,
14]. These standard methods are involve heating a sample under specific conditions and assessing the change in weight. For proximate analysis of biomass, experimentation includes using a simple oven to determine MC and a furnace to determine VM and ash contents. The experimental techniques employed to estimate these parameters often consume considerable time, are financially demanding, and carry an increased likelihood of experimental error. Additionally, considering the large number of samples needed to determine the proximate analysis data for biomass characterization, the process is both tedious and destructive in nature. Hence, various researchers have recommended the use of TGA for proximate analysis of biomass. For instance, Posom et al. (2020) recommend to use TGA to determine the proximate analysis of bamboo chip and leucaena pellets biomass [
15], Torquato et al. (2017) investigated the appropriate conditions for determining proximate parameters of lignocellulosic agricultural residue using TGA [
16], and García et al. (2013) analyzed 13 different types of biomass for proximate analysis using TGA [
8]. Similarly, previous research studies have demonstrated the potential of utilizing NIR Spectroscopy, as an alternative method, for assessing biomass proximate analysis parameters across the wavelength range between 780 and 2500 nm. Posom et al. (2022) developed a reliable and accurate model for online measurement of MC in sugarcane [
17]. Toscano et al. (2022) estimated the MC of industrial wood chip fuel using a portable NIR spectrometer [
18]. Sirisomboon et al. (2020) compared the performance of proximate data models, namely MC, VM, FC, and ash, through direct scanning of bamboo chips [
19]. Shrestha et al. (2018) evaluated MC in bamboo chips using diode array near-infrared spectroscopy [
20]. Posom et al. (2017) determined the HHV, VM, FC, and A of ground bamboo using FT−NIRS [
21]. Adnan et al. (2017) assessed the feasibility of NIRS and chemometrics for rapid and non−destructive prediction of MC in intact green coffee beans [
22]. The above research demonstrates the potential of TGA for determining proximate parameters and NIRS for rapidly predicting biomass proximate parameters. Therefore, this study integrates TGA and NIRS to develop calibration models using chemometircs, specifically PLSR.
Although NIRS is a rapid, reliable, and non-destructive analytical method, it requires the development of individual calibration models based on spectral data and each reference parameter. This process can be time-consuming and costly. However, in the long term, it proves to be beneficial for the accurate assessment of proximate analysis parameters. To the best of our knowledge, there have been no publications reporting the development of a global NIRS model for proximate parameters using fast-growing trees and agricultural residue which can be catagorized into wood and non−wood biomass. In this work, we elucidate the rationale for combining both types of biomass materials for model development. Additionally, this study marks the first application of the recent multi-preprocessing methods, specifically the 5−range and 3−range methods, for the NIRS evaluation of proximate parameters in biomass. Therefore, the main aims of this study are as follows:
1. To determine the proximate analysis parameters, i.e., MC, VM, FC and ash content of biomass, using TGA.
2. To develop separate calibration models using Full−PLSR, GA−PLSR, SPA−PLSR, the multi−preprocessing 5 range−PLSR, and the multi−preprocessing 3 range−PLSR for the non−destructive assessment of each proximate analysis parameter in both chipped and ground biomass.
3. To compare and select the best−performing PLSR−based model for each proximate analysis parameter from chipped and ground biomass, and establish it as a rapid, reliable, non−destructive alternative method for assessment of proximate analysis parameters.
4. To determine the LOQ value for each proximate parameters using the calibration set of the proposed high-performance model, both for chip and ground biomass.
4. Conclusions
In this study, proximate parameters (MC, VM, FC, and ash content) were determined using a TGA. PLSR models were developed and compared utilizing NIR spectroscopy within a wavenumber range of 3594.87 to 12,489.48 cm−1 to assess these proximate analysis parameters, both in chip and ground biomass. These models were constructed from raw spectra, traditional preprocessing approaches, GA, SPA, and spectral MP with 5 range and 3 range methods.
The findings of this study underscore the potential of NIR spectroscopy as a promising alternative tool for the quantitative prediction of MC and FC in chip biomass. The selected model for MC and FC in chip biomass is suitable for most applications, including research, but should be used with caution. As for the remaining proximate parameters, i.e., MC and FC in ground biomass, and VM and ash content in both chip and ground biomass, the models exhibit fair performance and are primarily suitable for rough screening purposes. The models perform more accurately in chip biomass, highlighting the efficacy of diffuse reflectance sphere macro sample rotating mode compared to the transflectance mode of scanning in ground biomass. In addition, the chip biomass model can be preferred for proximate analysis instead of the ground biomass model, as it eliminates the need for biomass grounding, saving time, labor, and costs prior to scanning.
The model developed in this study proved to be more robust than those in previous studies. This is attributed to the wider variation in different types of biomass, including fast−growing trees and agricultural residues, whereas previous studies were focused on specific biomass species. The accuracy, robustness, and sensitivity of these models can be further enhanced by expanding the dataset with samples from diverse sources and validating their performance with unknown samples before industrial application. Implementing the selected models will benefit researchers, engineers, and industries aiming to design thermochemical conversion systems that can select suitable biomass and efficiently extract maximum energy from it, all while minimizing energy consumption and environmental impact.
Author Contributions
Conceptualization, B.S., J.P., P.S. and B.P.S; Methodology, B.S., J.P., P.S. and B.P.S; Software, B.S. and J.P.; Formal analysis, B.S. and J.P.; Investigation, B.S.; Resources, B.S. and B.P.S.; Data curation, B.S., J.P. and P.S.; Writing the original draft, B.S.; Writing–review & editing, B.S., J.P., P.S., B.P.S and A.F; Visualization, B.S.; Supervision, J.P., P.S. B.P.S and A.F; Project administration, P.S.; Funding acquisition, P.S.; Validation, B.P.S and A.F. All authors have read and agreed to the published version of the manuscript.
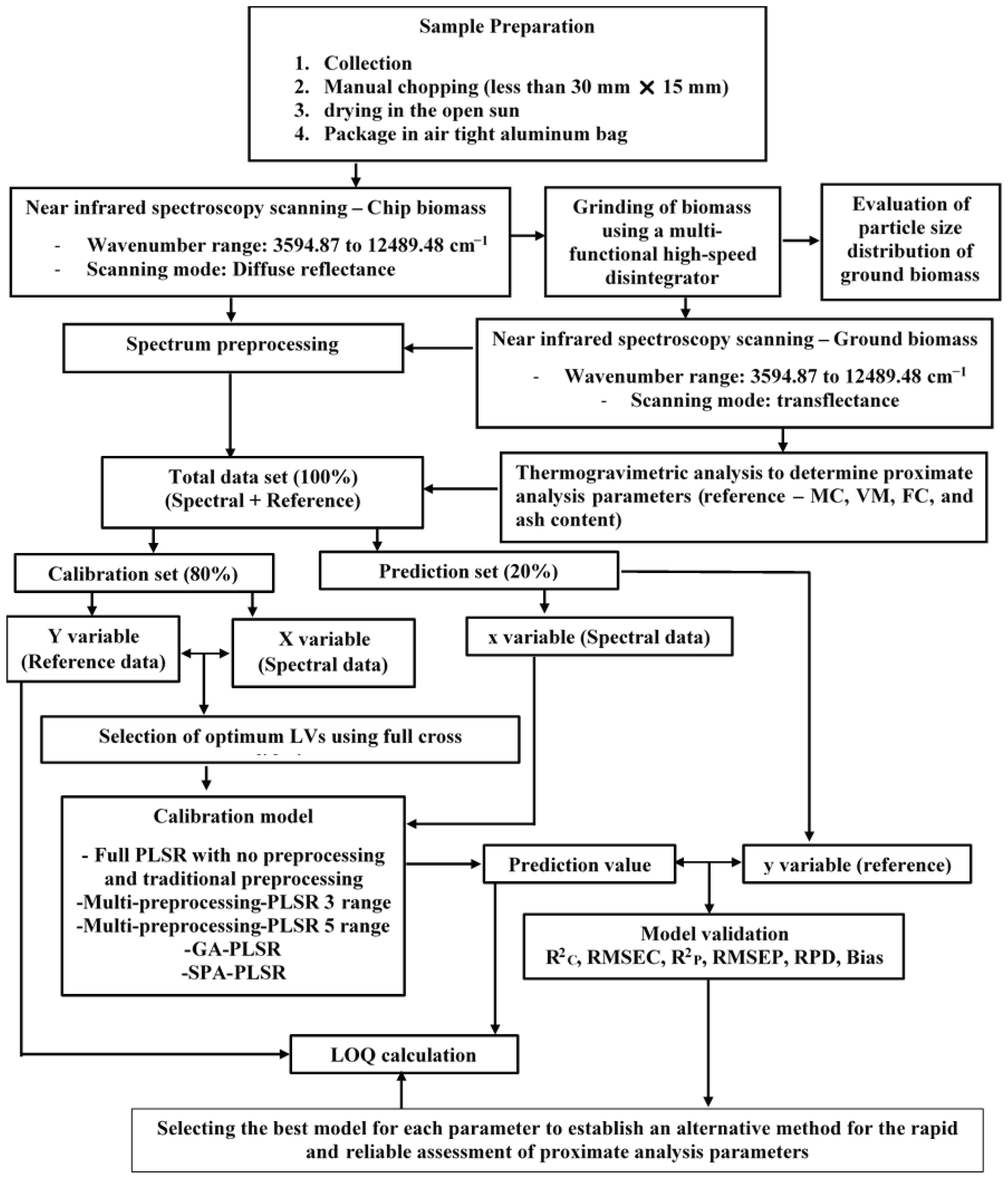
Figure 1.
Flowchart depicting the overall research methodology for the rapid assessment of biomass proximate analysis parameters using TGA, while using NIRS with PLSR is an alternative.
Figure 1.
Flowchart depicting the overall research methodology for the rapid assessment of biomass proximate analysis parameters using TGA, while using NIRS with PLSR is an alternative.
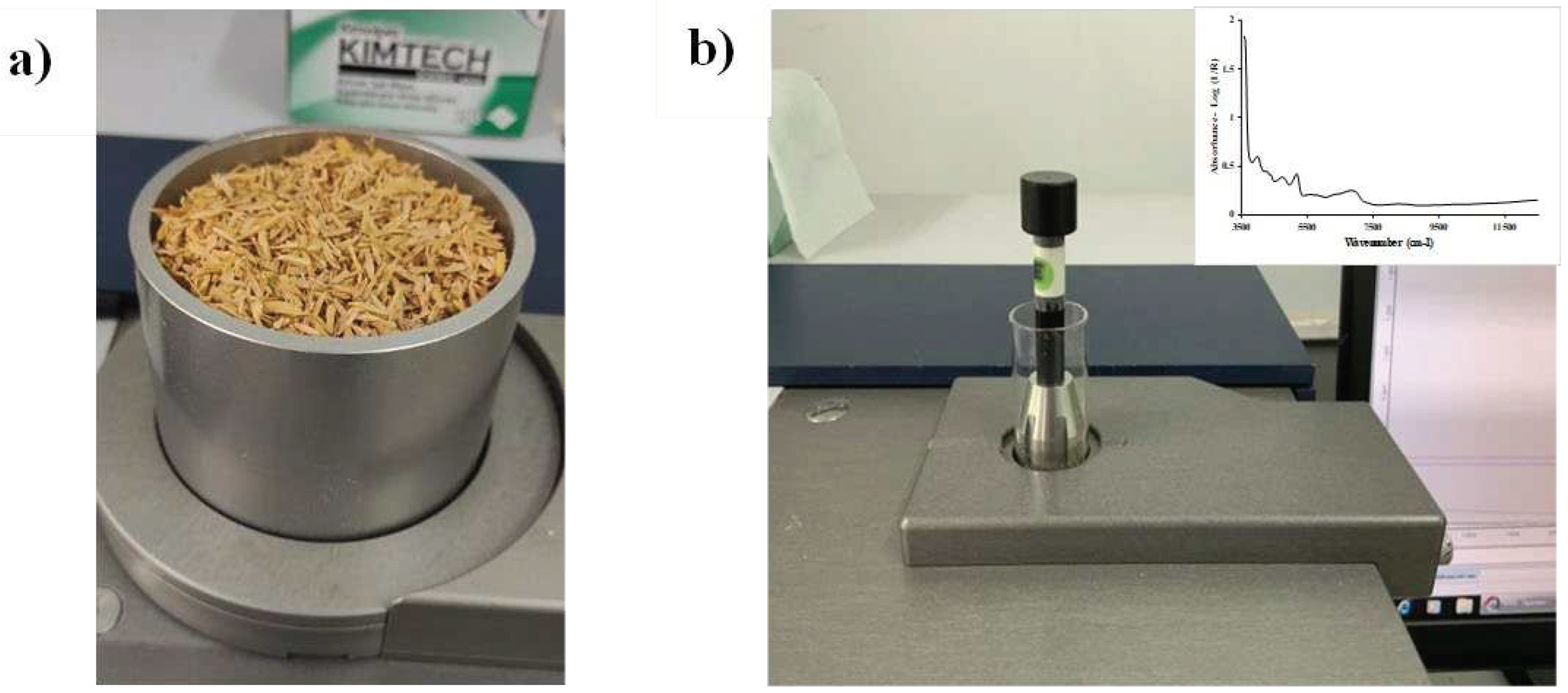
Figure 2.
FT−NIR spectrometer scanning of (a) chip biomass in diffuse reflectance and sphere macro sample rotating mode using a quartz bottom sample cup, and (b) ground biomass in transflectance mode using a glass vial.
Figure 2.
FT−NIR spectrometer scanning of (a) chip biomass in diffuse reflectance and sphere macro sample rotating mode using a quartz bottom sample cup, and (b) ground biomass in transflectance mode using a glass vial.
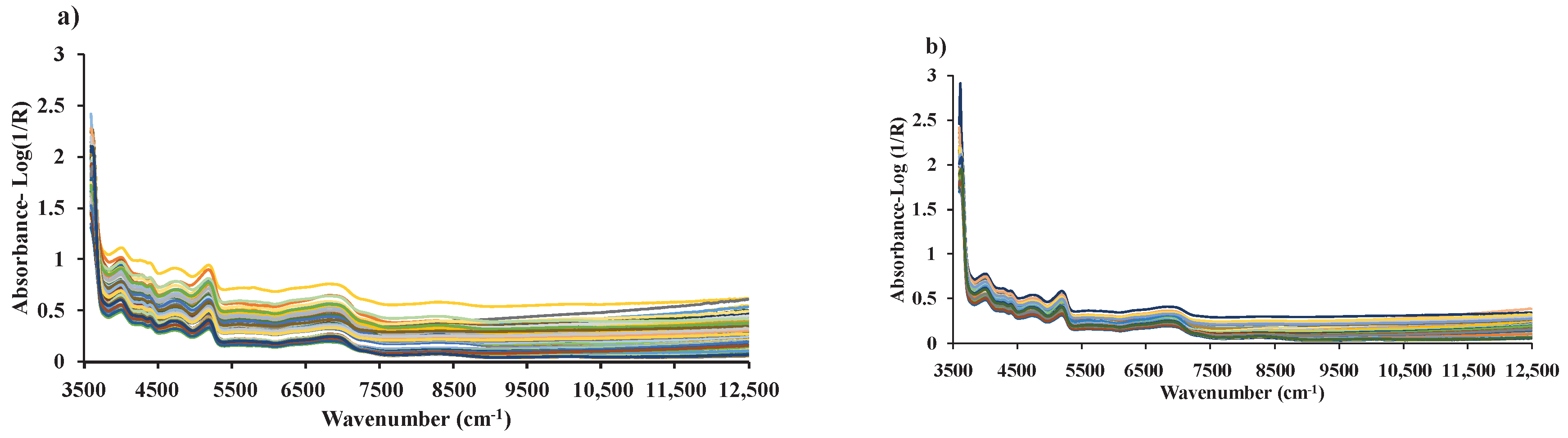
Figure 3.
Raw spectra of (a) chip biomass obtained from diffuse reflectance mode of FT−NIRS scanning and (b) ground biomass obtained from transflectance mode of FT−NIRS scanning across the entire wavenumber range of 3594.87 to 12,489.48 cm−1.
Figure 3.
Raw spectra of (a) chip biomass obtained from diffuse reflectance mode of FT−NIRS scanning and (b) ground biomass obtained from transflectance mode of FT−NIRS scanning across the entire wavenumber range of 3594.87 to 12,489.48 cm−1.
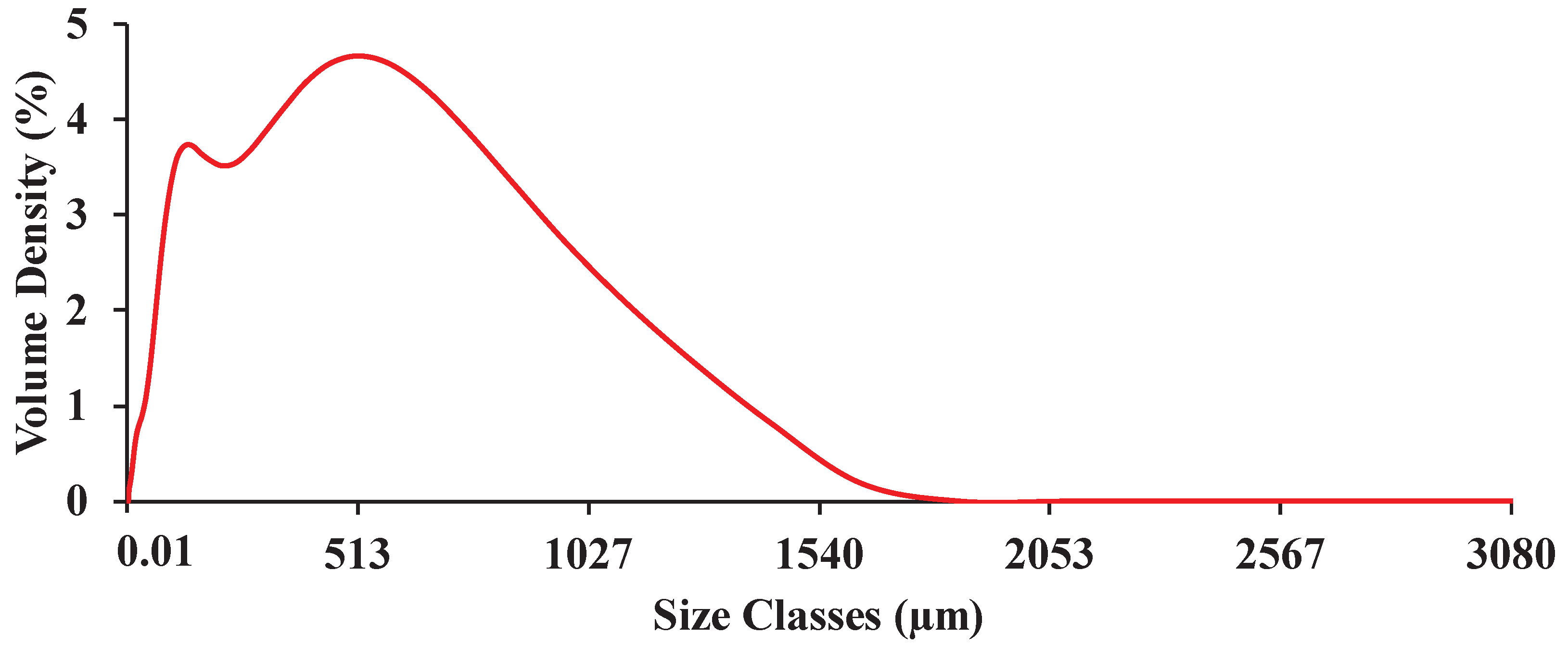
Figure 4.
Representative particle size distribution of the ground biomass ranging from 0.01 to 3080 µm.
Figure 4.
Representative particle size distribution of the ground biomass ranging from 0.01 to 3080 µm.
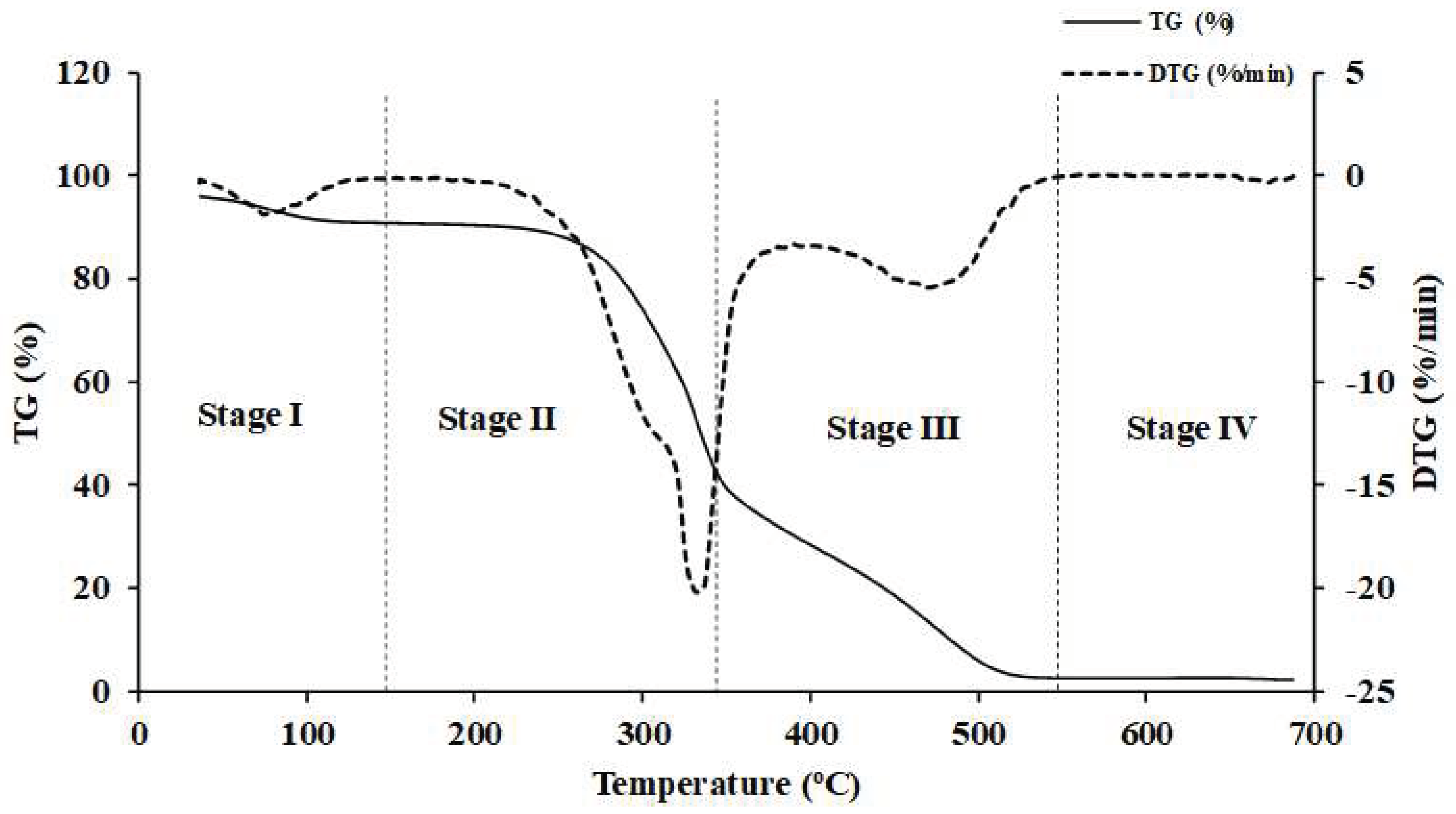
Figure 5.
The TG and DTG curves obtained from TGA analysis of Nepalese biomass.
Figure 5.
The TG and DTG curves obtained from TGA analysis of Nepalese biomass.
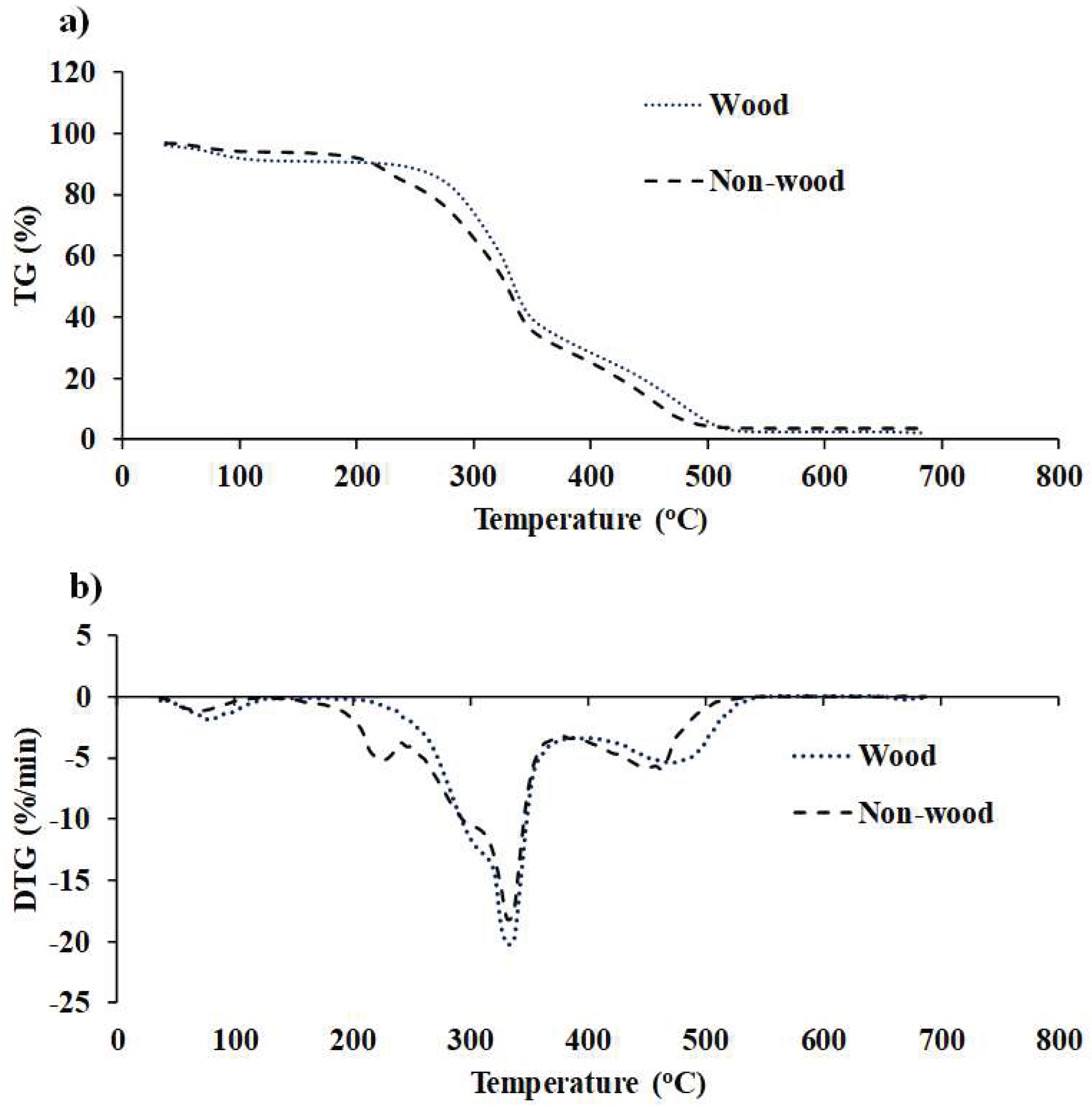
Figure 6.
(a) TG and (b) DTG profiles of fast−growing tree and agricultural residue biomass obtained using TGA within the temperature range of 35 to 700 °C with a heat flow rate of 10°C/min.
Figure 6.
(a) TG and (b) DTG profiles of fast−growing tree and agricultural residue biomass obtained using TGA within the temperature range of 35 to 700 °C with a heat flow rate of 10°C/min.
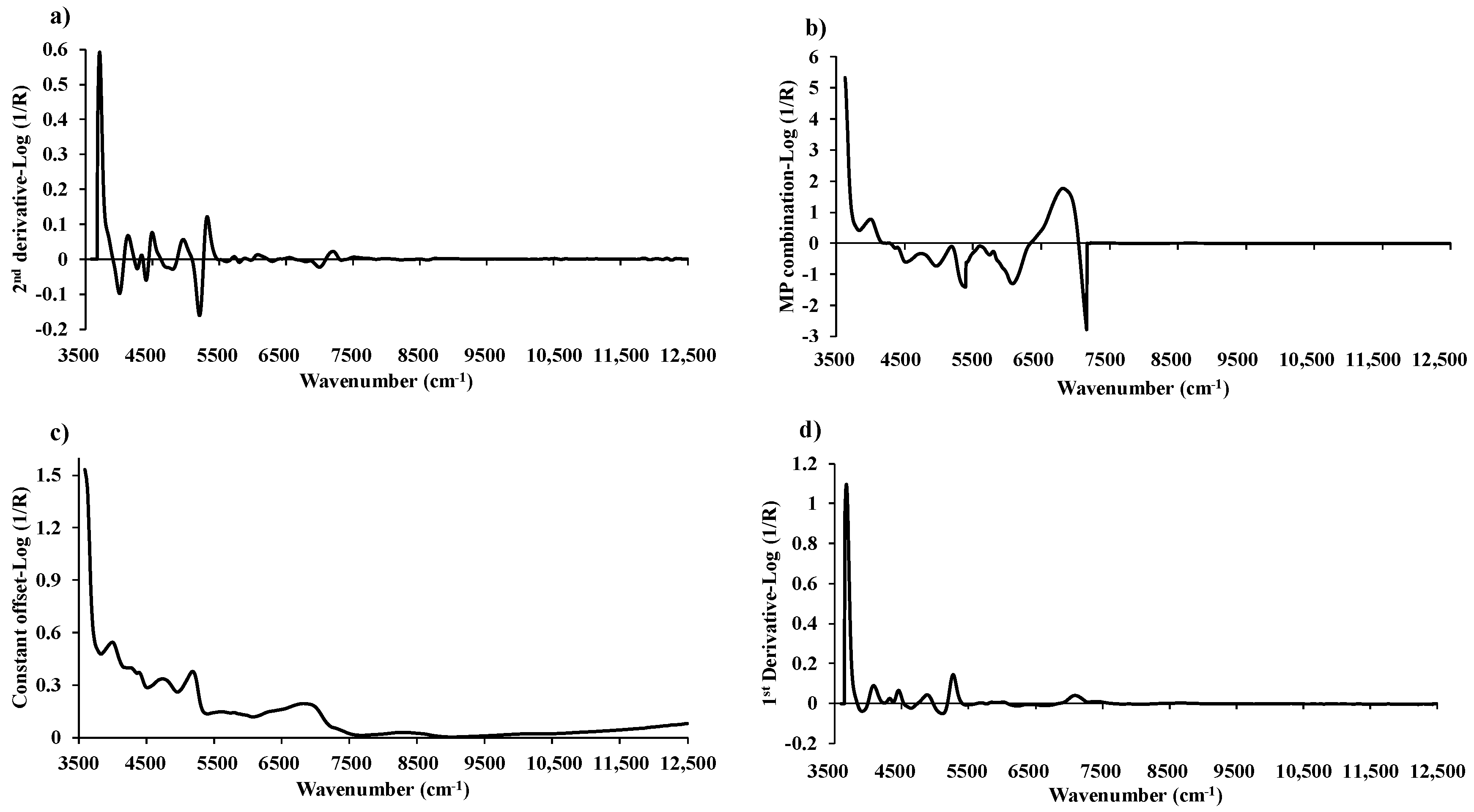
Figure 7.
The average pretreated spectra for: (a) MC with the second derivative, (b) VM with a multi-preprocessing combination set of 4, 4, 3, 2, and 0, (c) FC with a constant offset, and (d) ash content with the first derivative.
Figure 7.
The average pretreated spectra for: (a) MC with the second derivative, (b) VM with a multi-preprocessing combination set of 4, 4, 3, 2, and 0, (c) FC with a constant offset, and (d) ash content with the first derivative.
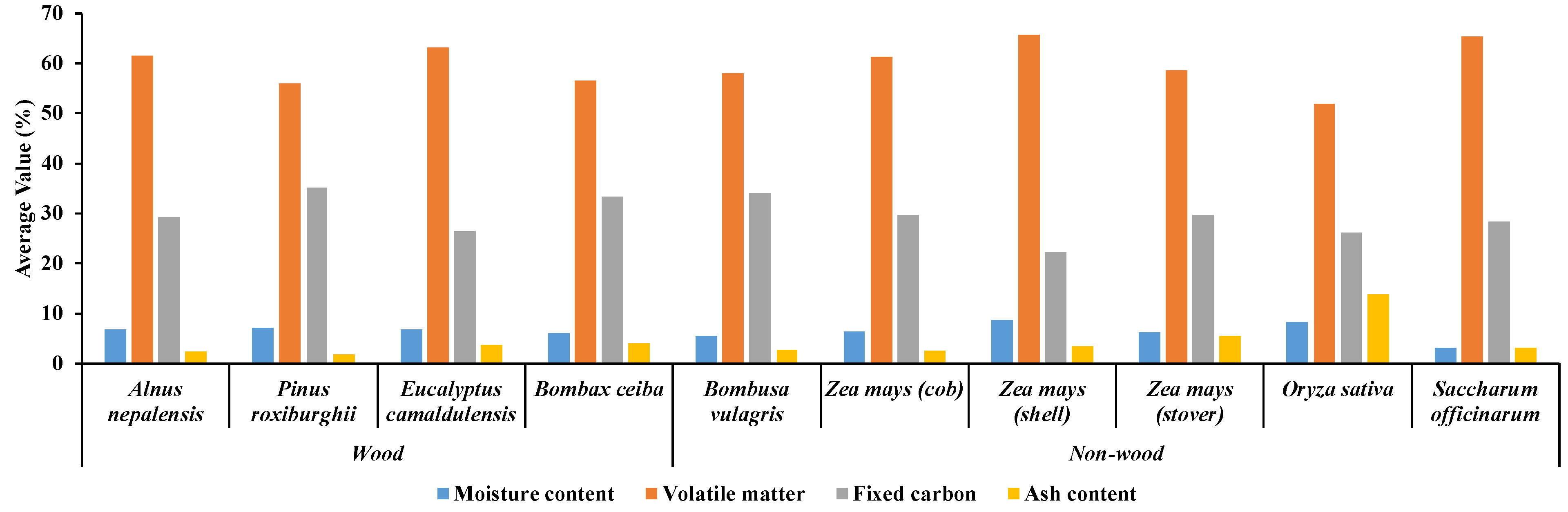
Figure 8.
Overall proximate analysis results of ten different biomass varieties, including four wood varieties of fast−growing trees and six non−wood varieties (five agricultural residue and 1 fast−growing tree) obtained from TGA analysis.
Figure 8.
Overall proximate analysis results of ten different biomass varieties, including four wood varieties of fast−growing trees and six non−wood varieties (five agricultural residue and 1 fast−growing tree) obtained from TGA analysis.
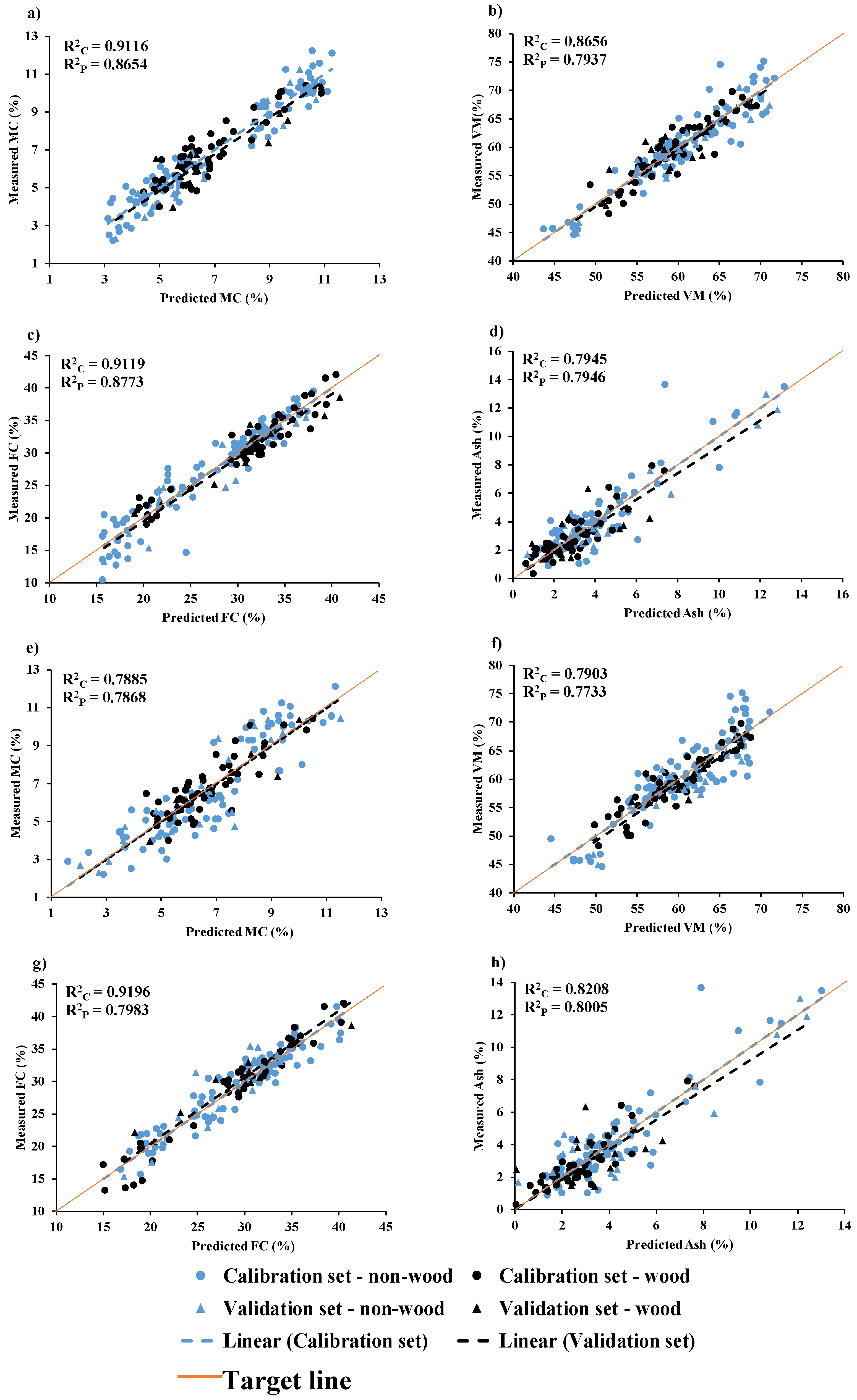
Figure 9.
Measured versus predicted value in calibration set and validation set for chip biomass (a) MC, (b) VM, (c) FC, and (d) Ash and for ground biomass (e) MC, (f) VM, (g) FC, and (h) Ash.
Figure 9.
Measured versus predicted value in calibration set and validation set for chip biomass (a) MC, (b) VM, (c) FC, and (d) Ash and for ground biomass (e) MC, (f) VM, (g) FC, and (h) Ash.
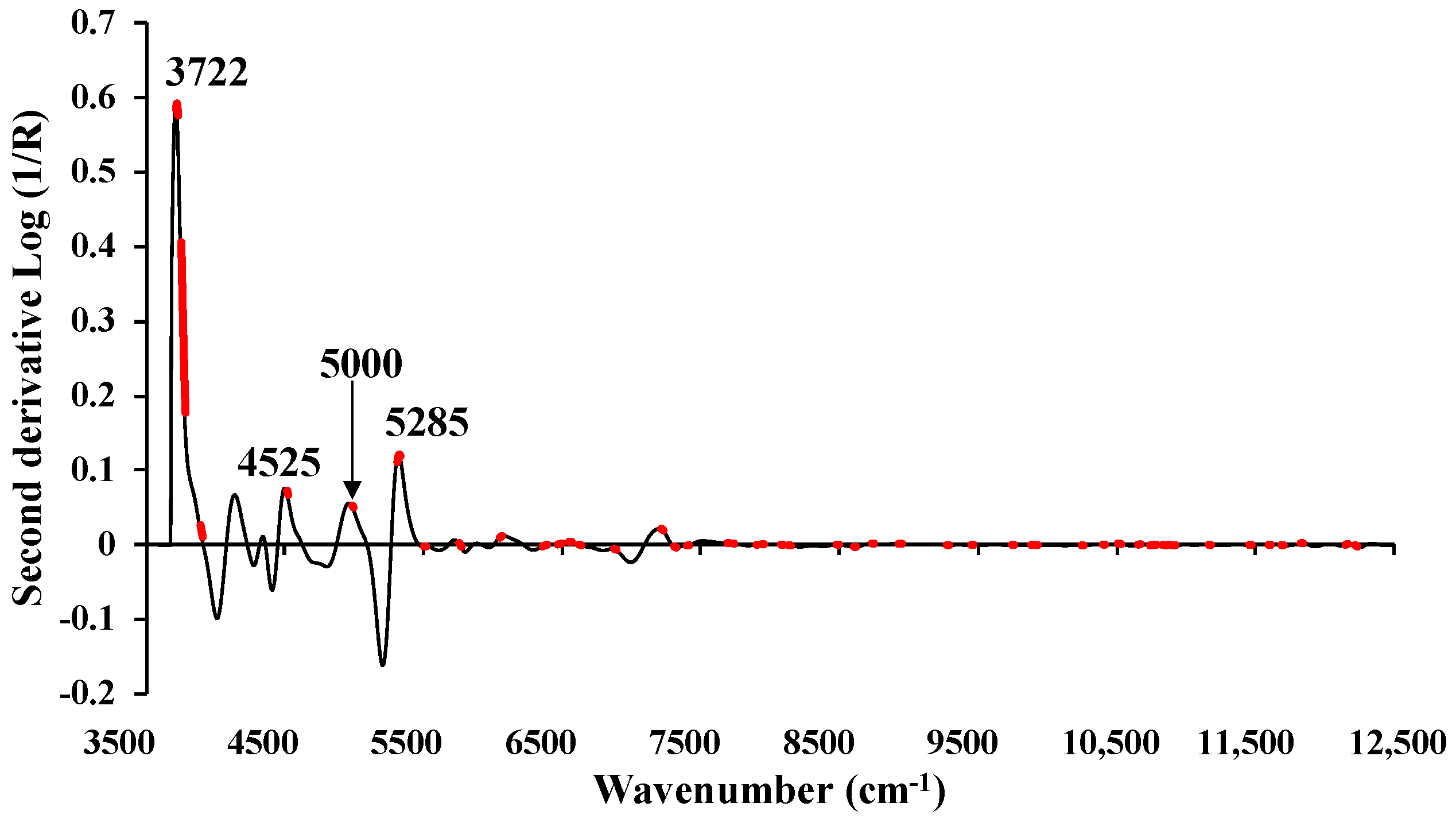
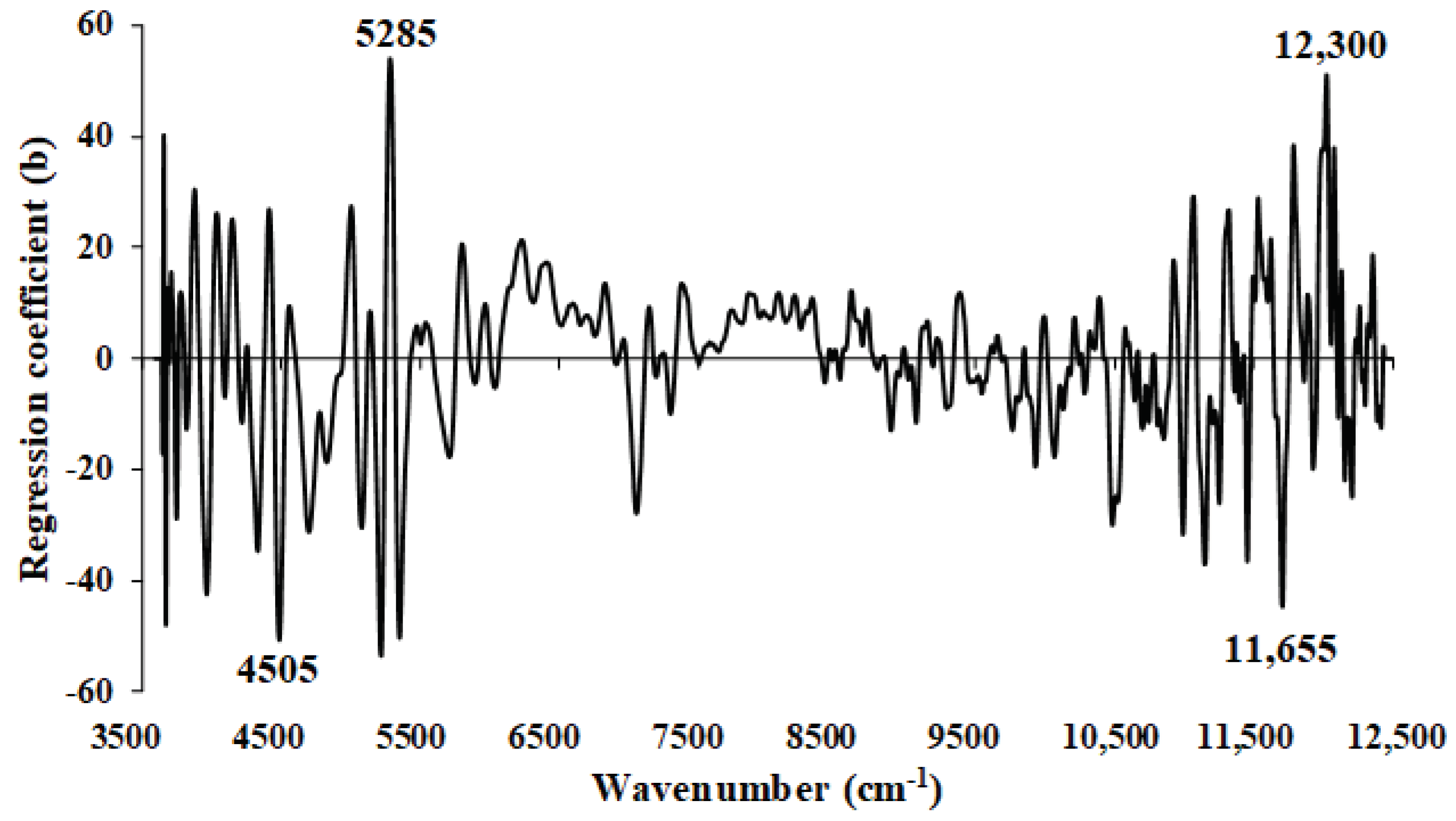
Figure 10.
The average absorbance value of MC (%) of chip biomass obtained using second derivative preprocessing with a selection of important wavenumbers obtained from GA, within full wavenumber range of 3594.87–12,489.5 cm−1.
Figure 10.
The average absorbance value of MC (%) of chip biomass obtained using second derivative preprocessing with a selection of important wavenumbers obtained from GA, within full wavenumber range of 3594.87–12,489.5 cm−1.
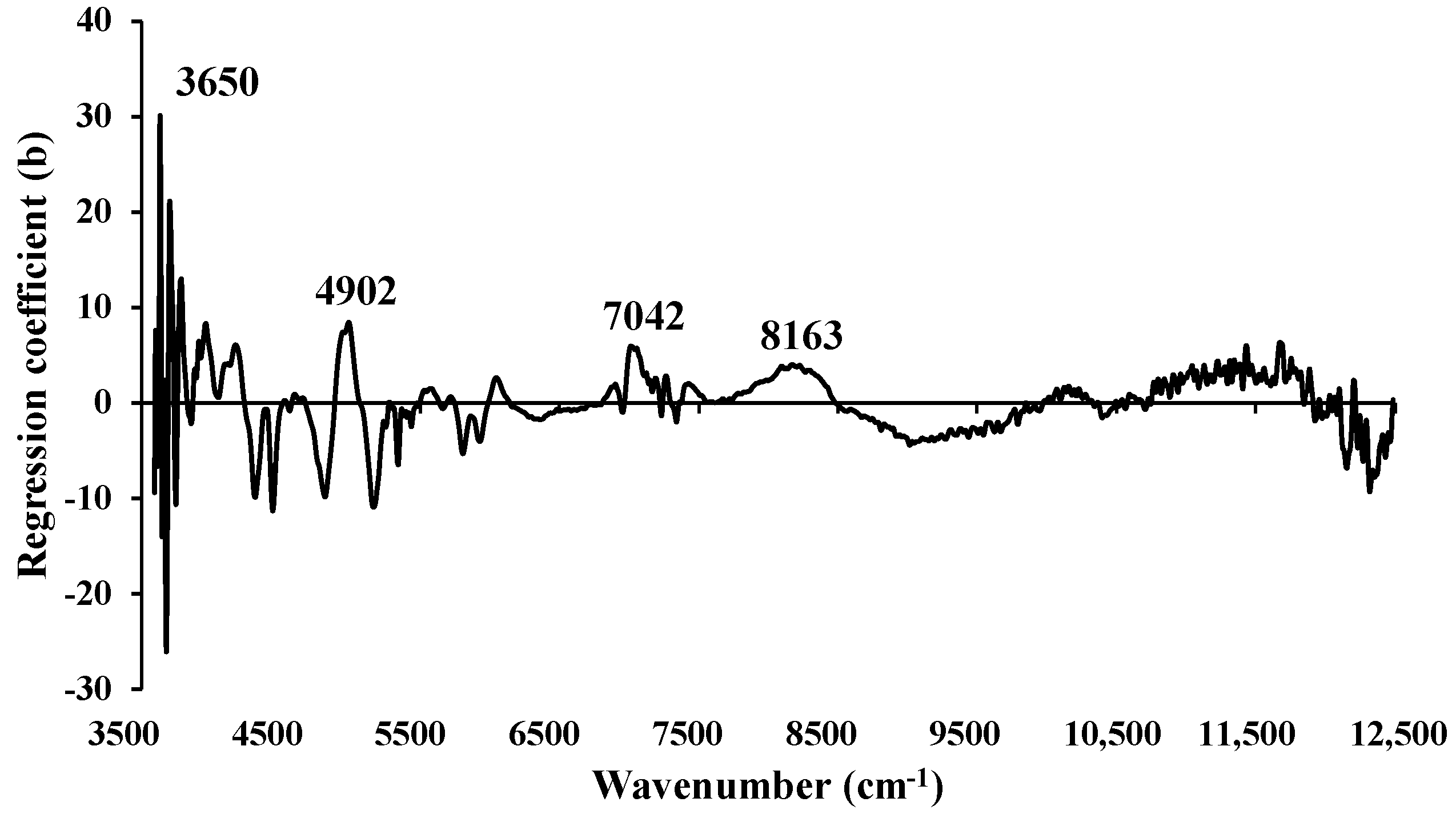
Figure 11.
The regression coefficient for the MC (%) of ground biomass using the Full−PLSR with spectral preprocessing of mean centering.
Figure 11.
The regression coefficient for the MC (%) of ground biomass using the Full−PLSR with spectral preprocessing of mean centering.
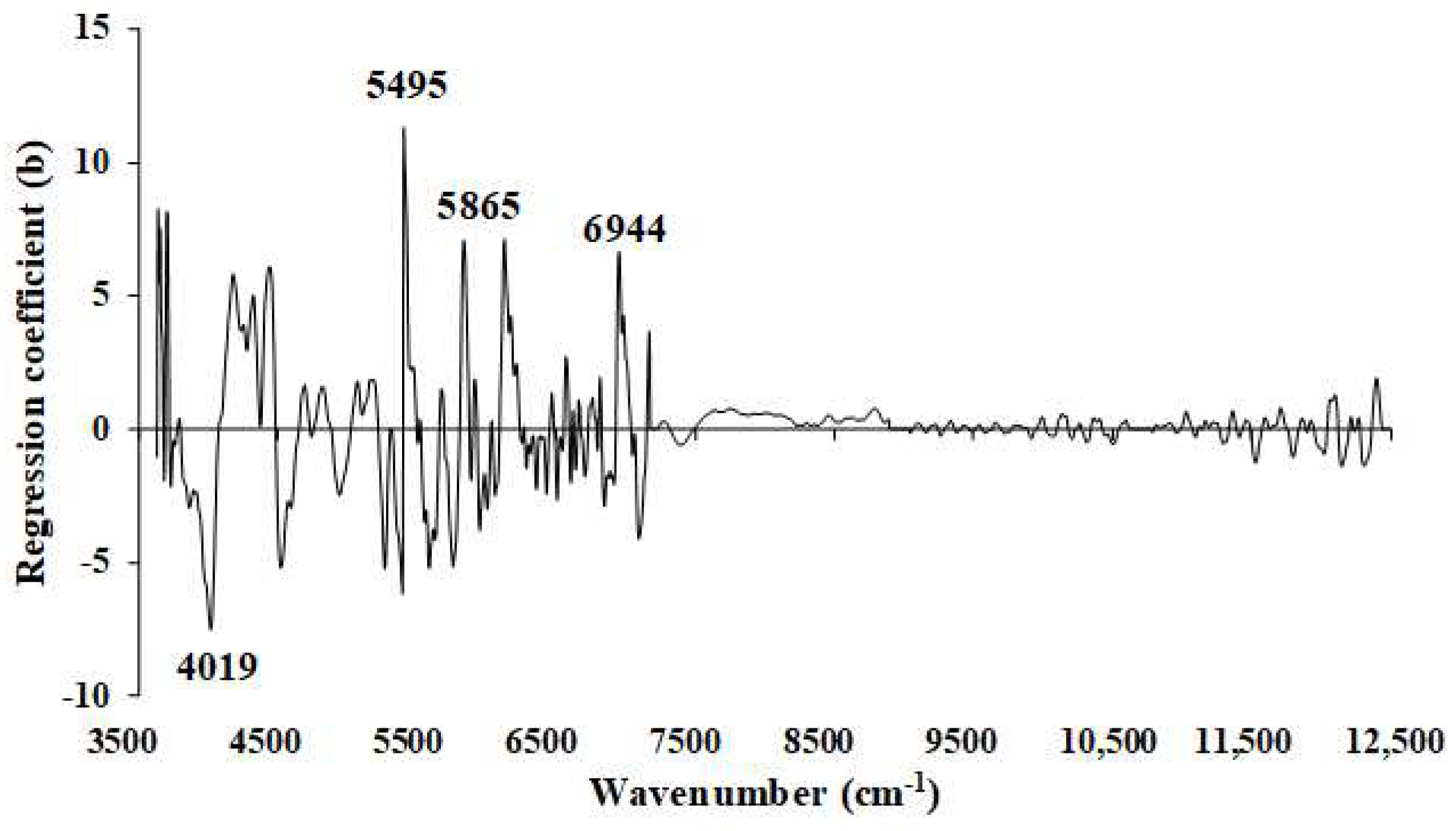
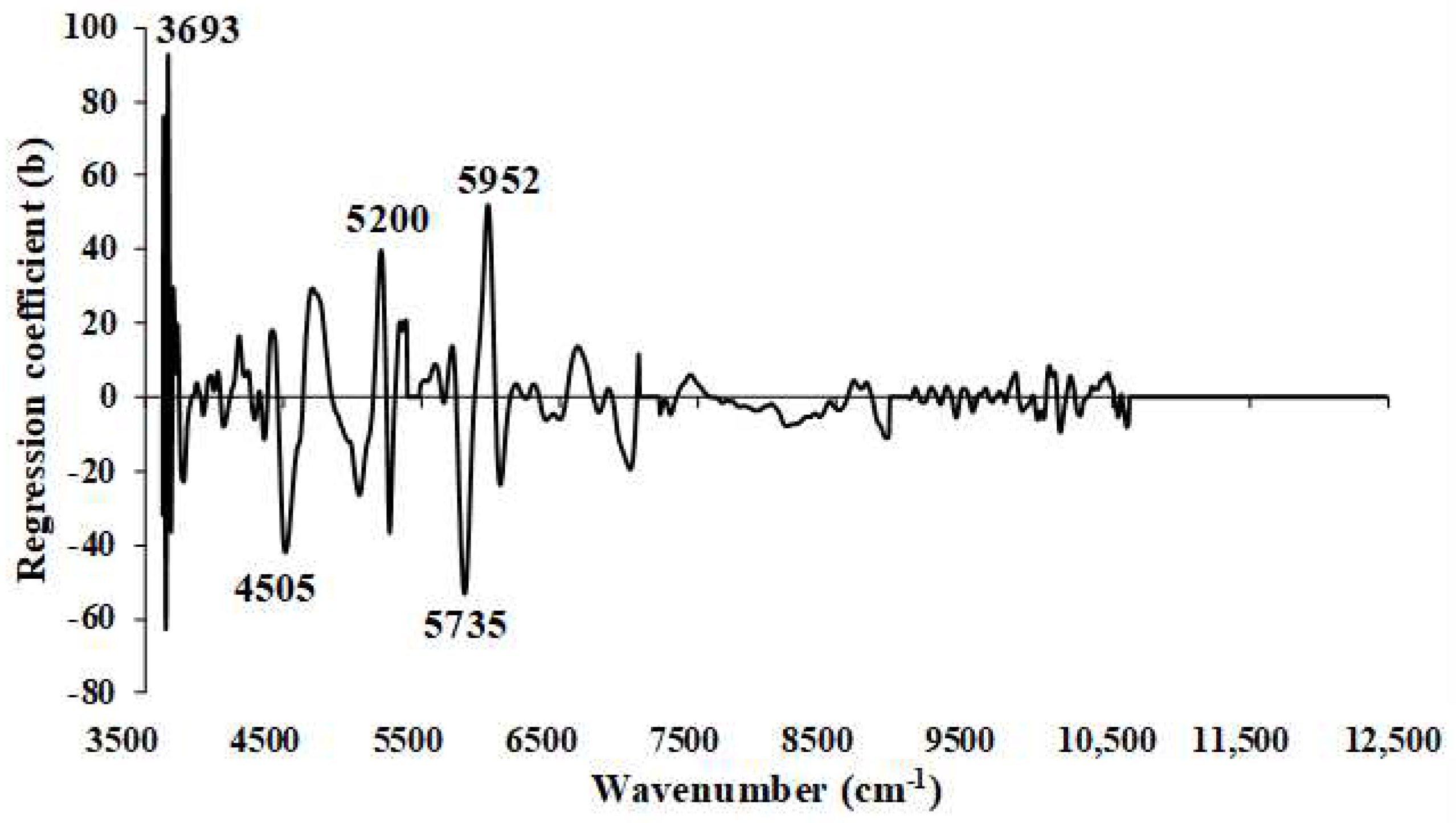
Figure 12.
Regression coefficient for the % of VM in chip biomass using the MP PLSR 5−range method with a spectral multi-preprocessing combination set of 4, 4, 3, 2, 0.
Figure 12.
Regression coefficient for the % of VM in chip biomass using the MP PLSR 5−range method with a spectral multi-preprocessing combination set of 4, 4, 3, 2, 0.
Figure 13.
Regression coefficient for the % of VM in ground biomass using the MP PLSR 5−range method with a spectral multi-preprocessing combination set of 3, 5, 4, 5, and 0.
Figure 13.
Regression coefficient for the % of VM in ground biomass using the MP PLSR 5−range method with a spectral multi-preprocessing combination set of 3, 5, 4, 5, and 0.
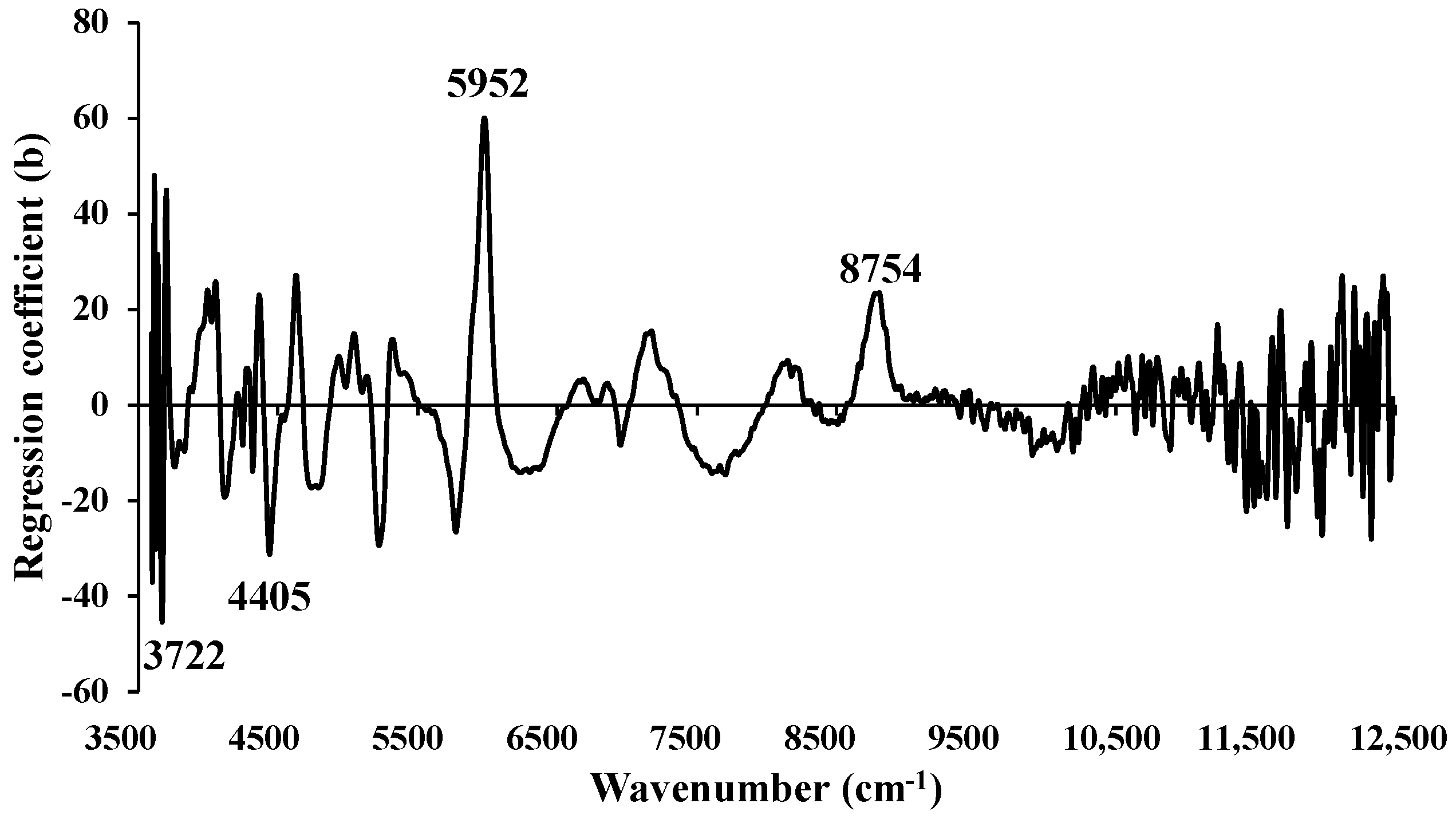
Figure 14.
Regression coefficient for the % of FC in chip biomass using Full−PLSR method with a spectral preprocessing of constant offset.
Figure 14.
Regression coefficient for the % of FC in chip biomass using Full−PLSR method with a spectral preprocessing of constant offset.
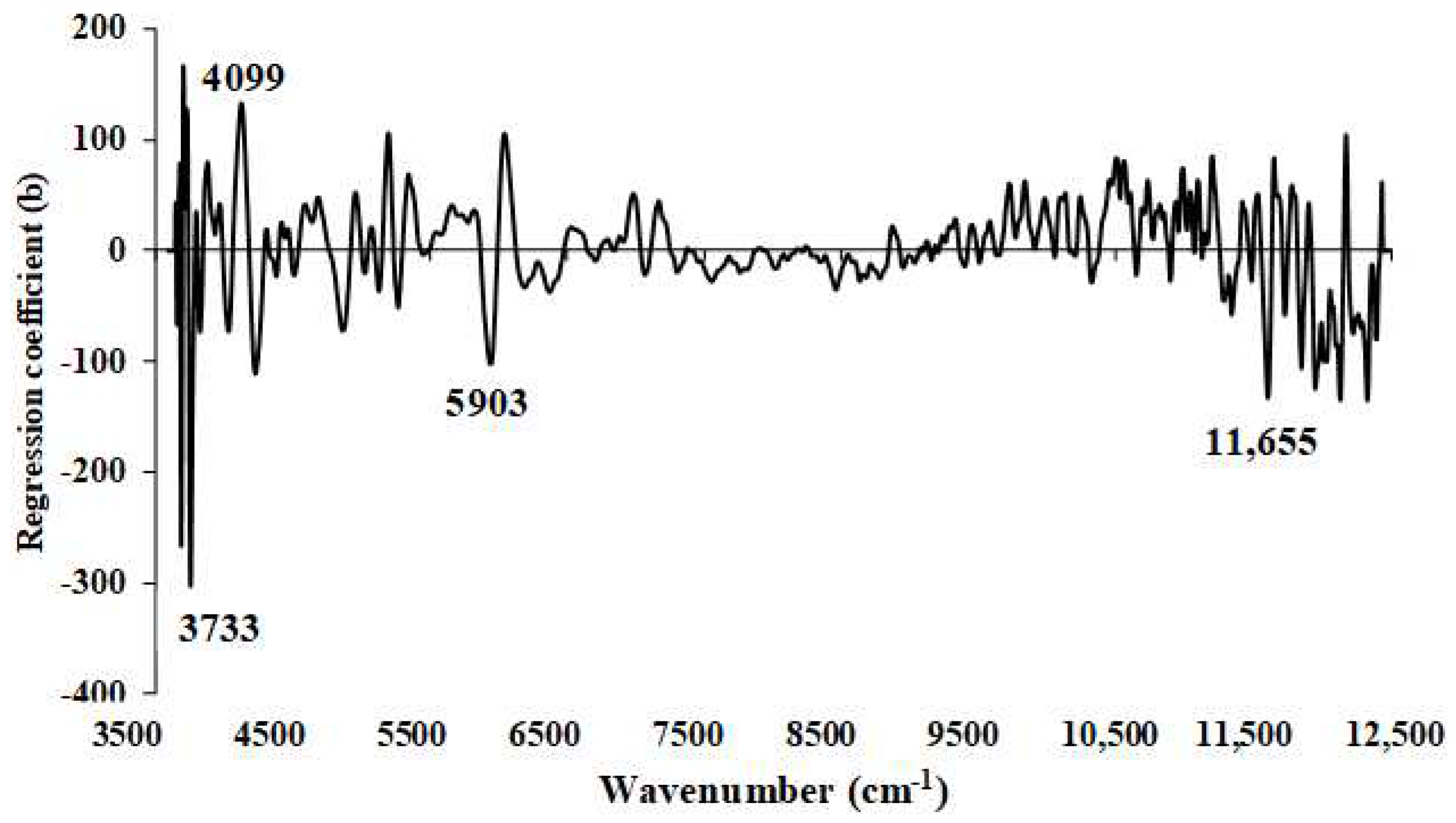
Figure 15.
Regression coefficient for the % of FC in ground biomass using Full−PLSR method with a spectral preprocessing of first derivative.
Figure 15.
Regression coefficient for the % of FC in ground biomass using Full−PLSR method with a spectral preprocessing of first derivative.
Figure 16.
Regression coefficient for the % of ash content in chip biomass using the MP PLSR 3−range method with a spectral multi−preprocessing combination set of 5, 4, and 4.
Figure 16.
Regression coefficient for the % of ash content in chip biomass using the MP PLSR 3−range method with a spectral multi−preprocessing combination set of 5, 4, and 4.
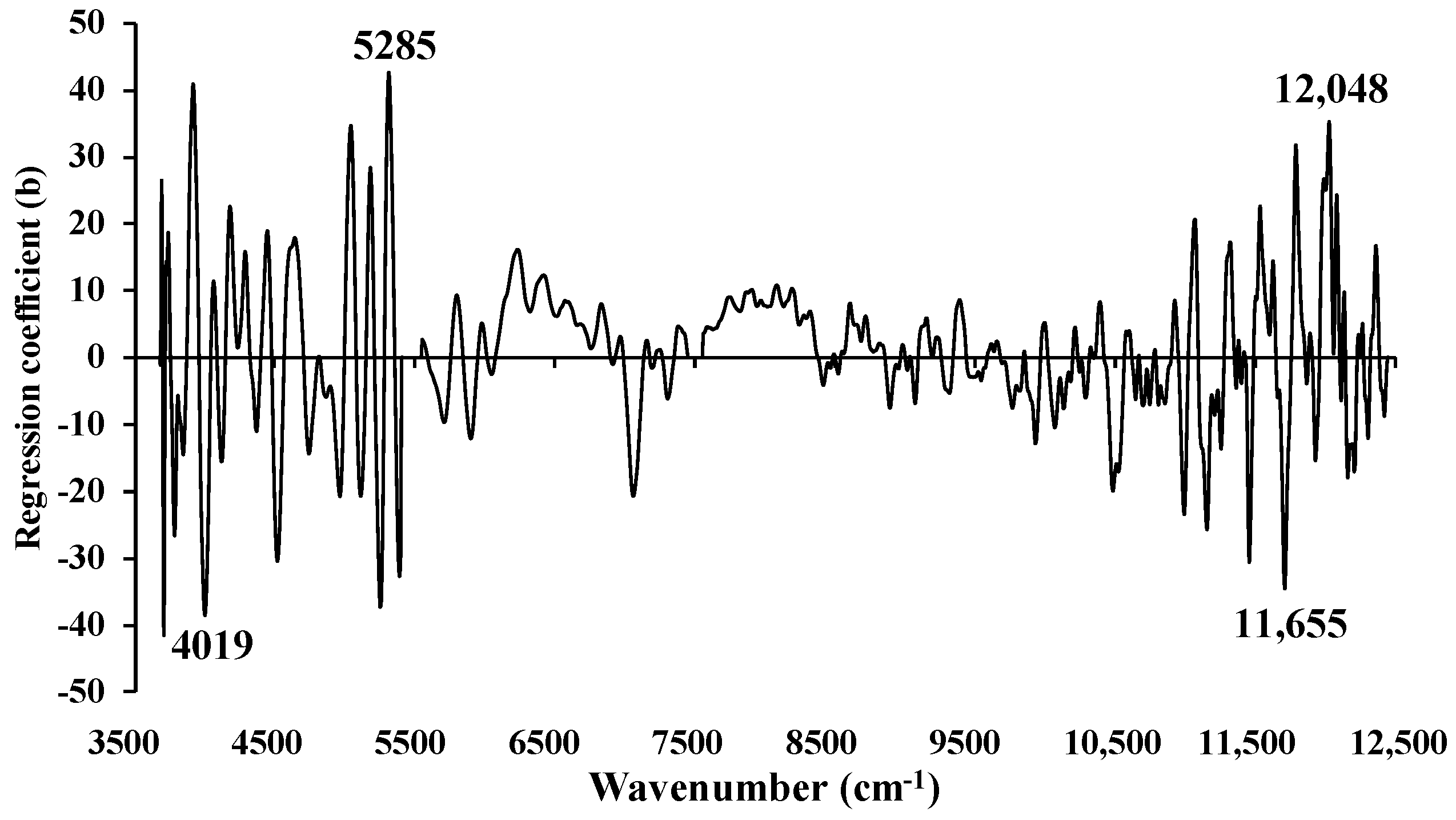
Figure 17.
Regression coefficient for the % of ash content in ground biomass using Full−PLSR method with a spectral preprocessing of D1.
Figure 17.
Regression coefficient for the % of ash content in ground biomass using Full−PLSR method with a spectral preprocessing of D1.
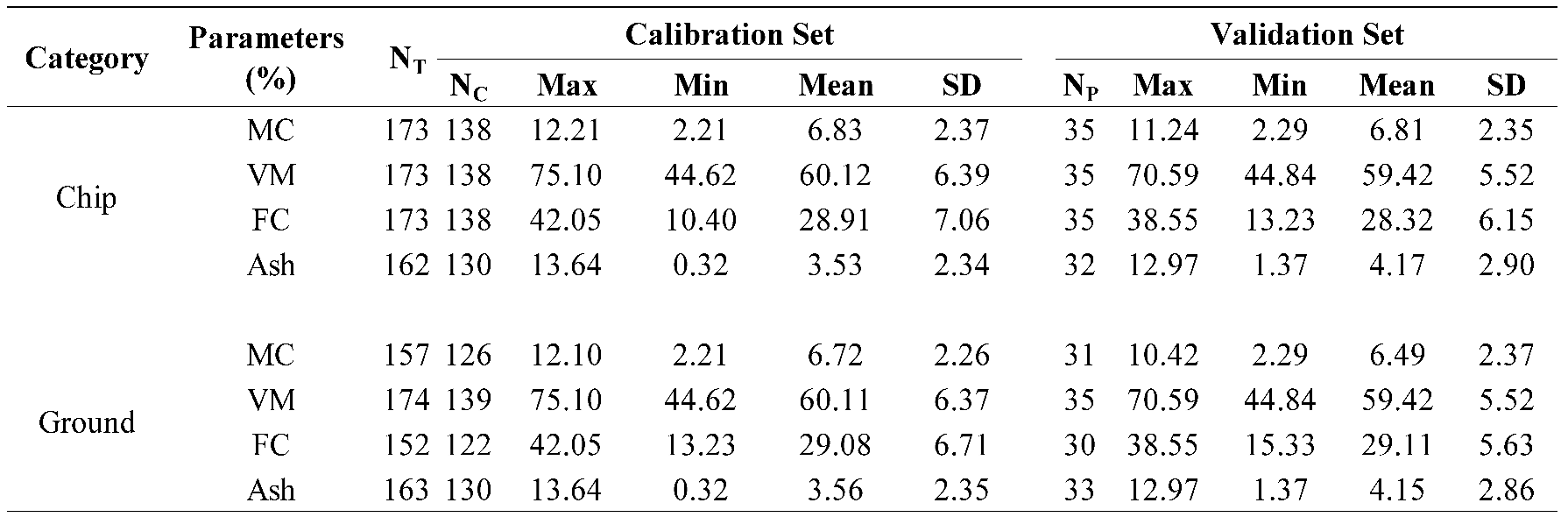
Table 1.
Statistical data of the proximate analysis parameters of the chip and ground biomass used in the PLSR model development.
Table 1.
Statistical data of the proximate analysis parameters of the chip and ground biomass used in the PLSR model development.
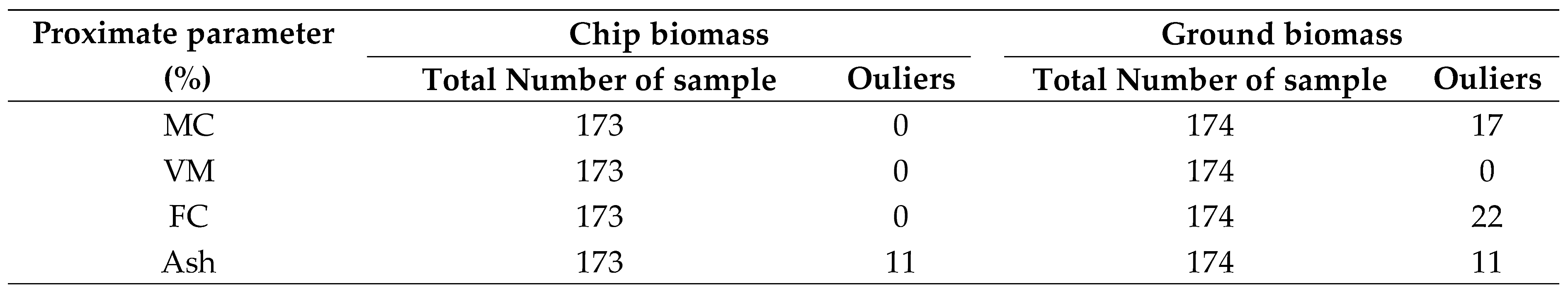
Table 2.
Table 2. Details on the number of identified outliers in chip and ground biomass evaluated before the development of the PLSR model.
Table 2.
Table 2. Details on the number of identified outliers in chip and ground biomass evaluated before the development of the PLSR model.
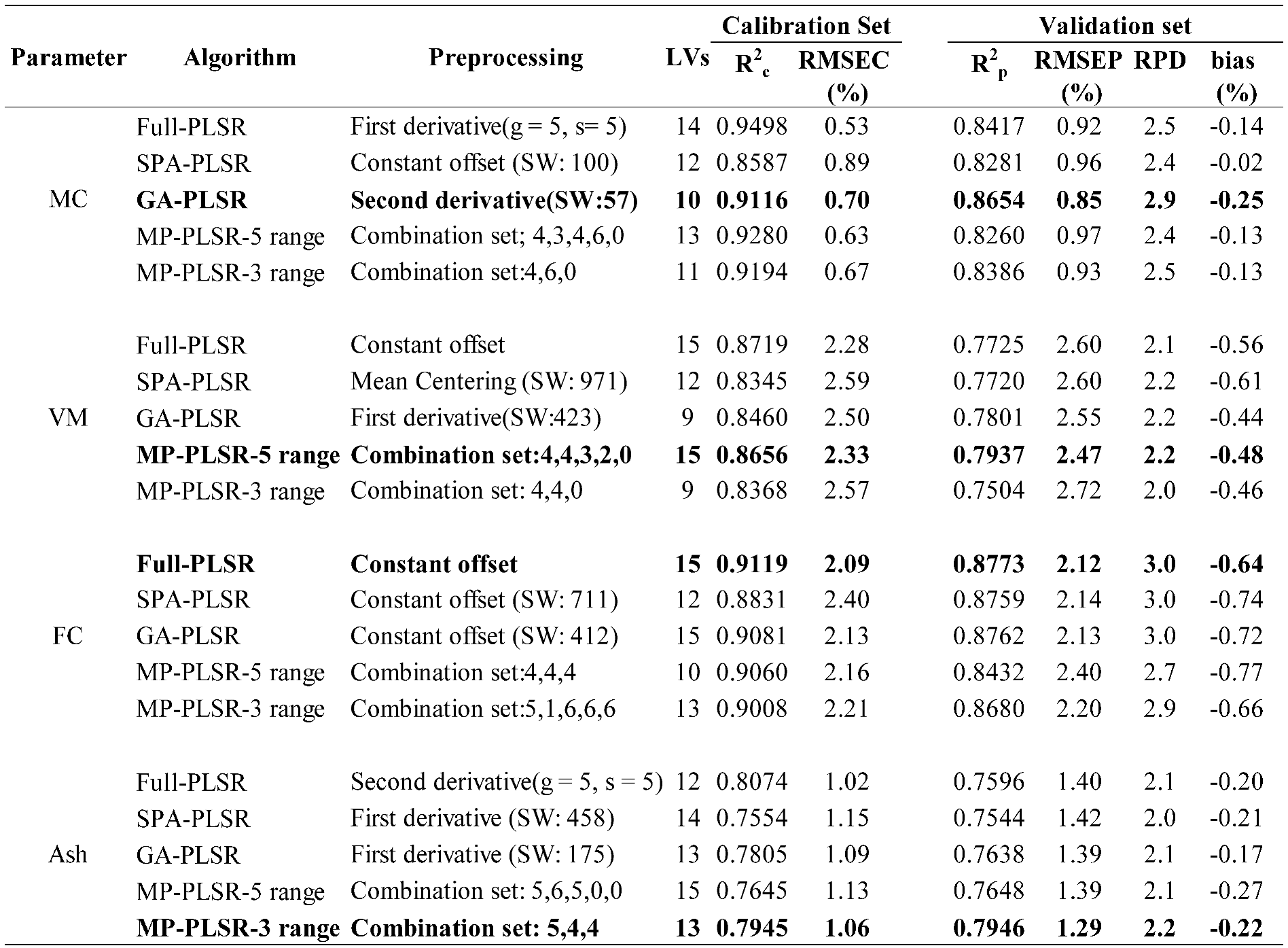
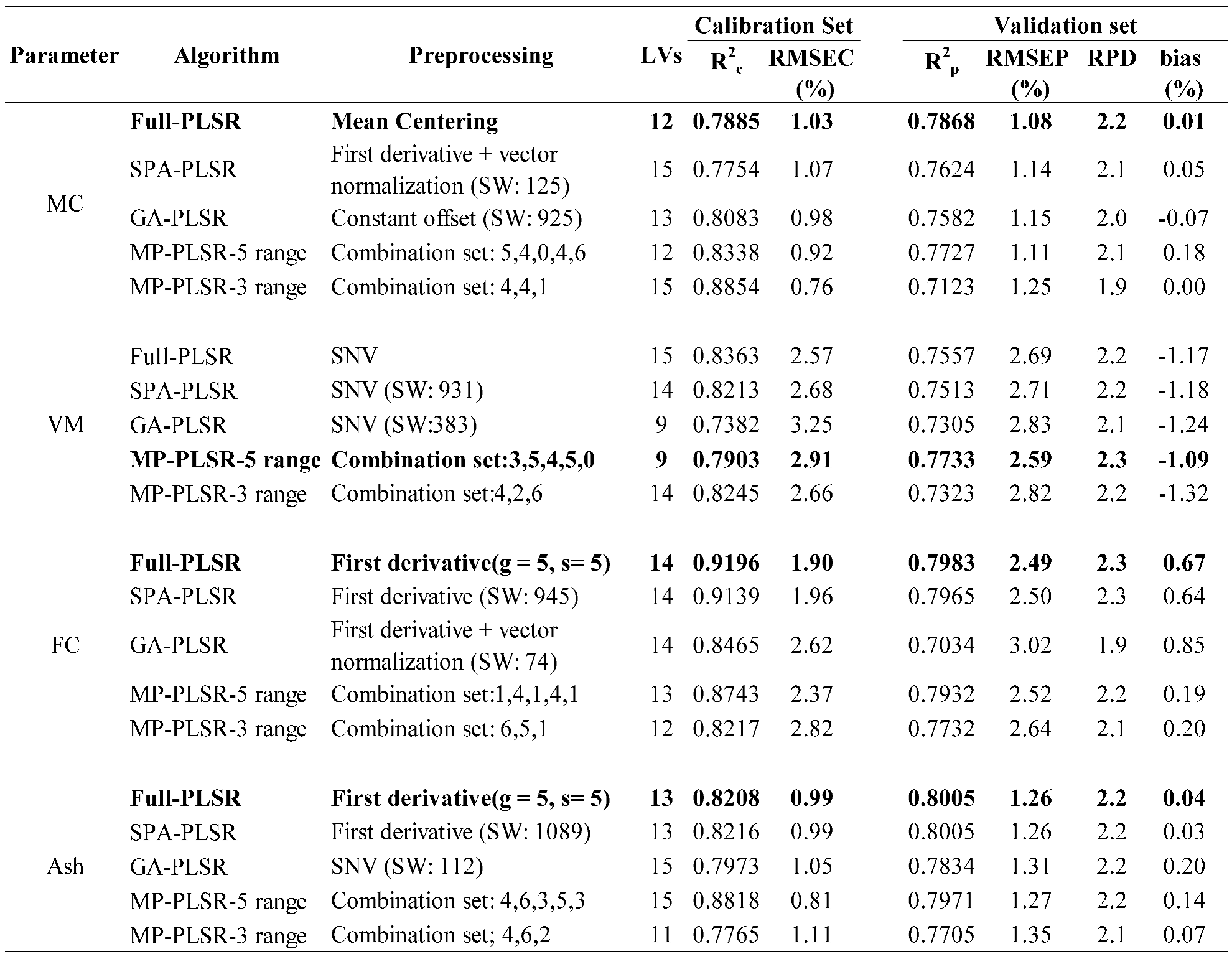
Table 3.
Results of the PLS regression based model for the proximate analysis (%) of chip biomass, bolded model showing the best performance.
Table 3.
Results of the PLS regression based model for the proximate analysis (%) of chip biomass, bolded model showing the best performance.
Table 4.
Results of the PLS regression based model for the proximate analysis (%) of ground biomass, bolded model showing the best performance.
Table 4.
Results of the PLS regression based model for the proximate analysis (%) of ground biomass, bolded model showing the best performance.
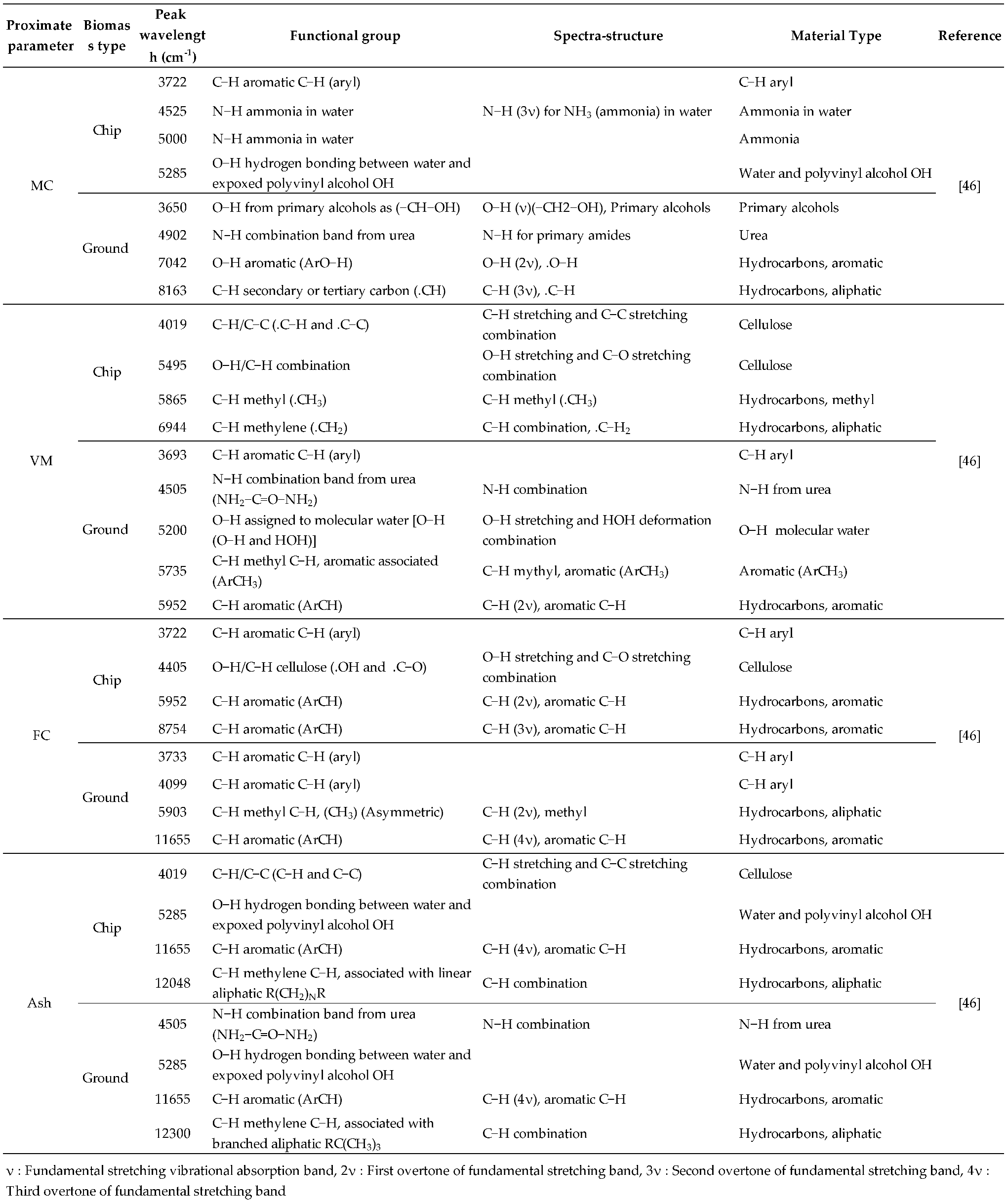
Table 5.
The dominant peaks on the regression coefficient plot obtained from the best−performing PLSR−based model.
Table 5.
The dominant peaks on the regression coefficient plot obtained from the best−performing PLSR−based model.
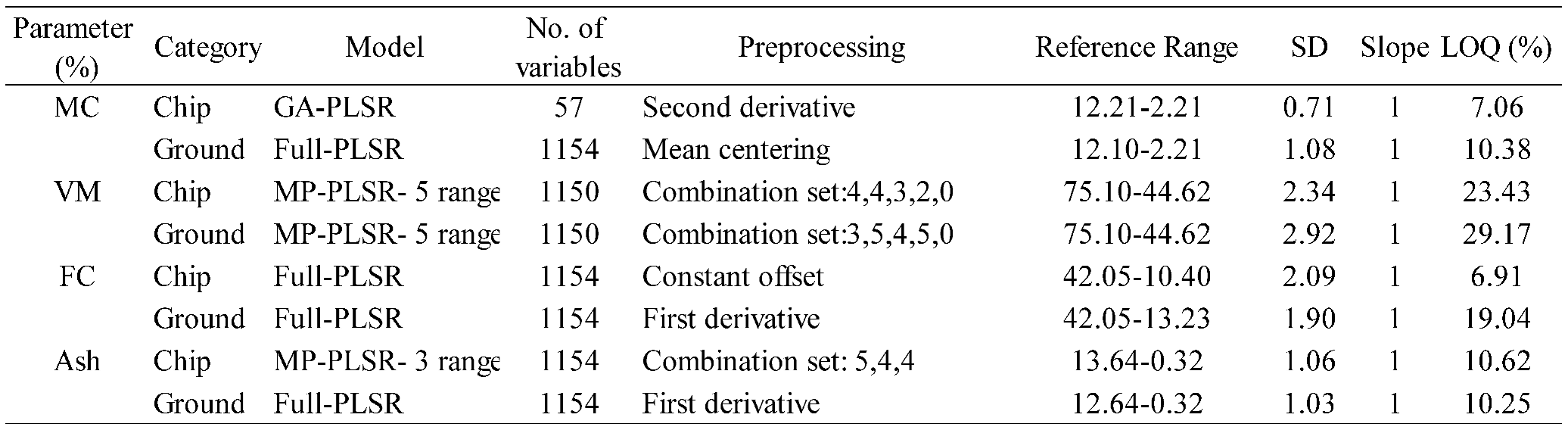
Table 6.
LOQ results of PLSR models for the % of MC, VM, FC, and ash content in chip and ground biomass.
Table 6.
LOQ results of PLSR models for the % of MC, VM, FC, and ash content in chip and ground biomass.
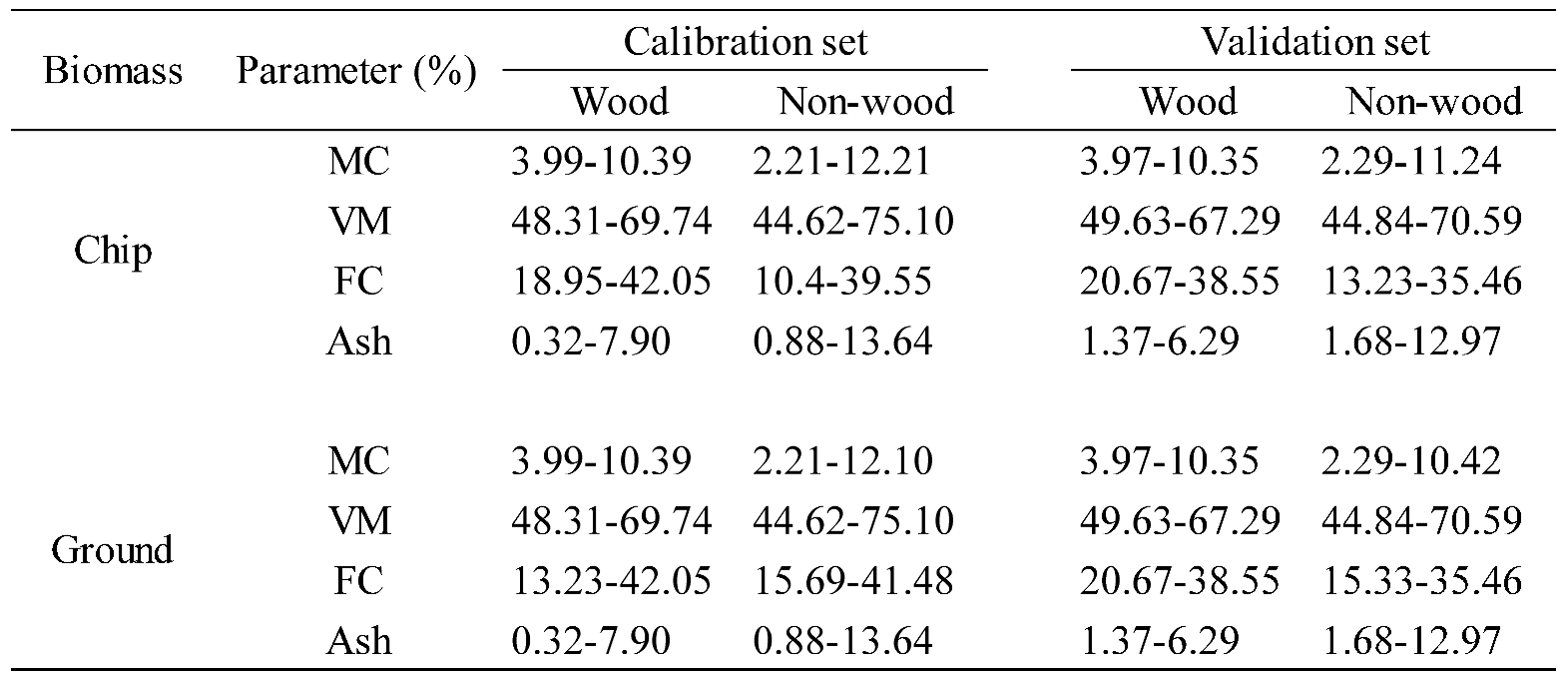
Table 7.
The range of % of MC, VM, FC, and Ash content of wood and non−wood samples in calibration and validation sets.
Table 7.
The range of % of MC, VM, FC, and Ash content of wood and non−wood samples in calibration and validation sets.