Submitted:
21 December 2023
Posted:
22 December 2023
You are already at the latest version
Abstract
Keywords:
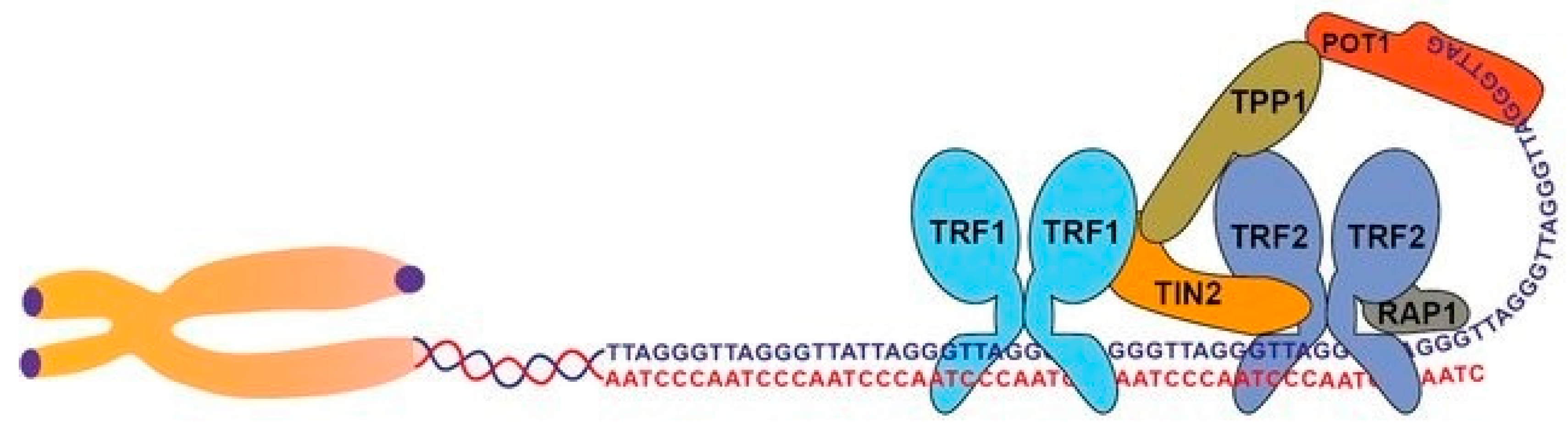
Challenges and Dynamics of Telomere Replication
Unwinding of G4 Structures for Telomere Replication
Overcoming Replication Challenges at Telomeres
Conclusion
Supplementary Materials
References
- de Lange, T. T-loops and the origin of telomeres. Nature reviews. Molecular cell biology 2004, 5, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Maestroni, L., Matmati, S., & Coulon, S. Solving the Telomere Replication Problem. Genes 2017, 8, 55. [CrossRef] [PubMed]
- de Lange, T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes & development 2005, 19, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Soudet, J., Jolivet, P., & Teixeira, M. T. Elucidation of the DNA end-replication problem in Saccharomyces cerevisiae. Molecular cell 2014, 53, 954–964. [CrossRef] [PubMed]
- Nandakumar, J. , Bell, C. F., Weidenfeld, I., Zaug, A. J., Leinwand, L. A., & Cech, T. R. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 2012, 492, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Chen, L. Y. , Redon, S., & Lingner, J. The human CST complex is a terminator of telomerase activity. Nature 2012, 488, 540–544. [Google Scholar] [CrossRef]
- Herbig, U. , Jobling, W. A., Chen, B. P., Chen, D. J., & Sedivy, J. M. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Molecular cell 2004, 14, 501–513. [Google Scholar] [CrossRef]
- Miller, K. M. , Rog, O., & Cooper, J. P. Semi-conservative DNA replication through telomeres requires Taz1. Nature 2006, 440, 824–828. [Google Scholar] [CrossRef]
- Sfeir, A. , Kosiyatrakul, S. T., Hockemeyer, D., MacRae, S. L., Karlseder, J., Schildkraut, C. L., & de Lange, T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 2009, 138, 90–103. [Google Scholar] [CrossRef]
- Arnoult, N. , Schluth-Bolard, C., Letessier, A., Drascovic, I., Bouarich-Bourimi, R., Campisi, J., Kim, S. H., Boussouar, A., Ottaviani, A., Magdinier, F., Gilson, E., & Londoño-Vallejo, A. Replication timing of human telomeres is chromosome arm-specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS genetics 2010, 6, e1000920. [Google Scholar] [CrossRef]
- Chang, Y. T. , Moser, B. A., & Nakamura, T. M. Fission yeast shelterin regulates DNA polymerases and Rad3(ATR) kinase to limit telomere extension. PLoS genetics 2013, 9, e1003936. [Google Scholar] [CrossRef] [PubMed]
- Tarsounas, M. , & Tijsterman, M. Genomes and G-quadruplexes: for better or for worse. Journal of molecular biology 2013, 425, 4782–4789. [Google Scholar] [CrossRef] [PubMed]
- Leman, A. R. , & Noguchi, E. Local and global functions of Timeless and Tipin in replication fork protection. Cell cycle (Georgetown Tex.), 2012, 11, 3945–3955. [Google Scholar] [CrossRef] [PubMed]
- Leman, A. R. , Dheekollu, J., Deng, Z., Lee, S. W., Das, M. M., Lieberman, P. M., & Noguchi, E. Timeless preserves telomere length by promoting efficient DNA replication through human telomeres. Cell cycle (Georgetown Tex.), 2012, 11, 2337–2347. [Google Scholar] [CrossRef]
- Huber, M. D. , Lee, D. C., & Maizels, N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic acids research 2002, 30, 3954–3961. [Google Scholar] [CrossRef] [PubMed]
- [16] Arnoult, N. , Saintome, C., Ourliac-Garnier, I., Riou, J. F., & Londoño-Vallejo, A. Human POT1 is required for efficient telomere C-rich strand replication in the absence of WRN. Genes & development 2009, 23, 2915–2924. [Google Scholar] [CrossRef] [PubMed]
- Safa, L. , Gueddouda, N. M., Thiébaut, F., Delagoutte, E., Petruseva, I., Lavrik, O., Mendoza, O., Bourdoncle, A., Alberti, P., Riou, J. F., & Saintomé, C. 5' to 3' Unfolding Directionality of DNA Secondary Structures by Replication Protein A: G-QUADRUPLEXES AND DUPLEXES. The Journal of biological chemistry 2016, 291, 21246–21256. [Google Scholar] [CrossRef]
- Wang, H. , Nora, G. J., Ghodke, H., & Opresko, P. L. Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding. The Journal of biological chemistry 2011, 286, 7479–7489. [Google Scholar] [CrossRef] [PubMed]
- Déjardin, J. , & Kingston, R. E. Purification of proteins associated with specific genomic Loci. Cell 2009, 136, 175–186. [Google Scholar] [CrossRef]
- Zimmermann, M. , Kibe, T., Kabir, S., & de Lange, T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes & development 2014, 28, 2477–2491. [Google Scholar] [CrossRef]
- Vannier, J. B. , Sarek, G., & Boulton, S. J. RTEL1: functions of a disease-associated helicase. Trends in cell biology 2014, 24, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R. , Zhang, J., Bochman, M. L., Zakian, V. A., & Ha, T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife 2014, 3, e02190. [Google Scholar] [CrossRef]
- Paeschke, K. , Bochman, M. L., Garcia, P. D., Cejka, P., Friedman, K. L., Kowalczykowski, S. C., & Zakian, V. A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 2013, 497, 458–462. [Google Scholar] [CrossRef]
- Rodriguez, R. , Miller, K. M., Forment, J. V., Bradshaw, C. R., Nikan, M., Britton, S., Oelschlaegel, T., Xhemalce, B., Balasubramanian, S., & Jackson, S. P. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nature chemical biology 2012, 8, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Wallgren, M. , Mohammad, J. B., Yan, K. P., Pourbozorgi-Langroudi, P., Ebrahimi, M., & Sabouri, N. G-rich telomeric and ribosomal DNA sequences from the fission yeast genome form stable G-quadruplex DNA structures in vitro and are unwound by the Pfh1 DNA helicase. Nucleic acids research 2016, 44, 6213–6231. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C. , Pourmal, S., & Pavletich, N. P. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. eLife 2015, 4, e09832. [Google Scholar] [CrossRef] [PubMed]
- Sarek, G. , Vannier, J. B., Panier, S., Petrini, J. H. J., & Boulton, S. J. TRF2 recruits RTEL1 to telomeres in S phase to promote t-loop unwinding. Molecular cell 2015, 57, 622–635. [Google Scholar] [CrossRef]
- Vannier, J. B., Sandhu, S., Petalcorin, M. I., Wu, X., Nabi, Z., Ding, H., & Boulton, S. J. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science 2013, 342, 239–242. [CrossRef]
- Ghosh, A. K. , Rossi, M. L., Singh, D. K., Dunn, C., Ramamoorthy, M., Croteau, D. L., Liu, Y., & Bohr, V. A. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. The Journal of biological chemistry 2012, 287, 196–209. [Google Scholar] [CrossRef]
- Vannier, J. B. , Pavicic-Kaltenbrunner, V., Petalcorin, M. I., Ding, H., & Boulton, S. J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 2012, 149, 795–806. [Google Scholar] [CrossRef]
- Ye, J. , Lenain, C., Bauwens, S., Rizzo, A., Saint-Léger, A., Poulet, A., Benarroch, D., Magdinier, F., Morere, J., Amiard, S., Verhoeyen, E., Britton, S., Calsou, P., Salles, B., Bizard, A., Nadal, M., Salvati, E., Sabatier, L., Wu, Y., Biroccio, A., … Gilson, E. TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 2010, 142, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C. M. , Reichenbach, P., Khoriauli, L., Giulotto, E., & Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Costantino, L. , & Koshland, D. The Yin and Yang of R-loop biology. Current opinion in cell biology 2015, 34, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Rippe, K. , & Luke, B. TERRA and the state of the telomere. Nature structural & molecular biology 2015, 22, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R. L. , Cox, K. E., Jeitany, M., Wakimoto, H., Bryll, A. R., Ganem, N. J., Bersani, F., Pineda, J. R., Suvà, M. L., Benes, C. H., Haber, D. A., Boussin, F. D., & Zou, L. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 2015, 347, 273–277. [Google Scholar] [CrossRef]
- Arora, R. , Lee, Y., Wischnewski, H., Brun, C. M., Schwarz, T., & Azzalin, C. M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nature communications 2014, 5, 5220. [Google Scholar] [CrossRef]
- Clynes, D. , Jelinska, C., Xella, B., Ayyub, H., Scott, C., Mitson, M., Taylor, S., Higgs, D. R., & Gibbons, R. J. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nature communications 2015, 6, 7538. [Google Scholar] [CrossRef]
- Chawla, R. , Redon, S., Raftopoulou, C., Wischnewski, H., Gagos, S., & Azzalin, C. M. Human UPF1 interacts with TPP1 and telomerase and sustains telomere leading-strand replication. The EMBO journal 2011, 30, 4047–4058. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C. M. , & Lingner, J. Telomere functions grounding on TERRA firma. Trends in cell biology 2015, 25, 29–36. [Google Scholar] [CrossRef]
- Teasley, D. C. , Parajuli, S., Nguyen, M., Moore, H. R., Alspach, E., Lock, Y. J., Honaker, Y., Saharia, A., Piwnica-Worms, H., & Stewart, S. A. Flap Endonuclease 1 Limits Telomere Fragility on the Leading Strand. The Journal of biological chemistry 2015, 290, 15133–15145. [Google Scholar] [CrossRef]
- Benarroch-Popivker, D. , Pisano, S., Mendez-Bermudez, A., Lototska, L., Kaur, P., Bauwens, S., Djerbi, N., Latrick, C. M., Fraisier, V., Pei, B., Gay, A., Jaune, E., Foucher, K., Cherfils-Vicini, J., Aeby, E., Miron, S., Londoño-Vallejo, A., Ye, J., Le Du, M. H., Wang, H., … Giraud-Panis, M. J. TRF2-Mediated Control of Telomere DNA Topology as a Mechanism for Chromosome-End Protection. Molecular cell 2016, 61, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Nagai, S. , Heun, P., & Gasser, S. M. Roles for nuclear organization in the maintenance of genome stability. Epigenomics 2010, 2, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Steglich, B. , Strålfors, A., Khorosjutina, O., Persson, J., Smialowska, A., Javerzat, J. P., & Ekwall, K. The Fun30 chromatin remodeler Fft3 controls nuclear organization and chromatin structure of insulators and subtelomeres in fission yeast. PLoS genetics 2015, 11, e1005101. [Google Scholar] [CrossRef] [PubMed]
- Kaminker, P. G., Kim, S. H., Desprez, P. Y., & Campisi, J. A novel form of the telomere-associated protein TIN2 localizes to the nuclear matrix. Cell cycle (Georgetown Tex.) 2009, 8, 931–939. [CrossRef] [PubMed]
- Walker, J. R. , & Zhu, X. D. Post-translational modifications of TRF1 and TRF2 and their roles in telomere maintenance. Mechanisms of ageing and development 2012, 133, 421–434. [Google Scholar] [CrossRef]
- Greider C., W. Regulating telomere length from the inside out: the replication fork model. Genes & development 2016, 30, 1483–1491. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



