Submitted:
12 December 2023
Posted:
13 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Fungal Material
2.2. DNA Extraction and Purification
2.3. DNA Library Preparation and Sequencing on the Oxford Nanopore Technologies and Illumina platforms
2.4. Genome Assembly
2.5. Genome Analysis
3. Results
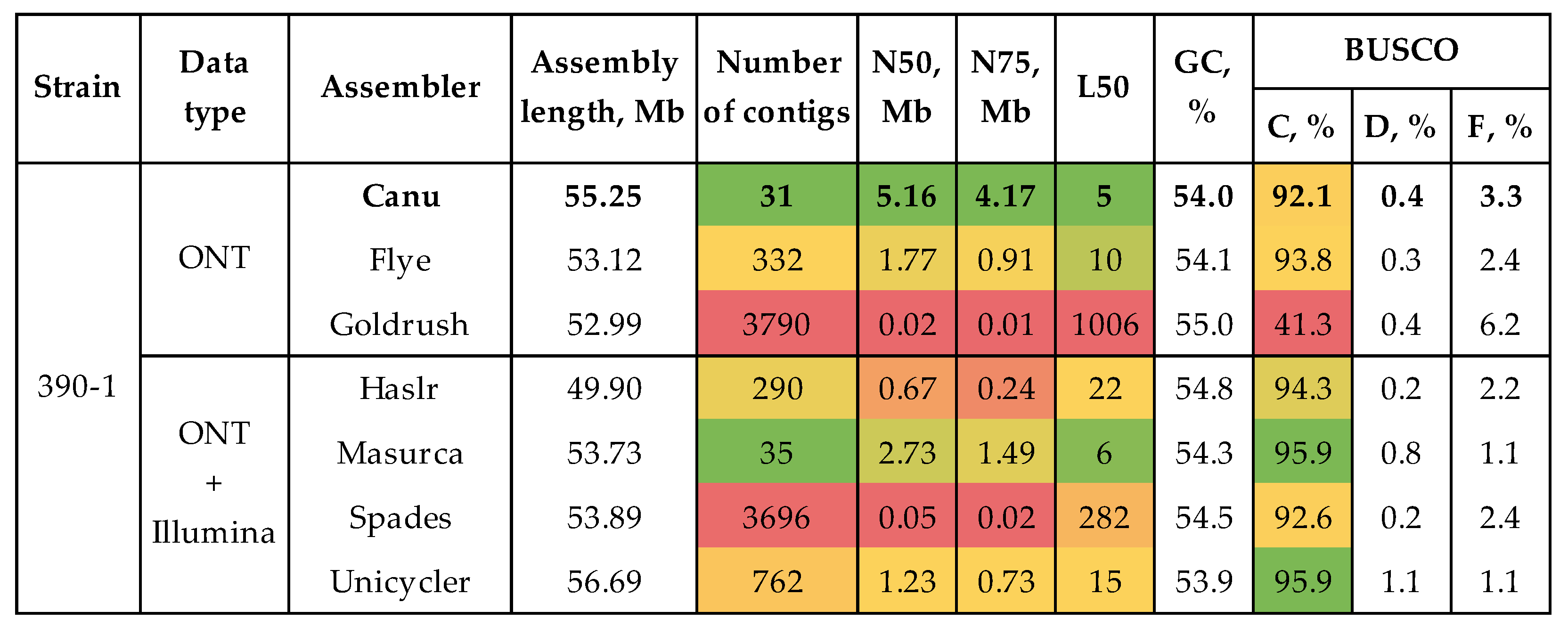
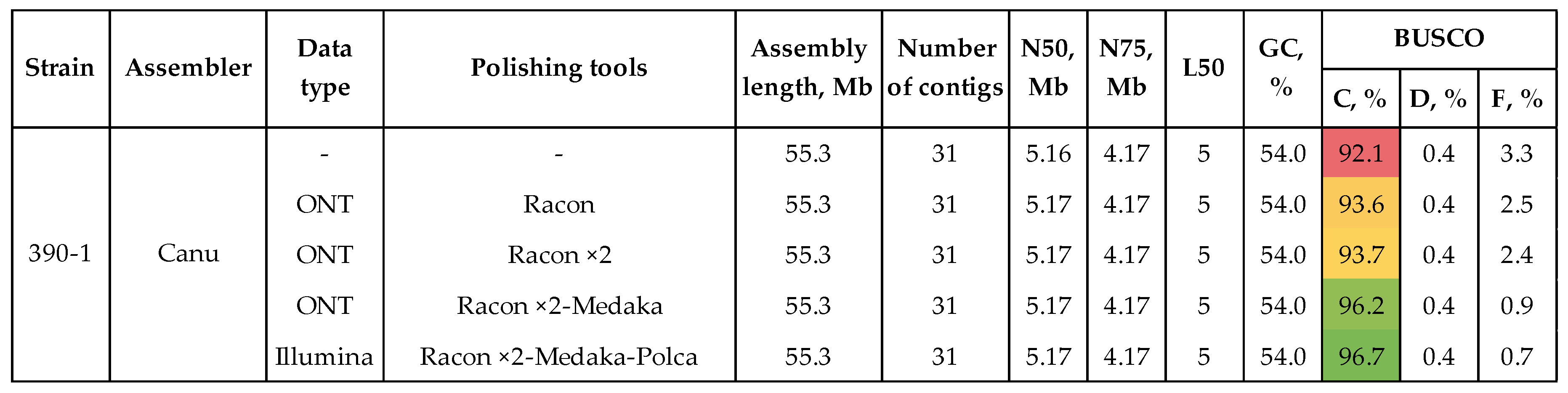
3.1. Genome Assembly and Polishing
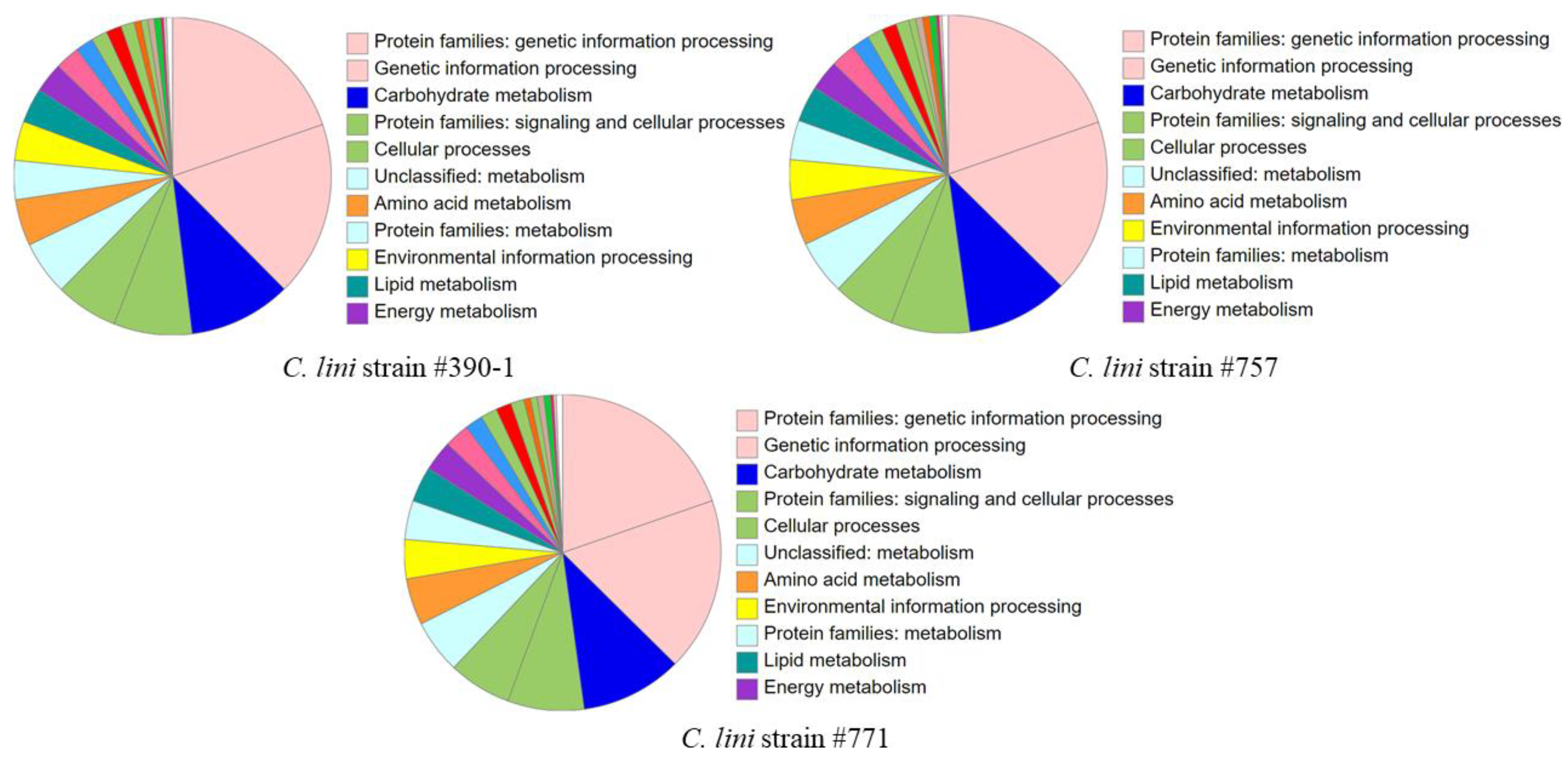
3.2. Genome Annotation and Search for Effector Proteins
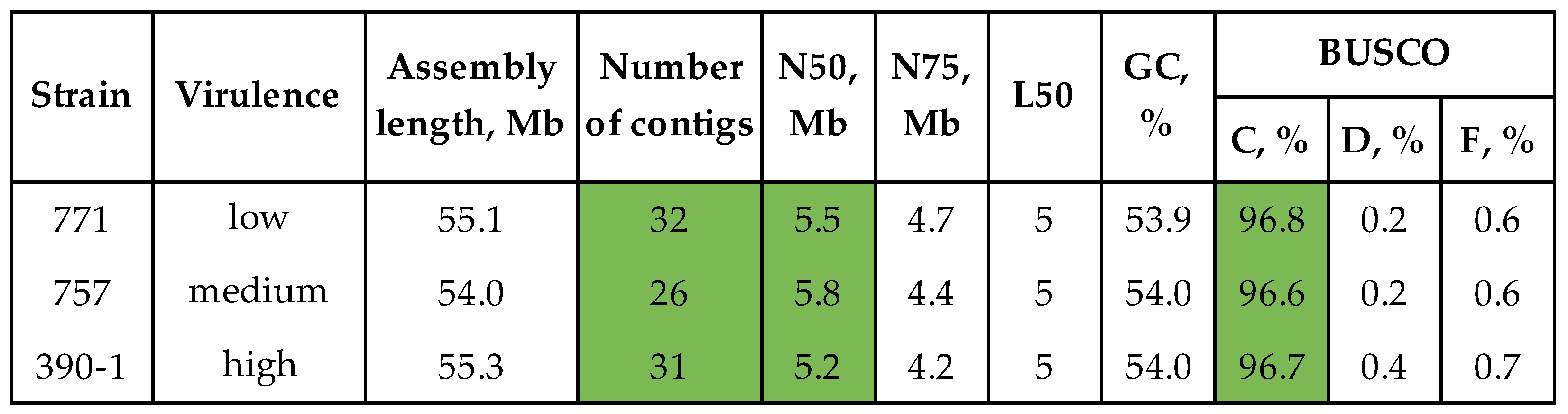
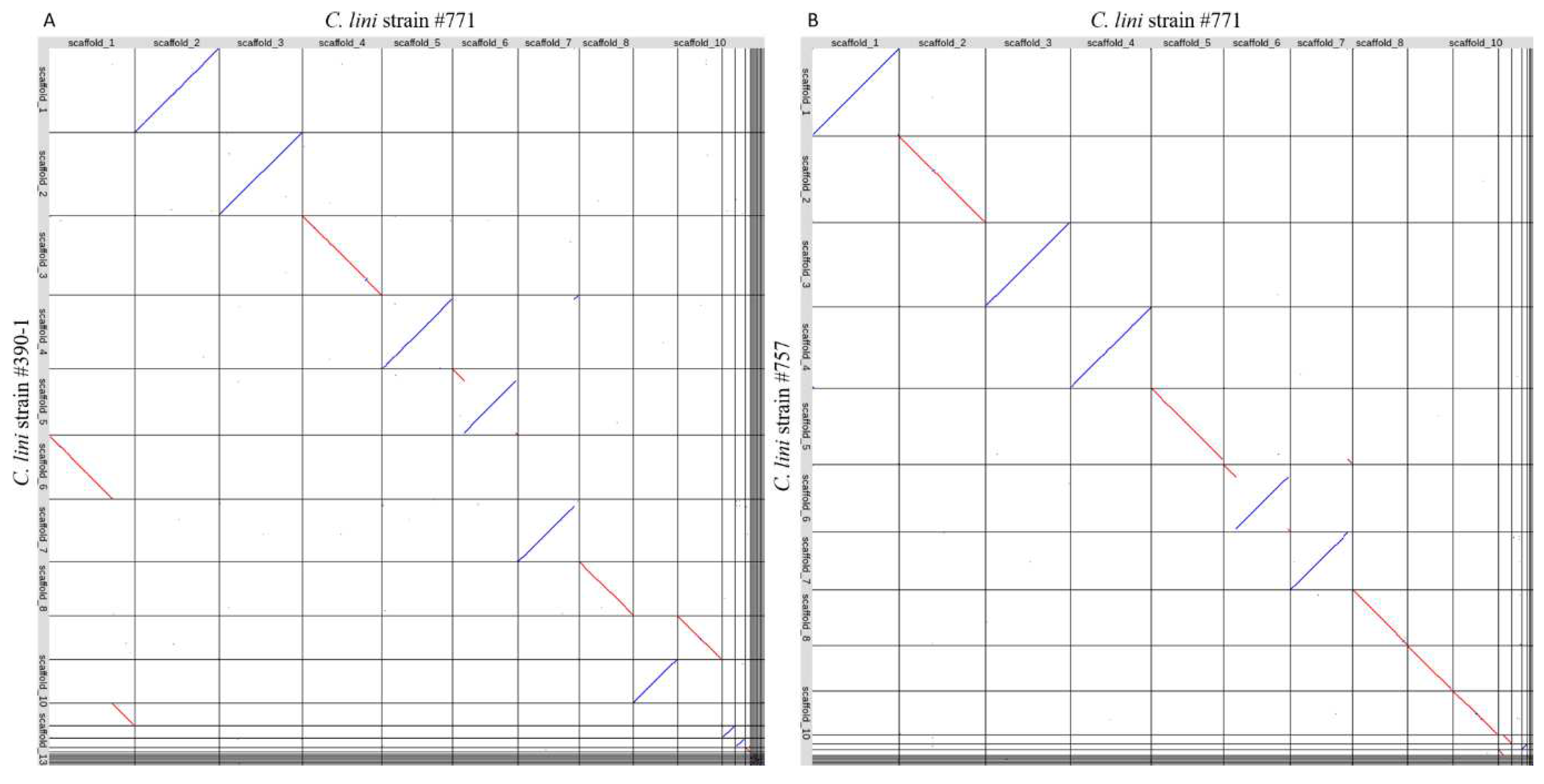
3.3. Comparative Genomic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, R.; Kumar, A.; Dhaka, R.K.; Chahar, M.; Malyan, S.K.; Singh, A.P.; Rana, A. Climate change and agriculture: impact assessment and sustainable alleviation approach using rhizomicrobiome. In Bioinoculants: Biological Option for Mitigating global Climate Change; Springer: 2023; pp. 87-114.
- Sembiring, H.; A. Subekti, N.; Erythrina; Nugraha, D.; Priatmojo, B.; Stuart, A.M. Yield gap management under seawater intrusion areas of Indonesia to improve rice productivity and resilience to climate change. Agriculture 2020, 10, 1. [Google Scholar] [CrossRef]
- Różewicz, M.; Wyzińska, M.; Grabiński, J. The most important fungal diseases of cereals—problems and possible solutions. Agronomy 2021, 11, 714. [Google Scholar] [CrossRef]
- Zakaria, L. Diversity of Colletotrichum species associated with anthracnose disease in tropical fruit crops—a review. Agriculture 2021, 11, 297. [Google Scholar] [CrossRef]
- Talhinhas, P.; Loureiro, A.; Oliveira, H. Olive anthracnose: a yield- and oil quality-degrading disease caused by several species of Colletotrichum that differ in virulence, host preference and geographical distribution. Molecular Plant Pathology 2018, 19, 1797–1807. [Google Scholar] [CrossRef]
- Singh, S.; Prasad, D.; Singh, V.P. Evaluation of fungicides and genotypes against anthracnose disease of mungbean caused by Colletotrichum lindemuthianum. International Journal of Bio-resource and Stress Management 2022, 13, 448–453. [Google Scholar] [CrossRef]
- Gruzdeviene, E.; Dabkevicius, Z. The control of flax anthracnose [Colletotrichum lini [West.] Toch.] by fungicidal seed treatment. Journal of plant protection research 2003, 43. [Google Scholar]
- Nyvall, R.F. Diseases of flax. In Field Crop Diseases Handbook; Nyvall, R.F., Ed.; Springer US: Boston, MA, 1989; pp. 251–264. [Google Scholar]
- De Silva, D.D.; Crous, P.W.; Ades, P.K.; Hyde, K.D.; Taylor, P.W.J. Life styles of Colletotrichum species and implications for plant biosecurity. Fungal Biology Reviews 2017, 31, 155–168. [Google Scholar] [CrossRef]
- Münch, S.; Lingner, U.; Floss, D.S.; Ludwig, N.; Sauer, N.; Deising, H.B. The hemibiotrophic lifestyle of Colletotrichum species. Journal of Plant Physiology 2008, 165, 41–51. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.L.; Moreno, H.L.A.; Correia, H.L.N.; Santana, M.F.; de Queiroz, M.V. Colletotrichum: species complexes, lifestyle, and peculiarities of some sources of genetic variability. Applied Microbiology and Biotechnology 2020, 104, 1891–1904. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Udayanga, D.; Cai, L.; Chukeatirote, E.; Hyde, K.D. Endophytic Colletotrichum from tropical grasses with a new species C. endophytica. Fungal Diversity 2013, 61, 107–115. [Google Scholar] [CrossRef]
- Zheng, H.; Yu, Z.; Jiang, X.; Fang, L.; Qiao, M. Endophytic Colletotrichum species from aquatic plants in southwest China. Journal of fungi 2022, 8. [Google Scholar] [CrossRef]
- Talukdar, R.; Padhi, S.; Rai, A.K.; Masi, M.; Evidente, A.; Jha, D.K.; Cimmino, A.; Tayung, K. Isolation and characterization of an endophytic fungus Colletotrichum coccodes producing tyrosol from Houttuynia cordata Thunb. using ITS2 RNA secondary structure and molecular docking study. Frontiers in bioengineering and biotechnology 2021, 9, 650247. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Shim, S.H. The fungus Colletotrichum as a source for bioactive secondary metabolites. Archives of pharmacal research 2019, 42, 735–753. [Google Scholar] [CrossRef] [PubMed]
- Vieira, W.; Bezerra, P.A.; Silva, A.C.D.; Veloso, J.S.; Camara, M.P.S.; Doyle, V.P. Optimal markers for the identification of Colletotrichum species. Molecular phylogenetics and evolution 2020, 143, 106694. [Google Scholar] [CrossRef]
- Bhunjun, C.S.; Phukhamsakda, C.; Jayawardena, R.S.; Jeewon, R.; Promputtha, I.; Hyde, K.D. Investigating species boundaries in Colletotrichum. Fungal Diversity 2021, 107, 107–127. [Google Scholar] [CrossRef]
- Van Hemelrijck, W.; Debode, J.; Heungens, K.; Maes, M.; Creemers, P. Phenotypic and genetic characterization of Colletotrichum isolates from Belgian strawberry fields. Plant Pathology 2010, 59, 853–861. [Google Scholar] [CrossRef]
- Liu, F.; Ma, Z.Y.; Hou, L.W.; Diao, Y.Z.; Wu, W.P.; Damm, U.; Song, S.; Cai, L. Updating species diversity of Colletotrichum, with a phylogenomic overview. Studies in mycology 2022, 101, 1–56. [Google Scholar] [CrossRef]
- Crouch, J.; O’Connell, R.; Gan, P.; Buiate, E.; Torres, M.F.; Beirn, L.; Shirasu, K.; Vaillancourt, L. The genomics of Colletotrichum. In Genomics of Plant-Associated Fungi: Monocot Pathogens, Dean, R.A., Lichens-Park, A., Kole, C., Eds.; Springer Berlin Heidelberg: Berlin, Heidelberg, 2014; pp. 69–102. [Google Scholar]
- Gardiner, D.M.; McDonald, M.C.; Covarelli, L.; Solomon, P.S.; Rusu, A.G.; Marshall, M.; Kazan, K.; Chakraborty, S.; McDonald, B.A.; Manners, J.M. Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. 2012.
- Lelwala, R.V.; Korhonen, P.K.; Young, N.D.; Scott, J.B.; Ades, P.K.; Gasser, R.B.; Taylor, P.W.J. Comparative genome analysis indicates high evolutionary potential of pathogenicity genes in Colletotrichum tanaceti. PloS one 2019, 14, e0212248. [Google Scholar] [CrossRef]
- Wang, H.; Huang, R.; Ren, J.; Tang, L.; Huang, S.; Chen, X.; Fan, J.; Li, B.; Wang, Q.; Hsiang, T.; et al. The evolution of mini-chromosomes in the fungal genus Colletotrichum. mBio 2023, 14, e0062923. [Google Scholar] [CrossRef]
- Krasnov, G.S.; Pushkova, E.N.; Novakovskiy, R.O.; Kudryavtseva, L.P.; Rozhmina, T.A.; Dvorianinova, E.M.; Povkhova, L.V.; Kudryavtseva, A.V.; Dmitriev, A.A.; Melnikova, N.V. High-quality genome assembly of Fusarium oxysporum f. sp. lini. Frontiers in genetics 2020, 11, 959. [Google Scholar] [CrossRef] [PubMed]
- Sigova, E.A.; Pushkova, E.N.; Rozhmina, T.A.; Kudryavtseva, L.P.; Zhuchenko, A.A.; Novakovskiy, R.O.; Zhernova, D.A.; Povkhova, L.V.; Turba, A.A.; Borkhert, E.V.; et al. Assembling quality genomes of flax fungal pathogens from Oxford Nanopore Technologies data. Journal of fungi 2023, 9. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 2011, 17, 3. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome research 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nature Biotechnology 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Wong, J.; Coombe, L.; Nikolić, V.; Zhang, E.; Nip, K.M.; Sidhu, P.; Warren, R.L.; Birol, I. Linear time complexity de novo long read genome assembly with GoldRush. Nature Communications 2023, 14, 2906. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome research 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nature biotechnology 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Wong, J.; Coombe, L.; Nikolic, V.; Zhang, E.; Nip, K.M.; Sidhu, P.; Warren, R.L.; Birol, I. Linear time complexity de novo long read genome assembly with GoldRush. Nat Commun 2023, 14, 2906. [Google Scholar] [CrossRef]
- Haghshenas, E.; Asghari, H.; Stoye, J.; Chauve, C.; Hach, F. HASLR: Fast Hybrid Assembly of Long Reads. iScience 2020, 23, 101389. [Google Scholar] [CrossRef]
- Zimin, A.V.; Puiu, D.; Luo, M.C.; Zhu, T.; Koren, S.; Marcais, G.; Yorke, J.A.; Dvorak, J.; Salzberg, S.L. Hybrid assembly of the large and highly repetitive genome of Aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res 2017, 27, 787–792. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Current protocols in bioinformatics 2020, 70, e102. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS computational biology 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome research 2017, 27, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Sovic, I.; Nagarajan, N.; Sikic, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 2017, 27, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Zimin, A.V.; Salzberg, S.L. The genome polishing tool POLCA makes fast and accurate corrections in genome assemblies. PLoS computational biology 2020, 16, e1007981. [Google Scholar] [CrossRef]
- Li, H. New strategies to improve minimap2 alignment accuracy. Bioinformatics 2021, 37, 4572–4574. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. Journal of molecular biology 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Chen, N. Using Repeat Masker to identify repetitive elements in genomic sequences. Current protocols in bioinformatics 2004, 5, 4.10. 11–4.10. 14. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nature biotechnology 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Sperschneider, J.; Dodds, P.N. EffectorP 3.0: prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. Molecular plant-microbe interactions : MPMI 2022, 35, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Molecular biology and evolution 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Gan, P.; Hiroyama, R.; Tsushima, A.; Masuda, S.; Shibata, A.; Ueno, A.; Kumakura, N.; Narusaka, M.; Hoat, T.X.; Narusaka, Y.; et al. Telomeres and a repeat-rich chromosome encode effector gene clusters in plant pathogenic Colletotrichum fungi. Environmental Microbiology 2021, 23, 6004–6018. [Google Scholar] [CrossRef]
- Taga, M.; Tanaka, K.; Kato, S.; Kubo, Y. Cytological analyses of the karyotypes and chromosomes of three Colletotrichum species, C. orbiculare, C. graminicola and C. higginsianum. Fungal Genetics and Biology 2015, 82, 238–250. [Google Scholar] [CrossRef]
- Wang, H.; Huang, R.; Ren, J.; Tang, L.; Huang, S.; Chen, X.; Fan, J.; Li, B.; Wang, Q.; Hsiang, T.; et al. The evolution of mini-chromosomes in the fungal genus Colletotrichum. mBio 2023, 14, e00629–00623. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Wang, Y.; Hao, Y.; Yao, Y.; Wang, Y.; Zhang, K.; Tan, X.; Li, Z.; Wang, W. Genome sequence resource for Colletotrichum scovillei, the cause of anthracnose disease of chili. Molecular plant-microbe interactions : MPMI 2021, 34, 122–126. [Google Scholar] [CrossRef]
- Liang, X.; Cao, M.; Li, S.; Kong, Y.; Rollins, J.A.; Zhang, R.; Sun, G. Highly contiguous genome resource of Colletotrichum fructicola generated using long-read sequencing. Molecular plant-microbe interactions : MPMI 2020, 33, 790–793. [Google Scholar] [CrossRef]
- O'Connell, R.J.; Thon, M.R.; Hacquard, S.; Amyotte, S.G.; Kleemann, J.; Torres, M.F.; Damm, U.; Buiate, E.A.; Epstein, L.; Alkan, N.; et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nature genetics 2012, 44, 1060–1065. [Google Scholar] [CrossRef] [PubMed]
- Gan, P.; Ikeda, K.; Irieda, H.; Narusaka, M.; O'Connell, R.J.; Narusaka, Y.; Takano, Y.; Kubo, Y.; Shirasu, K. Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. The New phytologist 2013, 197, 1236–1249. [Google Scholar] [CrossRef] [PubMed]
- Gan, P.; Narusaka, M.; Kumakura, N.; Tsushima, A.; Takano, Y.; Narusaka, Y.; Shirasu, K. Genus-wide comparative genome analyses of Colletotrichum species reveal specific gene family losses and gains during adaptation to specific infection lifestyles. Genome biology and evolution 2016, 8, 1467–1481. [Google Scholar] [CrossRef]
- Gan, P.; Narusaka, M.; Tsushima, A.; Narusaka, Y.; Takano, Y.; Shirasu, K. Draft genome assembly of Colletotrichum chlorophyti, a pathogen of herbaceous plants. Genome announcements 2017, 5. [Google Scholar] [CrossRef]
- Gan, P.; Tsushima, A.; Narusaka, M.; Narusaka, Y.; Takano, Y.; Kubo, Y.; Shirasu, K. Genome sequence resources for four phytopathogenic fungi from the Colletotrichum orbiculare species complex. Molecular plant-microbe interactions : MPMI 2019, 32, 1088–1090. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Wang, B.; Dong, Q.; Li, L.; Rollins, J.A.; Zhang, R.; Sun, G. Pathogenic adaptations of Colletotrichum fungi revealed by genome wide gene family evolutionary analyses. PloS one 2018, 13, e0196303. [Google Scholar] [CrossRef]
- Jayawardena, R.; Bhunjun, C.S.; Gentekaki, E.; Hyde, K.; Promputtha, I. Colletotrichum: lifestyles, biology, morpho-species, species complexes and accepted species. mycosphere 2021, 12, 519–669. [Google Scholar] [CrossRef]
- Yan, Y.; Yuan, Q.; Tang, J.; Huang, J.; Hsiang, T.; Wei, Y.; Zheng, L. Colletotrichum higginsianum as a model for understanding host(-)pathogen interactions: a review. International journal of molecular sciences 2018, 19. [Google Scholar] [CrossRef]
- Abreha, K.B.; Ortiz, R.; Carlsson, A.S.; Geleta, M. Understanding the sorghum-Colletotrichum sublineola interactions for enhanced host resistance. Frontiers in plant science 2021, 12, 641969. [Google Scholar] [CrossRef]
- Boufleur, T.R.; Ciampi-Guillardi, M.; Tikami, I.; Rogerio, F.; Thon, M.R.; Sukno, S.A.; Massola Junior, N.S.; Baroncelli, R. Soybean anthracnose caused by Colletotrichum species: Current status and future prospects. Mol Plant Pathol 2021, 22, 393–409. [Google Scholar] [CrossRef]
- Dallery, J.F.; Lapalu, N.; Zampounis, A.; Pigne, S.; Luyten, I.; Amselem, J.; Wittenberg, A.H.J.; Zhou, S.; de Queiroz, M.V.; Robin, G.P.; et al. Gapless genome assembly of Colletotrichum higginsianum reveals chromosome structure and association of transposable elements with secondary metabolite gene clusters. BMC genomics 2017, 18, 667. [Google Scholar] [CrossRef] [PubMed]
- Plaumann, P.L.; Schmidpeter, J.; Dahl, M.; Taher, L.; Koch, C. A dispensable chromosome is required for virulence in the hemibiotrophic plant pathogen Colletotrichum higginsianum. Frontiers in microbiology 2018, 9, 1005. [Google Scholar] [CrossRef] [PubMed]
- Bhadauria, V.; MacLachlan, R.; Pozniak, C.; Cohen-Skalie, A.; Li, L.; Halliday, J.; Banniza, S. Genetic map-guided genome assembly reveals a virulence-governing minichromosome in the lentil anthracnose pathogen Colletotrichum lentis. The New phytologist 2019, 221, 431–445. [Google Scholar] [CrossRef]
- Novakovskiy, R.O.; Dvorianinova, E.M.; Rozhmina, T.A.; Kudryavtseva, L.P.; Gryzunov, A.A.; Pushkova, E.N.; Povkhova, L.V.; Snezhkina, A.V.; Krasnov, G.S.; Kudryavtseva, A.V.; et al. Data on genetic polymorphism of flax (Linum usitatissimum L.) pathogenic fungi of Fusarium, Colletotrichum, Aureobasidium, Septoria, and Melampsora genera. Data in brief 2020, 31, 105710. [Google Scholar] [CrossRef]
- Latunde-Dada, A.O.; Lucas, J.A. Localized hemibiotrophy in Colletotrichum: cytological and molecular taxonomic similarities among C. destructivum, C. linicola and C. truncatum. Plant Pathology 2007, 56, 437–447. [Google Scholar] [CrossRef]
- Crouch, J.A.; O'Connell, R.; Gan, P.; Buiate, E.; Torres, M.; Beirn, L.; Shirasu, K.; Vaillancourt, L. The Genomics of Colletotrichum; 2014.
- Wang, Y.; Xu, W.-T.; Lu, R.-S.; Chen, M.; Liu, J.; Sun, X.-Q.; Zhang, Y.-M. Genome sequence resource for Colletotrichum gloeosporioides, an important pathogenic fungus causing anthracnose of Dioscorea alata. Plant Disease 2023, 107, 893–895. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, G.S.; Pushkova, E.N.; Novakovskiy, R.O.; Kudryavtseva, L.P.; Rozhmina, T.A.; Dvorianinova, E.M.; Povkhova, L.V.; Kudryavtseva, A.V.; Dmitriev, A.A.; Melnikova, N.V. High-quality genome assembly of Fusarium oxysporum f. sp. lini. Frontiers in genetics 2020, 11, 959. [Google Scholar] [CrossRef]
- Sigova, E.A.; Pushkova, E.N.; Rozhmina, T.A.; Kudryavtseva, L.P.; Zhuchenko, A.A.; Novakovskiy, R.O.; Zhernova, D.A.; Povkhova, L.V.; Turba, A.A.; Borkhert, E.V.; et al. Assembling Quality Genomes of Flax Fungal Pathogens from Oxford Nanopore Technologies Data. Journal of fungi 2023, 9, 301. [Google Scholar] [CrossRef]
- Becerra, S.; Baroncelli, R.; Boufleur, T.R.; Sukno, S.A.; Thon, M.R. Chromosome-level analysis of the Colletotrichum graminicola genome reveals the unique characteristics of core and minichromosomes. Frontiers in microbiology 2023, 14, 1129319. [Google Scholar] [CrossRef]
- Ma, W.; Wang, Y.; McDowell, J. Focus on effector-triggered susceptibility. Molecular Plant-Microbe Interactions® 2018, 31, 5–5. [Google Scholar] [CrossRef]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: from pathogen perception to robust defense. Annual Review of Plant Biology 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, D.-K.; Chuang, S.-C.; Chen, C.-Y.; Chao, Y.-T.; Lu, M.-Y.J.; Lee, M.-H.; Shih, M.-C. Comparative genomics of three Colletotrichum scovillei strains and genetic analysis revealed genes involved in fungal growth and virulence on chili pepper. Frontiers in microbiology 2022, 13, 818291. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




