Submitted:
04 July 2023
Posted:
05 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
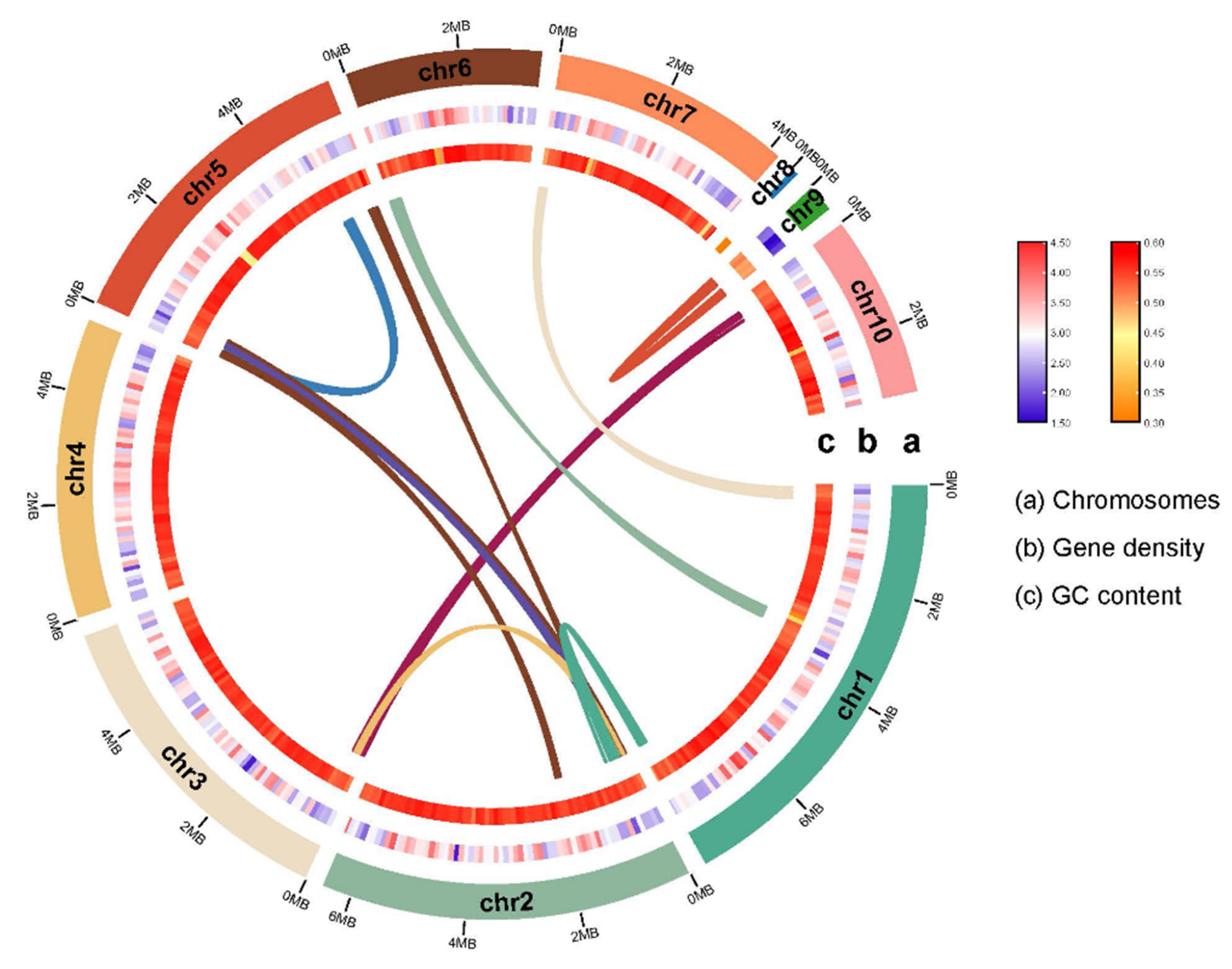
2.1. Genome Sequencing, Assembly, and Annotation
2.2. Phylogenetic analysis and Comparative genomics analysis

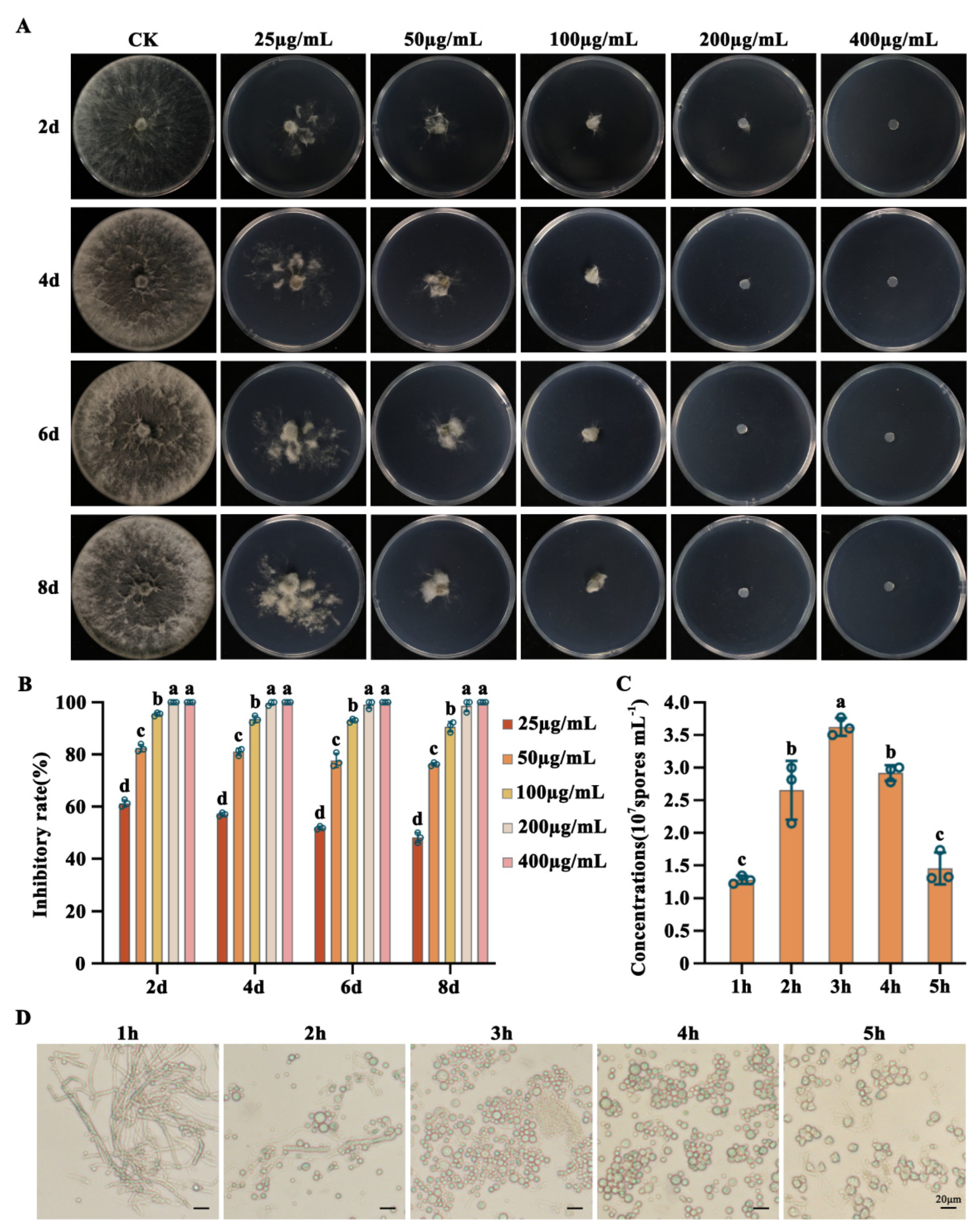
2.3. Sensitivity of L. theobromae to neomycin (G-418) and Protoplast Preparation
2.4. Screening and Detection of Transformant
3. Discussion
4. Materials and Methods
4.1. Fungal strain, culture conditions and neomycin (G-418) sensitivity determination
4.2. DNA extraction, genome sequencing, assembly
4.3. Genome annotation
4.4. Phylogenetic Analysis
4.5. Construction of recombinant plasmids
4.6. Fluorescent protein labeling strain construction
4.7. Microscopic Examinations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ma, L.; Lin, J.; Li, Q.; Zhang, L.; Chen, A. Antifungal constituents from the husk of Carya cathayensis. Scientia Silvae Sinicae. 2009, 45, 90–94. [Google Scholar]
- Zhu, C.; Deng, X.; Shi, F. Evaluation of the antioxidant activity of Chinese hickory (Carya cathayensis) kernel ethanol extraction. African Journal of Biotechnology. 2008, 7, 44–45. [Google Scholar]
- Wang, Q.W.; Zhang, C.Q. q-LAMP Assays for the Detection of Botryosphaeria dothidea causing Chinese hickory canker in trunk, water, and air samples. Plant Disease. 2019, 103, 3142–3149. [Google Scholar] [CrossRef]
- Zhang, C.Q.; Xu, B.C. First report of canker on Chinese hickory ( Carya cathayensis ) caused by Botryosphaeria dothidea in China. Plant Disease. 2011, 95, 1319. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.J.; Wang, H.D.; Wang, Y.P.; Zhang, C.Q. Management of Chinese hickory (Carya cathayensis) trunk canker through effective fungicide application programs and baseline sensitivity of Botryosphaeria dothidea to trifloxystrobin. Australasian Plant Pathology. 2017, 46, 1–8. [Google Scholar] [CrossRef]
- Zhuang, C.; Wang, Q.; Wu, Q.; Qiu, Z.; Xu, B.; Zhang, C. Diversity of Botryosphaeriaceae species associated with Chinese hickory tree (Carya cathayensis) trunk cankers. Plant Disease. 2021, S2210289R. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Ozbudak, E.; Liu, G.; Hosmani, P.S.; Saha, S.; Flores-Gonzalez, M.; Mueller, L.A.; Rodrigues-Stuart, K.; Dewdney, M.M.; Lin, Y.; et al. Draft genome sequence resource of the citrus stem-end rot fungal pathogen Lasiodiplodia theobromae CITRA15. Phytopathology 2021, 111, 761–764. [Google Scholar] [CrossRef]
- Ali, S.S.; Asman, A.; Shao, J.; Balidion, J.F.; Bailey, B.A. Genome and transcriptome analysis of the latent pathogen Lasiodiplodia theobromae, an emerging threat to the cacao industry. Genome. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.; Di Lonardo, D.P.; Bodelier, P.L. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiology Ecology. 2017, 93. [Google Scholar] [CrossRef]
- Bochner, B.R.; Gadzinski, P.; Panomitros, E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Research. 2001, 11, 1246–1255. [Google Scholar] [CrossRef]
- Bochner, B.R. Innovations: New technologies to assess genotype–phenotype relationships. Nature Reviews Genetics. 2003, 4, 309–314. [Google Scholar] [CrossRef]
- Bao, J.; Wu, Q.; Huang, J.; Zhang, C.Q. High-quality genome assembly and annotation resource of Botryosphaeria dothidea strain BDLA16-7, causing trunk canker disease on Chinese hickory. Plant Disease. 2022, 106, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, Y.; Hu, S.; Huang, J.; Zhang, C. High-quality genome assembly and annotation resource of three Botryosphaeria pathogens causing Chinese hickory canker. Molecular Plant-Microbe Interactions 2022, 35, 941–943. [Google Scholar] [CrossRef]
- Muniz, C.R.; Da, S.G.; Souza, M.J.; Freire, F.C.; Kema, G.H.; Guedes, M.I. Agrobacterium tumefaciens-mediated transformation of Lasiodiplodia theobromae, the causal agent of gummosis in cashew nut plants. Genetics and Molecular Research. 2014, 13, 2906–2913. [Google Scholar] [CrossRef]
- Li, D.; Tang, Y.; Lin, J.; Cai, W. Methods for genetic transformation of filamentous fungi. Microbial Cell Factories. 2017, 16, 168. [Google Scholar] [CrossRef]
- Bao, J.; Chen, M.; Zhong, Z.; Tang, W.; Lin, L.; Zhang, X.; Jiang, H.; Zhang, D.; Miao, C.; Tang, H.; et al. PacBio sequencing reveals transposable elements as a key contributor to genomic plasticity and virulence variation in Magnaporthe oryzae. Molecular Plant. 2017, 10, 1465–1468. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Research. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. STRIDE: Species tree root inference from gene duplication events. Molecular Biology and Evolution. 2017, 34, 3267–3278. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology. 2019, 20, 238. [Google Scholar] [CrossRef]
- Gomez-Lunar, Z.; Hernandez-Gonzalez, I.; Rodriguez-Torres, M.D.; Souza, V.; Olmedo-Alvarez, G. Microevolution analysis of Bacillus coahuilensis unveils differences in phosphorus acquisition strategies and their regulation. Frontiers in Microbiology. 2016, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Kim, J.M.; Kim, H.G.; Kim, B.T.; Kim, D.H. Protoplast-mediated transformation of the filamentous fungus Cladosporium phlei: evidence of tandem repeats of the integrative transforming vector. Plant Pathology Journal. 2009, 25, 179–183. [Google Scholar] [CrossRef]
- Men, P.; Wang, M.; Li, J.; Geng, C.; Lu, X. Establishing an efficient genetic manipulation system for sulfated echinocandin producing fungus Coleophoma empetri. Frontiers in Microbiology. 2021, 12, 734780. [Google Scholar] [CrossRef] [PubMed]
- Miga, K.H.; Koren, S.; Rhie, A.; Vollger, M.R.; Gershman, A.; Bzikadze, A.; Brooks, S.; Howe, E.; Porubsky, D.; Logsdon, G.A.; et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020, 585, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Bruna, T.; Hoff, K.J.; Lomsadze, A.; Stanke, M.; Borodovsky, M. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genomics and Bioinformatics. 2021, 3, a108. [Google Scholar] [CrossRef] [PubMed]
- Robert-Siegwald, G.; Vallet, J.; Abou-Mansour, E.; Xu, J.; Rey, P.; Bertsch, C.; Rego, C.; Larignon, P.; Fontaine, F.; Lebrun, M.H. Draft genome sequence of Diplodia seriata F98.1, a fungal species involved in grapevine trunk diseases. Genome Announcements 2017, 5. [Google Scholar] [CrossRef] [PubMed]



| Features | L. theobromae (stain LTTK16-3) |
|---|---|
| GWH accession 1 | GWHBEBO00000000 |
| ONT Reads (Gb) | 5.96 |
| Reads Coverage (×) | 139 |
| Assembly size (Mb) | 42.82 |
| Contig number | 10 |
| Contig N50 (Mb) | 5.67 |
| Contig L50 | 4 |
| Maximum contig length (Mb) | 7.93 |
| Telomeric repeats (TTAGGG)n 2 | 6/5:3 |
| GC content | 54.57% |
| Repeat sequences | 3.37% |
| BUSCO completeness | 98.71% |
| Protein-coding genes | 12,516 |
| Pathogen-host interaction genes | 2,457 |
| Carbohydrate active enzymes | 237 |
| Cytochrome P450 enzymes | 190 |
| Putative secreted proteins | 715 |
| SMBGCs 3 | 51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





