1. Introduction
Breast cancer has the highest incidence among all types of solid cancers among women worldwide in 2020, leading to the highest mortality [
1]. To reduce mortality, mammography was introduced for breast screening in many countries since the early 2000s. Mammography remains the most common imaging technique for breast cancer diagnosis in most countries and a standard screening mammogram consists of x-ray imaging with 2 views on each breast in the mediolateral oblique (MLO) and craniocaudal (CC) projection. Mammographic images in these two views are interpreted by radiologists and other readers to determine whether the screening case is negative for breast cancer, or the woman needs to be recalled for further imaging and/or testing. The mortality for women with breast cancer from European populations has reduced by over 31% as attributed to population-based programs using mammography [
2]. Women diagnosed with abnormal mammograms are recommended for further testing, which can include additional images or biopsy. Over 60% of these biopsies are diagnosed as cancer free [
3].
Although the sensitivity (>86%) and specificity (>96%) [
4] of screening mammography to detect breast cancer for women with almost entirely fatty breasts is relatively high, a major challenge in mammography screening involves women with dense breasts, as breast cancer can be masked by glandular tissue. Tissue superposition occurs in mammography when there are overlapping layers of breast tissue that can obscure small or subtle abnormalities, making it difficult for radiologists to accurately interpret the images [
5]. This issue has been partially mitigated by digital breast tomosynthesis (DBT), which is an advanced mammographic technology that captures three-dimensional images of the breast, allowing for a more detailed and layered view of breast tissue. However, the larger volume of images generated by DBT necessitates more time for both image acquisition and interpretation [
6].
Over the past decade, Artificial Intelligence (AI) has garnered extensive attention in medical imaging for its promising advancements in diagnostic accuracy of interpretative tasks related to various organs like the brain, liver, breast, and lung [
7,
8,
9,
10,
11,
12,
13,
14,
15,
16,
17,
18,
19]. Particularly, deep learning methods applied to diagnose breast cancer through mammographic images have captivated extensive interest [
10,
12,
16,
17]. The effective training of AI models for clinical application demands a vast amount of data containing precise lesion locations. However, the acquisition of these extensive sets of images with lesion locations significantly increases the workload for radiologists and physicians. To mitigate some of these workload challenges, transfer learning [
20], involving the use of pre-trained AI models in different settings, has emerged as a potential solution.
Breast screening with AI models can assist radiologists in interpreting mammograms, especially in distinguishing between normal and abnormal cases [
21] The Globally-aware Multiple Instance Classifier (GMIC) [
16] AI model was designed to classify mammographic cases as benign or malignant. Furthermore, the Global-Local Activation Maps (GLAM) [
17] AI model extended GMIC to classify mammographic cases as benign or malignant by generating multiple scale saliency maps. The I&H AI model [
10] used deep neural networks to assist radiologists interpret screening mammograms. End2End AI model [
12] demonstrated a method of breast screening on mammograms using deep neural networks. All these four AI models used Residual Networks (ResNet) architecture [
22] in the training and testing processes. For completeness of the paper, a detailed review of these methods is given in the methods section.
This paper investigates the performance of these four publicly available state-of-the-art AI models: GMIC, GLAM, I&H and End2End, on a screening mammographic database of Australian women. This study's primary goals include:
- (1)
Comparing the performance of these models on an Australian dataset, which differs from their original training data (both in terms of population characteristics and the types of mammography machines (vendors) used), highlighting the influence of dataset variations on predictions.
- (2)
Investigating the potential improvement of model performance through transfer learning, and hence the value of tailoring the AI models for other nationalities' context.
- (3)
Examining the impact of image enhancement techniques on model predictions to assess their potential to enhance diagnostic accuracy.
- (4)
Exploring the association between the AI models' malignancy probability outputs and histopathological features, offering insights into the models' predictive accuracy and its potential clinical relevance, aiding further treatment/triaging decision-making.
2. Materials and Methods
Four state-of-the-art AI models involving deep neural networks were used to test an Australian mammographic database. Transfer learning of the four pre-trained AI models was conducted on the database to update these AI models. Since the images in our dataset were obtained from different vendors, they exhibited significantly different histograms and dynamic ranges. Therefore, we applied the Contrast Limited Adaptive Histogram Equalization (CLAHE) algorithm [
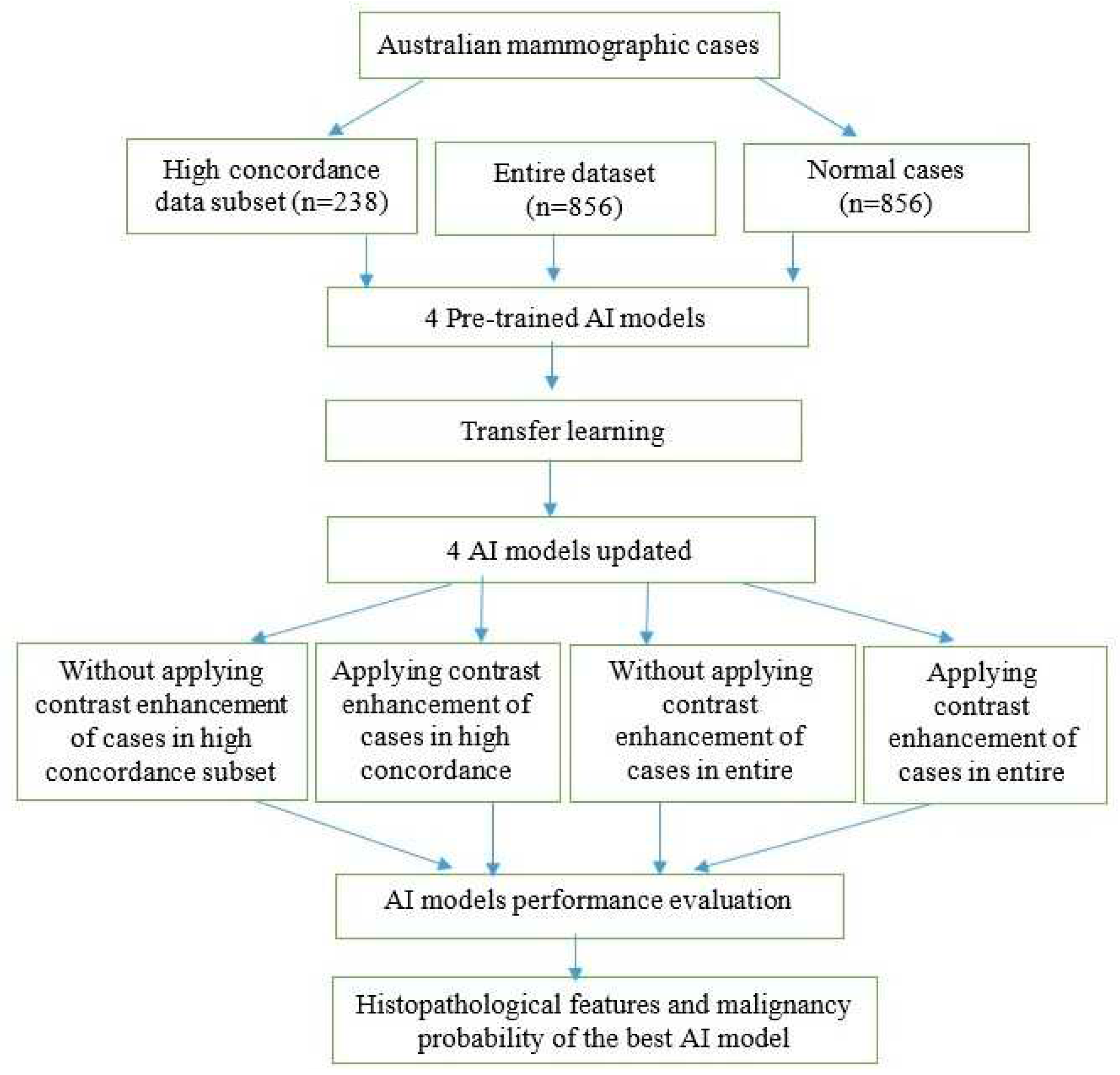
23] to enhance the contrast of mammographic cases and evaluated its impact on the performance of AI models. The receiver operating characteristic curve (ROC) and the area under the ROC curve (AUC) metrics were used to evaluate the performance of the four AI models in different scenarios. Histopathological features were analyzed with the malignancy probabilities of mammographic cases to provide the best AI model in terms of AUC values. Our method consisted of several steps as illustrated in
Figure 1.
2.1. Data Acquisition
After ethics approval from the University of XXXX, we used screening mammograms collected from the Australian mammographic database called
XXX to assess the performance of the four AI models. The XXX database consists of 1712 mammographic cases (856 normal cases and 856 malignant cases). Each malignant case was confirmed by the reports of follow-up biopsies. Each case had four mammographic views: right MLO, Left MLO, Right CC, and Left CC views. Mammograms were acquired from mammography machines manufactured by five different vendors, including Fuji Film (32% of cases), Konica (4% of cases) Siemens (34% of cases), Hologic (19%), and Sectra (11% of cases). Each case was annotated by two radiologists and recorded as box regions on the mammographic images.
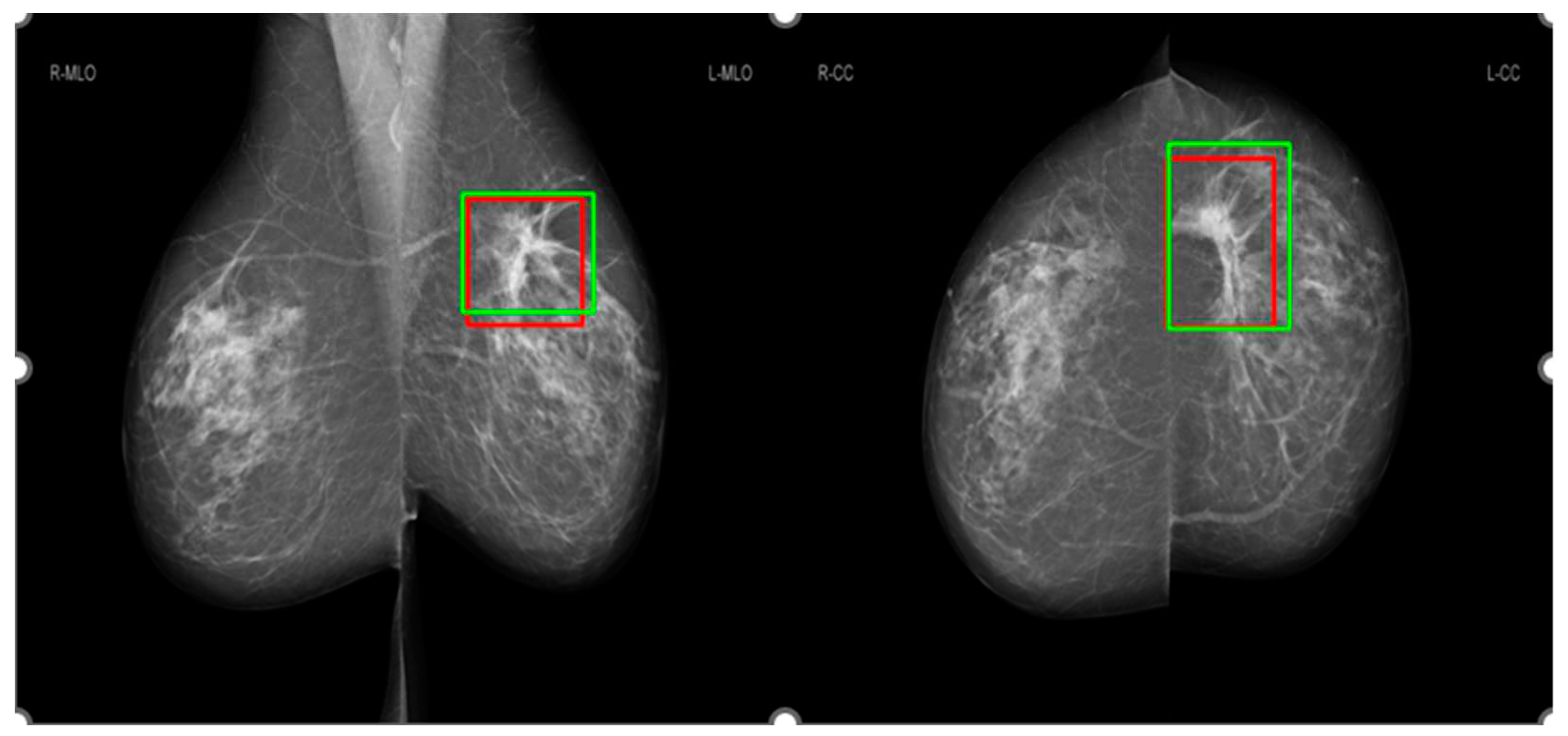
Figure 2 shows an example for the annotations of two radiologists on a mammographic case, with red boxes from Radiologist A and green boxes from Radiologist B. Concordance levels were constructed by analyzing Lin’s concordance correlation coefficient (CCC) [
24] between the annotations of two radiologists on mammograms according to McBride's interpretation guide [
25]. Lin’s CCC was computed based on the corners of two overlapped boxes of annotations on the same mammographic image. Intersection over Union [
26] metric was used to determine whether two boxes overlapped or not, with a value greater than 0 indicating the overlapping of two boxes. Mammographic images were classified as four concordance levels: ‘almost perfect’ at >0.99 (238 cases), ‘substantial’ at <0.95 to 0.95 (222 cases), ‘moderate’ at 0.95 to 0.90 (202 cases), and ‘poor’ at <0.90 (194 cases).
The training and testing mammographic cases of our database had an equal representation of breast density. Two image sets were developed: the first subset included cases rated with ‘almost perfect’ agreement between radiologists (termed ‘high-concordance subset’ in this paper), and the second dataset included all cases that have been marked with cancers with ‘no concordance threshold’ applied (termed ‘entire dataset’ in this paper).
2.2. AI models
The GLAM, GMIC, I&H and End2End models were evaluated in this study. These four models were selected as each model provided promising results in diagnosing cancers on mammographic images with high AUC values. The GMIC model combined the global and local context in the decision-making process [
16]. To obtain additional details of the local context, the GLAM incorporated zoom functionality for the local context, hence it is a similar approach taken by radiologists interpreting mammographic images [
17]. To mimic radiologists interpreting mammographic images from different views, I&H fused each model trained on each view for the decision-making process [
10] as sometimes a mammographic image from a single view is not enough to determine whether the mammographic image has cancer. Instead of searching cancer signs in a direction from the global to the local on a mammographic image like GMIC and GLAM, End2End trained a local classifier and then expanded to a global classifier to determine whether the mammographic images showed signs of cancers. Although the AUC values reported previously for GMIC, GLAM, I&H and End2End using their original mammography databases were 0.909, 0.882, 0.895, and 0.88, respectively, these AI models have reportedly provided relatively low AUC values on other mammographic databases from different ethnicities and manufacturers [
27].
2.2.1. Globally-aware Multiple Instance Classifier (GMIC)
The GMIC firstly learned the global feature map of a mammographic image using a ResNet-22 network [
25]. The global feature map was convolved with a 1 x 1 filter and Sigmoid operation to generate a malignant map. The value of each pixel in the global feature map was [0,1], which indicated whether the presence of malignancy. The feature map was then scanned to get non-overlapping
K patches with largest total intensity inside the patches. As suggested in the original paper,
K was set as 3. Local features of patches were extracted using a ResNet-34 network and then combined with a gated attention network for computing weights of features. The final step combined the malignant map and local feature with weighted representation of all patches to predict malignancy probability. All the mammographic images for GMIC models were resized to a resolution of 1920 x 2944 pixels using bilinear interpolation [
28]. For the GMIC model, the source codes are publicly available on the GitHub at
https://github.com/nyukat/GMIC.git.
2.2.2. Global-local Activation Maps (GLAM)
The GLAM learned the global saliency map of a mammographic image using a convolutional neural network (CNN). To capture different sizes of malignancy, the global saliency map was generated at different scales. The second stage generated a set of patches from the feature map based on the local maximum of average intensity. In the last stage, each image patch was applied a ResNet-34 network [
22] to extract the local feature map, which was then assigned to the corresponding mammographic image. All feature maps of local patches were combined with the global feature map to predict the probability of malignancy on a mammographic image using binary cross-entropy function. All the mammographic images for GLAM models were also resized to a resolution of 1920 x 2944 pixels. For the GLAM model, the source codes are publicly available on the GitHub at
https://github.com/nyukat/GLAM.git.
2.2.3. I&H
I&H trained AI models based on MLO and CC views on each breast and concatenated representations from four views to predict the probability of malignancy in each mammographic image. A ResNet-22 was used for model training in a mammographic image of each view. The mammographic images in CC view for I&H model were resized to 2677 x 1942 and 2974 x 1748 in MLO view. For this model, we used the source codes published by the authors on the GitHub at
https://github.com/nyukat/ breast_cancer_classifier.git.
2.2.4. End2End
End2End converted a patch classifier to a whole mammographic image classifier by adding heatmaps and convolutional layers on the top of the neural network. These convolutions used two Visual Geometry Group (VGG) [
29] blocks with 3 x3 convolutions and batch normalization. All the mammographic images for End2End were resized to a resolution of 1152 x 896 pixels. For this model, we used the source codes published by the authors on the GitHub at
https://github.com/lishen/end2end-all-conv.git.
2.3. Image enhancement
Image enhancement techniques can be helpful to optimize contrast of mammographic images and one example is from Min et al. [
30], where the study presented pseudo-color mammogram generation to enhance mass-like feature in mammographic images. In this study, we used the CLAHE [
23] algorithm to enhance mammographic images because it is fast and produces promising contrast enhancement. The CLAHE algorithm firstly divided an image into un-overlapped tiles. In the second stage, it conducted histogram equalization for each tile. The histogram equalization used a pre-defined clip limit to redistribute the bins and then map to an improved tile. The last stage combined each improved tiles to generate an enhanced image using bilinear interpolation. For the parameters of the CLAHE algorithm, the clip Limit was set to 12 and tile Grid Size was set to (8, 8).
2.4. Transfer learning
Transfer learning of the four AI models was conducted on the XXX database, including 856 cancer cases and 856 normal cases. All DICOM images were downsampled to match the resolution of the input images for the models and converted to PNG format to reduce the computational time of the training process. We conducted a four-fold cross validation to train and test the four AI models on the database with transfer learning. The training set was further split into training and validation sets to refine the stopping criteria. This step involved an iterative process, assessing the AI models' accuracy in the current epoch against the previous one. The training concluded when the validation process callback showed no improvement in model accuracy, typically after a patience threshold of 3 epochs had been reached.
The transfer learning of each AI model was optimized using Adam algorithm [
31]. The loss function used the binary cross-entropy. As suggested in the original studies, the learning rates for the GMIC, GLAM and I&H were set as 10
-5 and End2End was set as 10
-4, respectively. For an equitable comparison of performance between transfer learning models and pre-trained models, the transfer learning approach employed the ResNet-22 network for the global module and the ResNet-34 network for the local module. These are the same networks utilized by the pre-trained GMIC and GLAM models. Additionally, I&H utilized the ResNet-22 network as its pre-trained model, while End2End employed the VGG network as its pre-trained model.
2.5. Evaluation metrics
The performance of four AI models in the classification of malignancy on mammographic images was evaluated using sensitivity, specificity, the area under receiver operating characteristic curve (AUC). An ANOVA test was conducted for each AI model between the two image sets, with the corresponding p-values as shown in the Results section. A threshold of statistical significance was set as 0.05. Bonferroni correction was used to adjust for multiple comparisons.
2.6. Association between the malignancy probability from the AI and histopathological features
We also employed the Kruskal-Wallis U-test to investigate potential differences in malignancy probability as predicted by the top-performing AI model across distinct categories based on pathology reports. We considered pathological factors including Estrogen Receptor (ER), Progesterone Receptor (PR), Breast Cancer Grade, Human Epidermal Growth Factor Receptor 2 (Her2), and the differentiation between Ductal Carcinoma In Situ (DCIS) and invasive cancer. Additionally, an analysis was conducted based on the size of cancers, with tumours classified into four groups (mm): (0.999, 10.0], (10.0, 15.0], (15.0, 25.0], and (25.0, 150.0] intervals. The Kruskal-Wallis U-test was utilized to assess the statistical significance of differences among these groups.
3. Results
3.1. The performances of four AI models
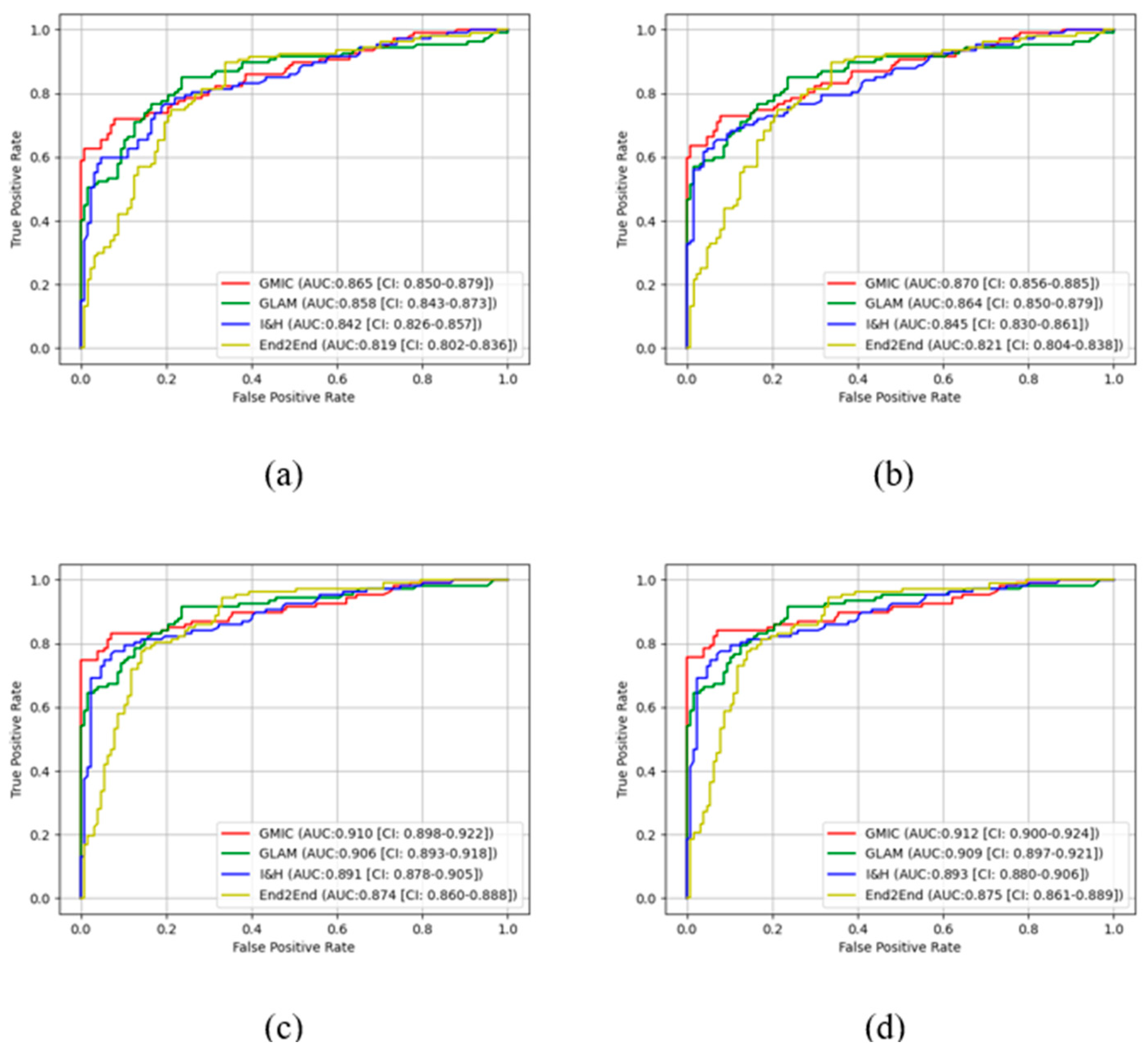
In the pre-trained stage, GMIC obtained significantly higher AUC score in both the high-concordance subset and entire dataset in original (0.865 and 0.824) and contrast-enhanced (0.870 and 0.836) modes, followed by the GLAM, I&H, and then End2End models (
Table 1). There were significant differences (P<0.05) (
Table 1) in the performances of these models between two datasets. The AUC values of the four AI models were higher when CLAHE image enhancement algorithm was applied, in comparison with the original mammograms (
Table 1) (e.g., 0.870 for GMIC + CLAHE vs. 0.865 for GMIC only in the high-concordance subset, and 0.836 for GMIC + CLAHE vs. 0.824 for GMIC only in the entire dataset).
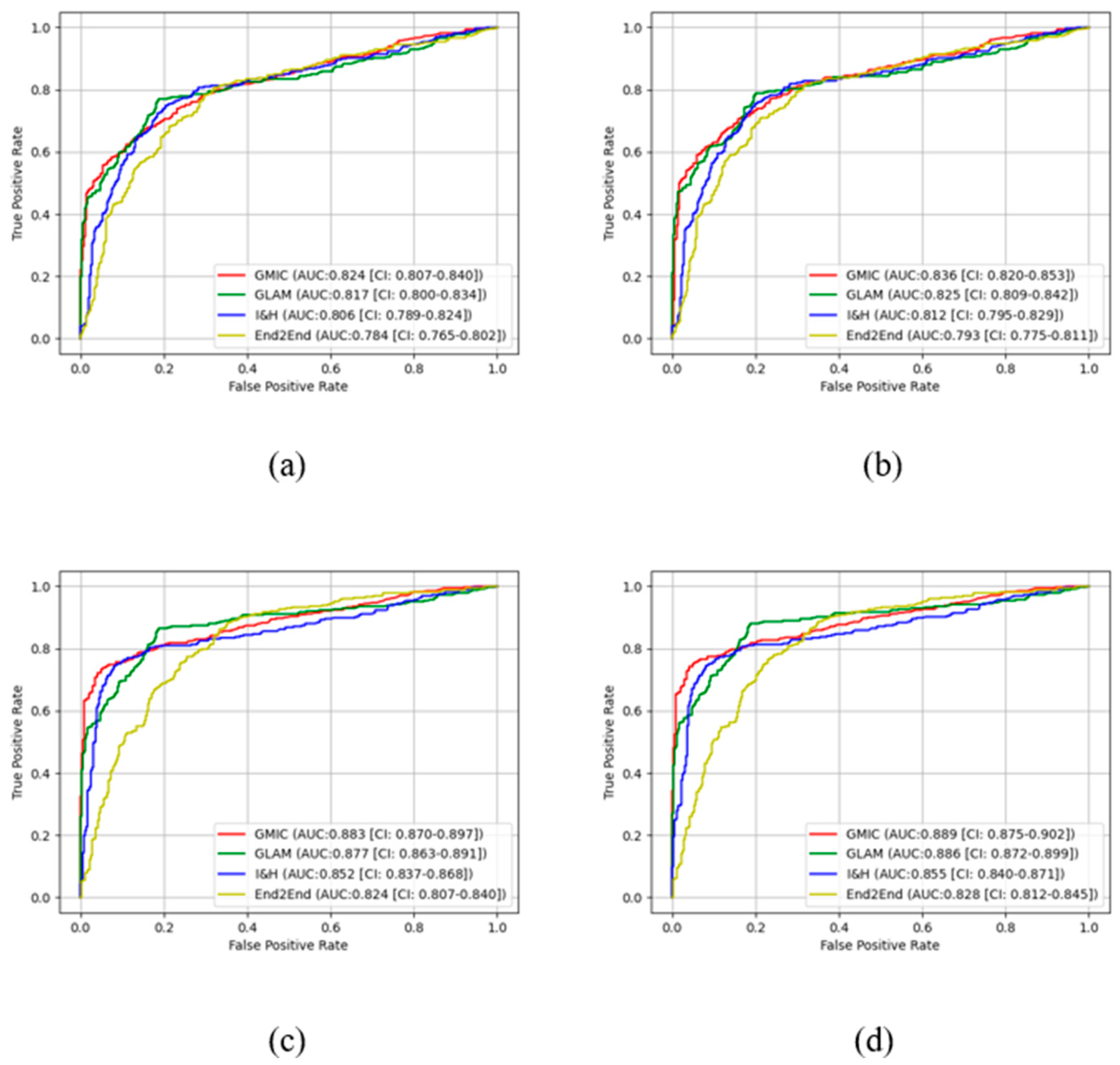
In the transfer learning stage, the highest AUC score was found with the GMIC for both the high-concordance subset and entire dataset (0.910 and 0.883) and again with the contrast-enhanced (0.912 and 0.889) mode, compared with the values generated by the GLAM, I&H, and then End2End’s models without contrast enhancement (
Table 1). Significantly higher AUC scores were also reported in the subset than the entire dataset across four models with and without contrast-enhancement (P<0.05) (
Table 1). There was an improvement in the AUC values of the four transfer learning AI models on the contrast-enhanced mammograms compared with the original mammograms in both datasets as shown in this table (e.g., 0.912 for GMIC + CLAHE vs. 0.910 for GMIC only in the high-concordance subset, and 0.889 for GMIC + CLAHE vs. 0.883 for GMIC only in the entire dataset).
Figure 3 and
Figure 4 show the comparison of ROC curves of the four AI models with and without transfer learning on high-concordance subset and entire dataset, respectively. The ROC curves in these figures show a clear improvement of performance among the four AI models with transfer learning (see
Figure 3 and 4 (a) and (c)) and CLAHE contrast enhancement (see
Figure 3 and 4 (b) and (d)).
Confidence intervals for the four AI models on high-concordance subset are shown in the legend of each subfigure.
Figure 3 and
Figure 4 also illustrate that the
receiver operating characteristic (ROC) curves of the four AI models, both with and without transfer learning and with and without contrast enhancement, exhibited superior performance in high-concordance compared to the entire dataset (e.g.,
Figure 3 (a) and
Figure 4 (a)). The ROC curves of the four AI transfer learning models shown more improvement on two datasets than those of the four pre-trained AI models (e.g.,
Figure 3 (a) and
Figure 4 (a) vs.
Figure 3 (c) and
Figure 4 (c)).
3.2. Pairwise Comparisons of four AI models
We conducted pair-wise comparisons among the models in various scenarios to explore if the difference in the performances were significant. In each scenario, six comparisons were made and the p-values were adjusted using Bonferroni correction. As shown in
Table 2, the differences were more significant when models were re-calibrated using transfer learning. This highlights the need of transfer learning to leverage the maximum added benefit of the model. The GMIC and GLAM models were not significantly different in the entire dataset because both models have similar architecture of networks and GLAM was an extended work of GMIC.
The I&H and GMIC or GLAM models were not significantly different when using the original or contrast-enhanced images in the entire dataset, but significant differences were observed when transfer learning models were used. The GMIC and End2End models were significantly different in both the high-concordance subset and the entire dataset due to different deep neural network architectures for the two models (one with ResNet and the other with VGG).
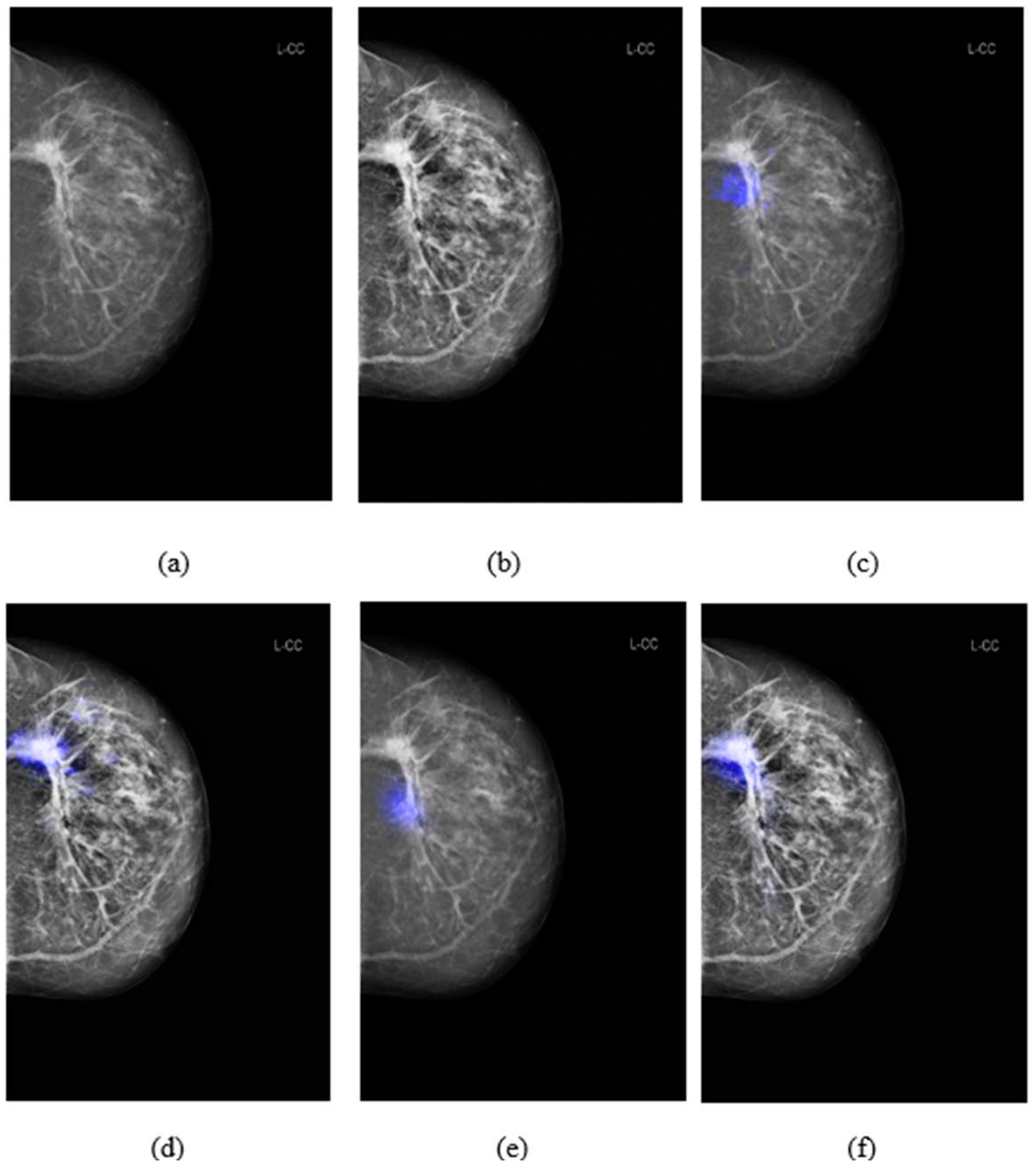
3.3. Comparison of salience maps on original and locally-enhanced mammographic images
Figure 5 shows the comparison of saliency maps generated from GLAM and GMIC on both an original mammographic image and with the applied CLAHE algorithm. The annotations of two radiologists on the same mammographic case were shown in the left CC view in
Figure 2. From
Figure 5 we can see that the saliency maps of GLAM (see
Figure 5(c)) and GMIC (see
Figure 5(e)) from original mammographic images deviated from the centroid of the radiologists’ annotations and occupied a smaller area of the annotations. However, the saliency maps of the two AI models from the contrast-enhanced image (see
Figure 5(d) and (f)) aligned with the centroid of radiologists’ annotations and occupied a larger area of the annotations.
3.4. Association between the malignancy probability from the AI and histopathological features
The outcomes of the Kruskal-Wallis tests, assessing the significance of differences in malignancy probability predicted by the highest-performing AI model (GMIC) across various pathological factors, revealed non-significant findings. The comparison based on ER, PR, and Her2 status yielded p-values of 0.342, 0.414, and 0.179 respectively. The examination of breast cancer grade resulted in a p-value of 0.169. Additionally, the differentiation between DCIS (503 cases) and invasive cancer (312 cases) exhibited a non-significant p-value of 0.152.
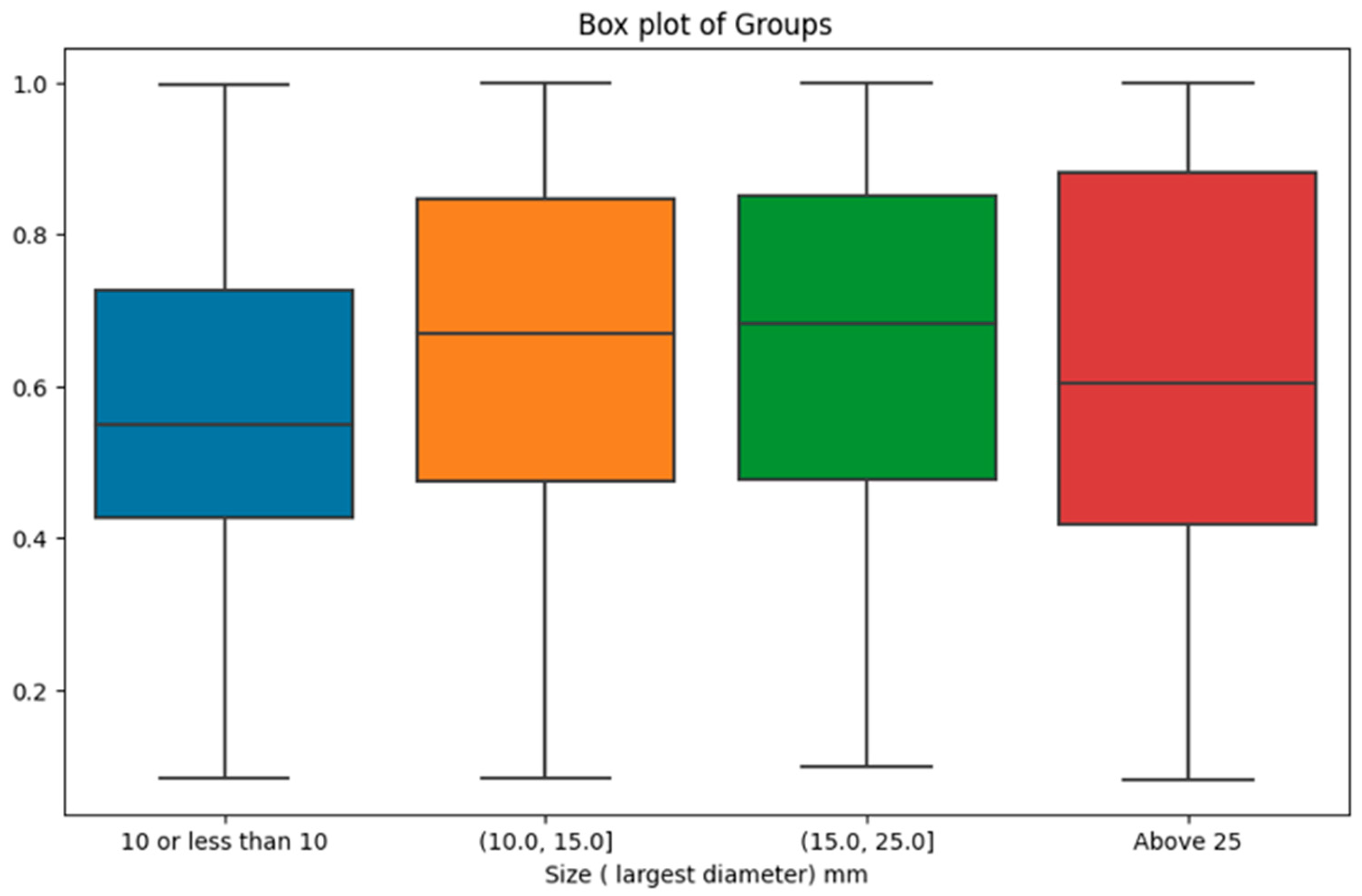
However, when investigating the impact of tumour size categories on malignancy probability, the results were statistically significant. There were 337 cases with tumor size in (0, 10.0mm], 174 cases in (10.0, 15.0mm], 179 cases in (15.0, 25.0mm], and 166 cases above 25mm. The analysis yielded a p-value of 0.0002, indicating that the distinct size groups indeed manifest significant differences in malignancy probability provided by the AI model. As shown in
Figure 6, the most prominent difference was observed between the first size category (i.e., lesions with a size of 10mm or less) with the lowest malignancy probability scores compared with the other size intervals.
4. Discussion
In previous studies, the mammograms for training and testing the GLAM, GMIC and I&H were conducted with the New York University Breast Cancer screening database, [
32] which included examinations from two manufacturers: Siemens and Hologic. The training and testing data for End2End were film-screen (FS) mammographic images from the Digital Database for Screening Mammography (DDSM) [
33]. Our dataset included digital mammographic images collected from a wider range of vendors such as Sectra, Fuji, Siemens, Hologic, GE Healthcare and Philips Healthcare. The mammographic images from the NYU and DDSM databases were obtained in the USA, whilst our dataset was obtained in Australia and could represent different populations, with the majority ethnicity group of our database unlikely to be matched with the USA databases. Previous research has shown an 8% difference in the AUC of an AI model on US screening mammograms and UK screening mammograms [
11].
Our results showed that transfer learning improved the performance of the four AI models in detecting cancer lesions on digital screening mammograms. As shown in
Table 1, the AUC of the transfer learning GMIC model increased from 0.865 for the pre-trained model to 0.910 in the high-concordance subset and from 0.824 to 0.883 in the entire dataset. Similar results were also found for GLAM, I&H and End2End. This indicates that transfer learning of the four models was influenced by the quality of the concordance levels, indicating that high quality data together with undertaking transfer learning are both important factors for training an effective AI model.
Applying image enhancement via CLAHE algorithm to our image set improved the performance of the AI models in detecting cancer lesions on screening mammograms. The AUC values of the four AI models were greater than those without enhanced mammographic images. Other image enhancement such as Pseudo-color mammogram generation and local histogram equalization algorithm [
34] may also improve the AUC performance of AI models and this could be a direction for future work.
We also explored the prediction of malignancy probability by the GMIC as the highest performing model across various pathological factors. Despite non-significant differences observed in the context of ER status, PR status, breast cancer grade, Her2 status, and the distinction between DCIS and invasive cancer, our investigation showed an association between tumour sizes and AI’s output. The exploration of tumour size categories revealed a highly significant variance in malignancy probability, with the most notable contrast emerging between the initial size category (tumours measuring 10mm or less) and the subsequent size intervals. This finding highlights the AI's potential limitation in confidently annotating malignancy in cases of small tumours and that radiologists should be mindful of the association between lower AI-assigned probability score and smaller tumor sizes. This insight reinforces the need for a nuanced understanding of AI results and their context in clinical practice.
To evaluate the four models, we investigated the performance of the AI models from the point of view of malignancy detection or reporting as a normal case. We did not include any cases with benign lesions in our Australian database, so the results cannot comment on the models’ ability to identify cases with benign features, and this may include cases that are benign but more challenging to AI and human readers. With transfer learning and contrast enhancement application, the AUC of GMIC with CLAHE in the high-concordance subset was 0.912 which is also the best model of four AI models on this study. It is imperative to engage in transfer learning when mammograms are gathered from distinct populations or various vendors as the performance of AI models can be influenced by the specific vendor or population, necessitating adaptation for optimal results.
5. Conclusion
In this paper, we presented the performance of four publicly available AI models for breast cancer detection in different situations such as concordance classification of annotations in the input data, the incorporation of contrast enhancement, and the application of transfer learning. The results showed that when tested on the high-concordance subset, these four AI models outperformed their performance on the entire datasets. Improvements in the performance of AI models were observed through the application of contrast enhancement to mammograms and the utilization of transfer learning. In addition, the AI models' malignancy probability scores were notably influenced by the sizes of tumors visible in the mammograms. Applying concordance classifications, transfer learning and contrast enhancement of mammograms to AI models is likely to provide an effective method for AI assisting decision-making when radiologists interpret mammographic images.
Author Contributions
Conceptualization, Z.J., Z.G., P.T., S.T., and S.L.; methodology, Z.J., Z.G., P.T., S.T., and S.L.; software, Z.J., formal analysis, Z.J., Z.G., P.T., S.T., and S.L.; data curation, Z.J., Z.G. and S.L.; writing—original draft preparation, Z.J.; writing—review and editing, all coauthors; visualization, Z.J.; supervision, S.L.; project administration, M.B., S.L.; funding acquisition, Z.G., P.T., S.T., and S.L.; All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Breast Cancer Foundation (NBCF) Australia.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by Human Ethics Research Committee of the University of Sydney (2019/1017).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data supporting this study’s findings are available on request from the corresponding authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA Cancer J Clin. 2021, 71, 209- 249.
- Paci, E.; Broeders, M.; Hofvind, S.; Puliti, D.; Duffy, S.W. European breast Cancer service screening outcomes: a first balance sheet of the benefits and harms, Cancer Epidemiol. Biomark. Prev. 2014, 23, 1159–1163. [Google Scholar] [CrossRef] [PubMed]
- Kopans, D.B. An open letter to panels that are deciding guidelines for breast cancer screening. Breast Cancer Research and Treatment 2015, 151, 19–25. [Google Scholar] [CrossRef] [PubMed]
- P.A. Carney, D.L. P.A. Carney, D.L. Miglioretti, B.C. Yankaskas, et al. Individual and combined effects of age, breast density, and hormone replacement therapy use on the accuracy of screening mammography, Ann Intern Med, 138, pp. 168-175, (2003).
- Al Mousa, D.S.; Brennan, P.C.; Ryan, E.A.; Lee, W.B.; Tan, J.; Mello-Thoms, C. How Mammographic Breast Density Affects Radiologists' Visual Search Patterns. Academic Radiology 2014, 21, 1386–93. [Google Scholar] [CrossRef] [PubMed]
- Chong. A.; Weinstein, S.P.; McDonald, E.S.; Conant, E.F. Digital Breast Tomosynthesis: Concepts and Clinical Practice. Radiology 2019, 292, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.Y.; Chao, H.S.; Chen, Y.M. Application of Artificial Intelligence in Lung Cancer. Cancers (Basel) 2022, 14(6), 1370. [Google Scholar] [CrossRef] [PubMed]
- Othman, E.; Mahmoud, M.; Dhahri, H.; Abdulkader, H.; Mahmood, A.; Ibrahim, M. Automatic Detection of Liver Cancer Using Hybrid Pre-Trained Models. Sensors 2022, 22(14), 5429. [Google Scholar] [CrossRef] [PubMed]
- Akinyelu, A. A.; Zaccagna, F.; Grist, J. T.; Castelli, M.; Rundo, L. Brain Tumor Diagnosis Using Machine Learning, Convolutional Neural Networks, Capsule Neural Networks and Vision Transformers, Applied to MRI: A Survey. Journal of Imaging 2022, 8(8), 205. [Google Scholar] [CrossRef] [PubMed]
- 10. Wu, N; Phang, J.; Park, J.; Shen, Y.; Huang, Z.; Zorin, M.; Jastrzebski, S.; Fevry T, T. et al., Deep Neural Networks Improve Radiologists' Performance in Breast Cancer Screening. IEEE Transactions on Medical Imaging 2020, 39, 1184-1194.
- McKinney, S. M.; Sieniek, V. M. Godbole et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Margolies, L.R.; Rothstein, J.H. et al. Deep Learning to Improve Breast Cancer Detection on Screening Mammography. Scientific Reports 2019, 9, 12495. [Google Scholar] [CrossRef]
- Al-Masni, M.; Al-Antari, M.; Park, J.M.; Gi, G.; Kim, T.Y.; Rivera, P.; Valarezo, E.; Choi, M.T.; Han, S.M.; Kim, T.S. Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Computer Methods and Programs in Biomedicine 2018, 157, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Dhungel, N.; Carneiro, G.; Bradley, A.P. A deep learning approach for the analysis of masses in mammograms with minimal user intervention, Med. Image Anal. 2017, 37, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Cao, Z.; Zhang, Y.; Tang, Y.; Lin, X.; Ouyang, R.; Wu, M.; Han, M.; Xiao, J.; Huang, L.; Wu, S.; Chang, P.; Ma, J. MommiNet-v2: Mammographic multi-view mass identification networks. Med. Image Anal. 2021, 73, 102204. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wu, N.; Phang, J.; Park, J.; Liu, K.; Tyagi, S.; Heacock, L.; Kim, S.G.; Moy, L.; Cho, K.; Geras, K.J. An interpretable classifier for high-resolution breast cancer screening images utilizing weakly supervised localization, Med. Image Anal. 2021, 68, 101908. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Shen, Y.; Wu, N.; Chledowski, J.; Fernandez-Granda, C.; Geras, K. Weakly-supervised High-resolution Segmentation of Mammography Images for Breast Cancer Diagnosis, Proceedings of Machine Learning Research 2021, 143, 268-285.
- Ueda, D. 2022, 17, e0265751.
- Yap, M.H.; Pons, G.; Marti, J.; Ganau, S.; Sentis, M.; Zwiggelaar, R.; Davison, A.K.; Marti, R.; Moi Hoon, Y.; Pons, G.; et al. Automated Breast Ultrasound Lesions Detection Using Convolutional Neural Networks. IEEE Journal of Biomedical and Health Informatics, 2018, 22, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A Comprehensive Survey on Transfer Learning, arXiv, 2019.
- Mina, L.M.; Mat Isa, N.A. Breast abnormality detection in mammograms using Artificial Neural Network, 2015 International Conference on Computer, Communications, and Control Technology (I4CT), Kuching, Malaysia, 2015.
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas, USA; 2016. [Google Scholar]
- Karel, Z. Contrast limited adaptive histogram equalization, Graphics Gems IV. Academic Press Professional, Inc.; USA, 1994. p. 474–85.
- Lin, L.I. K A concordance correlation coefficient to evaluate reproducibility. Biometrics, 1989, 45, 255–268. [Google Scholar] [CrossRef] [PubMed]
- GB, M. A proposal for strength of agreement criteria for lin’s concordance correlation coefficient. NIWA Client Report HAM2005-062, Hamilton, New Zealand: National Institute of Water & Atmospheric Research Ltd, 2005.
- Jaccard Index. Available online: https://en.wikipedia.org/wiki/Jaccard_index (accessed on 6 December 2023).
- Elbatel, M. Mammograms Classification: A Review, arXiv 2022, 2203, 1-6.
- OpenCV. Available online: https://docs.opencv.org (accessed on 6 December 2023).
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition, arXiv 2015, 1409, 1-14.
- Min, H.; Wilson, D.; Huang, Y.; Liu, S.; Crozier, S.; Bradley, A. ; Chandra., S. Fully Automatic Computer-aided Mass Detection and Segmentation via Pseudo-color Mammograms and Mask R-CNN, 17th International Symposium on Biomedical Imaging (ISBI), pp. 1111-1115, Iowa City, USA, 2020.
- Kingma. D.P.; Ba, J. Adam: A method for stochastic optimization, in International Conference on Learning Representations, pp. 1–15, San Diego, USA, 2015.
- Wu, N.; Phang, J.; Park, J.; Shen, Y.; Kim, S.G.; Heacock, L.; Moy, L.; Cho, K.; Geras, K.J. The NYU Breast Cancer Screening Dataset v1.0. Technical Report, 2019.
- Lee, R.S.; Gimenez, F.; Hoogi, A.; Rubin, D. Curated Breast Imaging Subset of DDSM, The Cancer Imaging Archive, 2016.
- Wang, Y.; Chen, Q.; Zhang, B. Image enhancement based on equal area dualistic sub-image histogram equalization method. IEEE Transactions Consumer Electronics 1999, 45, 68–75. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).