1. Introduction
Alzheimer's pathology is a neurological illness that worsens over time and mostly affects behavior, memory, and cognitive abilities. It is responsible for the development of dementia, a condition typified by a reduction in cognitive ability that impairs functioning and everyday living. Damaged brain cells and the loss of neural connections are signs of neurodegenerative brain illnesses, which severely limit a person's ability to carry out daily duties[
1]. Dementia is the sixth most common cause of mortality worldwide, with a new case reported every three seconds. About 23 million people are living with dementia in the Asia-Pacific area, 8.8 million in Europe, 5.8 million in the USA, and 5.3 million in India. The global figure is as high as 50 million. By 2050, this figure is expected to increase to 152 million[
2]. A global assessment of Alzheimer's reports from 2020 estimates that the overall projected cost of dementia worldwide is one trillion US dollars, and by 2030, that sum is expected to rise to two trillion US dollars [
3]. One of the main characteristics of Alzheimer's disease is the culmination of abnormal protein deposits within the brain. The illness is linked to two main types of these aberrant protein accumulations: tau tangles and beta-amyloid plaques. Clusters of beta-amyloid protein fragments that accumulate between nerve cells and impair cellular function are known as beta-amyloid plaques. On the other hand, tau tangles are made of twisted protein fibers that accumulate inside nerve cells, causing them to malfunction and ultimately die [
4]. Although the precise cause of Alzheimer's disease remains incompletely elucidated, it is believed to arise from genetic, environmental, and lifestyle factors contributing to its development[
5,
6].
A variety of symptoms are brought on by the advancement of Alzheimer's pathology and worsen with time. Memory disorders, disorientation, and trouble finishing familiar activities are common early signs. As the illness worsens, people may have altered personalities, trouble speaking, and poor judgment[
7,
8]. The physical alterations in the brain and the consequent reduction in cognitive function have a significant effect on the person's overall well-being. Although there is no known cure for Alzheimer's pathology, there are some interventions and therapies that can help control the symptoms and delay the illness's progression. These might include behavioral therapy and cognitive training as well as temporary pharmacological relief for cognitive symptoms and techniques to maintain memory and cognitive function. It has also been demonstrated that lifestyle changes including consistent physical activity, a balanced diet, social interaction, and mental stimulation have a good effect on general brain health [
9]. Alzheimer's patients and their families, carers, and society at large face several obstacles as a result of the condition. Certain determinants are similar to both conditions, however, the likelihood of developing Alzheimer's pathology and cancer is inversely correlated. Compared to control people, Alzheimer's patients had a 61 percent decreased risk of acquiring cancer[
10]
The inverse interaction between cancer and Alzheimer's is a perplexing phenomenon that has garnered substantial attention within medical research[
11,
12]. This intriguing observation suggests that individuals who develop Alzheimer's disease seem less likely to develop cancer. While the exact mechanisms behind this association remain unclear, several theories have been proposed to explain this phenomenon. One possible explanation is related to the role of the immune system. Cancer involves uncontrolled cell growth and division, which the immune system often recognizes as abnormal and attempts to suppress. In contrast, Alzheimer's pathology is typified by the accumulation of misfolded proteins and chronic inflammation. It is speculated that the immune response directed toward cancer cells might inadvertently help protect against Alzheimer's - related protein accumulation by stimulating immune mechanisms that target abnormal protein deposits[
13]. Another theory involves the influence of genetics and specific molecular pathways. Research has identified certain genetic factors associated with Cancer and Alzheimer's disease. Variations in these genes might affect an individual's susceptibility to either condition. Additionally, cellular growth, metabolism, and energy utilization pathways could shape the interaction between Alzheimer's and cancer [
13,
14]. While individual studies have reported on the inverse relationship involving Alzheimer's pathology and cancer, there is a critical gap in our comprehension of the specific molecular mechanisms that drive this association[
12]. The interaction linking Alzheimer's disease and cancer is not straightforward and can vary depending on the specific types of cancer and the stage of each disease. The exact nature of this association is still an active area of research, and further studies are required to fully comprehend the underlying mechanisms. Understanding why certain individuals are seemingly protected from one condition while experiencing an increased risk of the other holds immense potential for shedding light on shared pathways, regulatory networks, and cellular responses that underlie both Alzheimer's disease and cancer. To address this knowledge gap, comprehensive research is needed to unravel the intricate molecular interactions contributing to this inverse relationship. There has been few research conducted on the inverse interaction betwixt Alzheimer's disease and cancer, even though it offers an intriguing scientific challenge.
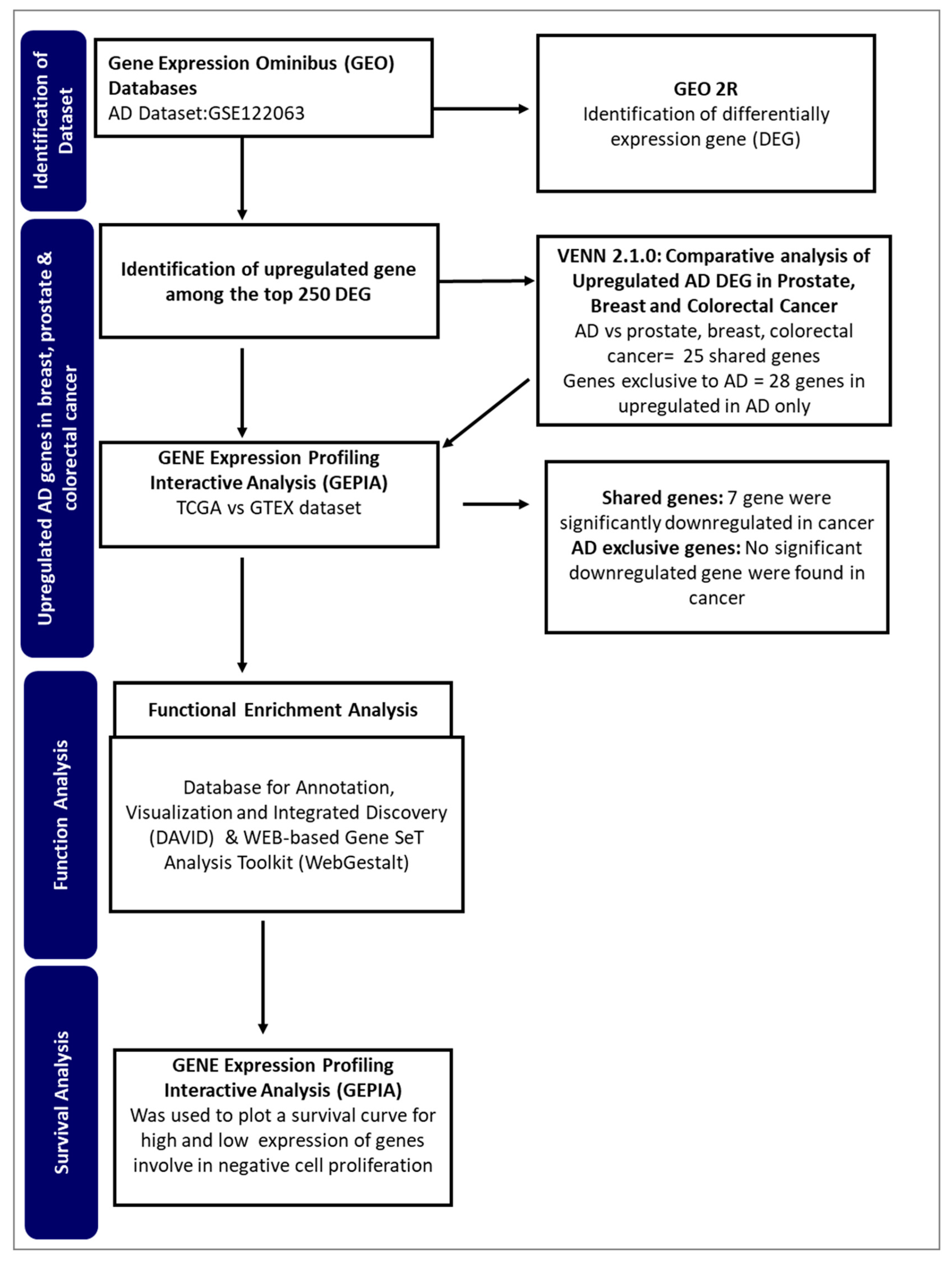
This study aims to identify potential genes from Alzheimer's disease that may be involved in the inverse interaction betwixt Alzheimer's pathology and cancer. By employing an integrated bioinformatics approach (
Figure 1), the study seeks to identify genes and regulatory networks that might provide insight into the observed phenomenon.
3. Discussion
The inverse interaction between Alzheimer's disease and cancer continues to be a complex and intriguing topic within the medical community. Cancer-Alzheimer's disease paradox suggests that individuals with a previous history of cancer may have a reduced risk of progressing to Alzheimer's pathology[
15]. Research into the mechanisms driving this relationship could provide valuable insights into the fundamental processes governing these two major diseases, potentially paving the way for innovative therapeutic strategies and a deeper understanding of human health and aging. In this study, we used an integrated computational approach to identify upregulated genes in Alzheimer's pathology that may also influence cancer suppression and progression. We hypothesized that these genes may potentially be associated with the inverse comorbidity betwixt Alzheimer's pathology and cancer.
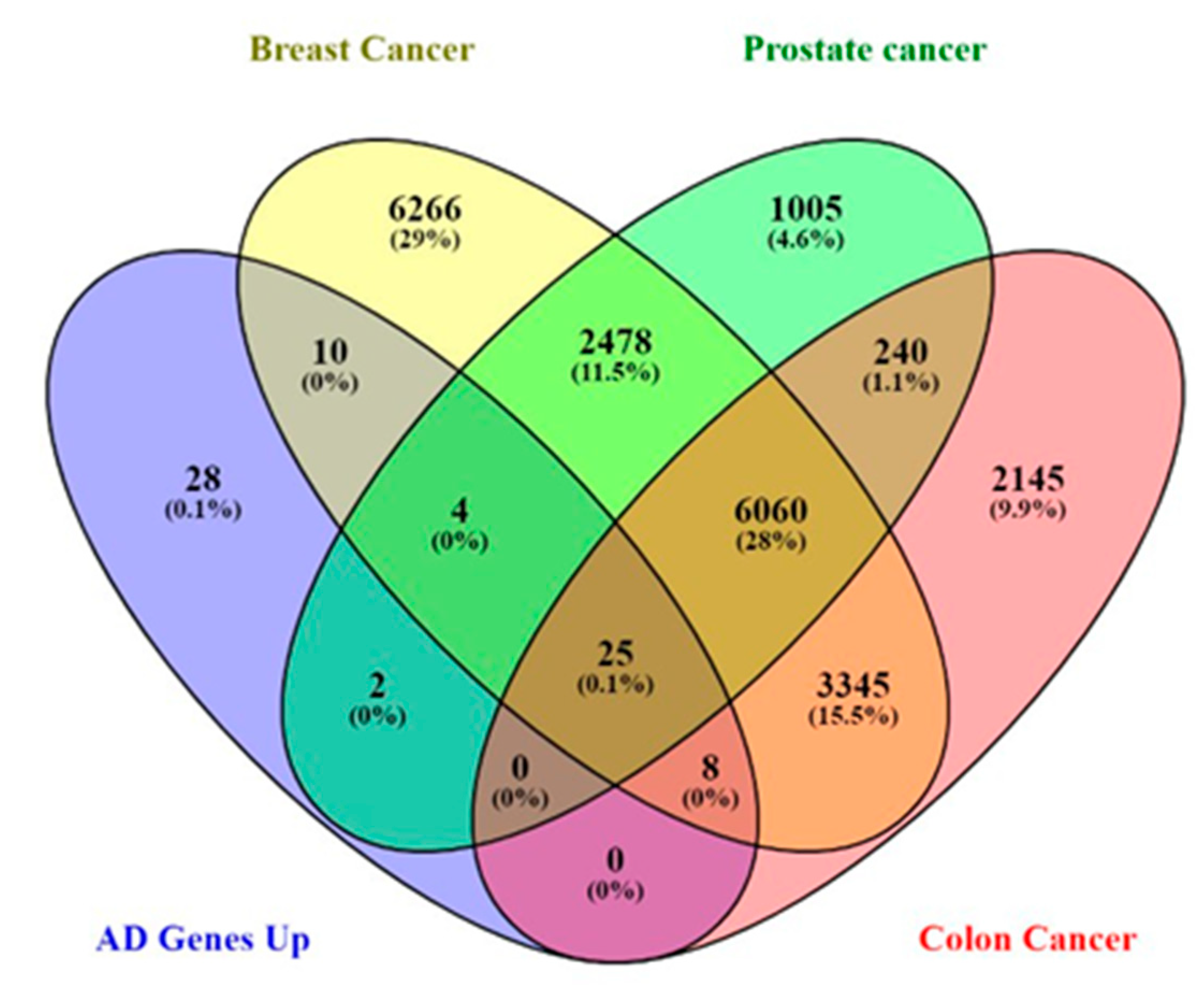
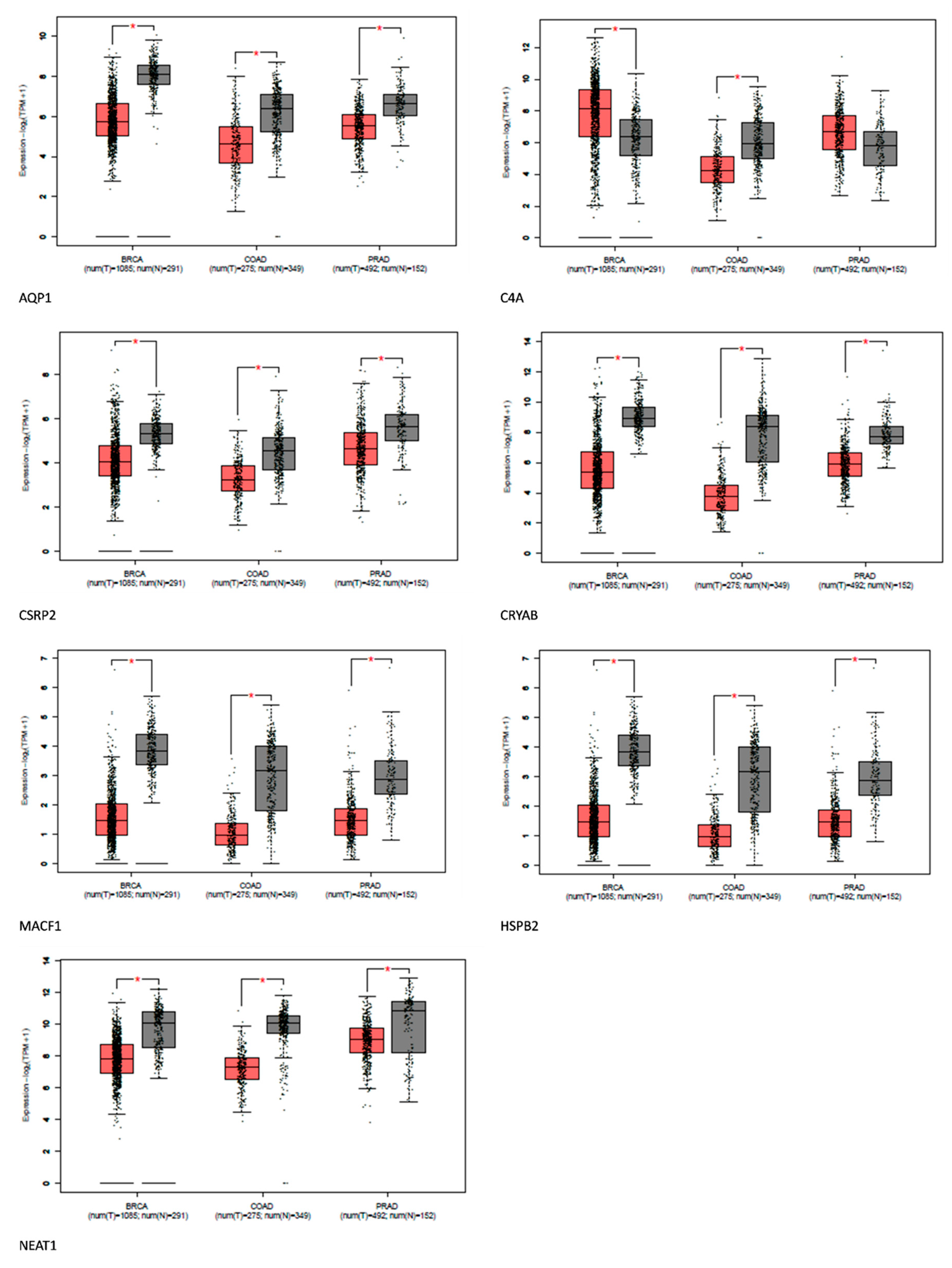
Comparative gene analysis reveals a similarity of 0.1% of the upregulated genes found in Alzheimer's disease-related genes and prostate, breast, and colorectal cancer. The gene expression analysis affirms that most of these genes were distinctively downregulated in the cancers. AQP1, C4A, CSRP2, CRYAB, MACF1, HSPB2, and NEAT1 were downregulated in some cancers. However, the role of these genes in tumor suppression and promotion is context-dependent.
HspB2 is a unique member of the small HSP family predominantly expressed in skeletal and heart muscles. Traditionally,
HSPS has been linked to cancer development, but
HspB2 contradicts this by acting as a tumor suppressor. This protein plays an anti-cancer role by partially restoring mutant p53 activity and promoting the expression of tumor-suppressor genes
RPRM,
BAI-1, and
TSAP6[
16]. In certain tumors, like pancreatic and esophageal cancers,
HSPB2 levels are reduced or absent.
HSPB2 also repairs p53 transcriptional activity, leading to the inhibition of pancreatic cancer cell progression. However, its role in breast cancer is debated, with some studies showing suppression [
17], while others indicating progression[
18]. Given these findings, further investigations into
HspB2 and its variants, especially in breast cancer, are vital to explain the role of
HspB2 in cancer and provide insight into its potential as a possible tumor suppressor in Alzheimer's disease.
CSRP2 is a multifunctional protein involved in cell proliferation, differentiation, and adhesion.
CSRP2 also suppresses colorectal cancer progression via the P130Cas/Rac1 signaling pathway[
19]. Again,
CSRP2 is thought to hinder the spread and metastasis of cancer cells in gastric cancer, whereas it plays a contrary function in breast cancer[
19,
20]. Therefore, the underlying mechanisms of
CSRP2 function in various cancers need to be carefully examined. The protein's function in cancer is complex, and understanding its intricacies could highlight its role in Alzheimer's disease.
CRYAB suppresses NPC tumor formation in mice and affects NPC progression-associated phenotypes[
21].
CRYAB inhibits migration and invasion through various mechanisms, such as inhibiting RAS activation and the Raf/MEK/ERK signaling pathway[
22]. While CRYAB has tumor-suppressive properties in the nasopharyngeal, and bladder, and it also promotes angiogenesis in some cancers. However, to elucidate the role of CRYAB in cancer and Alzheimer's pathology, there is a need to investigate various forms
of CRYAB present in Alzheimer's disease[
22,
23].
MACF1 is a multidomain cytoskeletal linker protein essential for cell migration, cytoskeletal organization, and intracellular transport. In cancer, it can both promote and inhibit progression[
24,
25]
MACF1 can suppress cancer cell motility by maintaining cytoskeletal integrity and may interact with tumor suppressor proteins [
25]. Though
MACF1 has a multifaceted role in cancer, it could be responsible for the inverse association between Alzheimer's disease and cancer since it can interact with tumor suppressor proteins.
Neat1 plays an important role in the growth and specialization of the mammary gland and corpus luteum. However, its impact on cancer has resulted in contradictory findings. While some research links
Neat1 to the advancement of tumors, recent studies have demonstrated its potential as a suppressor of tumor growth [
26]. The tumor-suppressive actions of NEAT1 have been attributed to several molecular pathways. These include preserving pre-messenger RNAs in the cell nucleus and promoting the development of pre-mRNAs and pre-microRNAs[
27,
28], protein sequestration into paraspeckles and removal from chromatin[
29], and controlling the expression of genes by making direct contacts with chromatin. It is s possible that NEAT1 suppresses tumors by influencing gene expression via a variety of methods simultaneously, or that its ability to operate as a tumor suppressor depends only on a subset of these processes [
27,
28,
30]. When exposed to nutlin or Adriamycin, increased levels of NEAT1 have been shown to inhibit the development of cells in lung cancer and osteosarcoma cell lines [
31]. Again, increased levels of
NEAT1 expression were linked to improved survival rates in cases of nasopharyngeal cancer. Additionally, elevated
NEAT1 levels were found to significantly suppress the growth of nasopharyngeal cancer cells in subcutaneous tumor experiments, functioning through a process that inhibits miR-101-3p[
32]. Research indicates that point mutations and deletions in the NEAT1 promoter are rather common in cases of liver and breast cancer. Moreover, mutations found in the NEAT1 promoter in breast cancer frequently result in decreased NEAT1 production. Although it is not shown that the particular mutations seen in breast cancer patients influence the p53 binding site, the deletions often damage the p53 response element[
33,
34,
35]. Furthermore, it was discovered that decreasing NEAT1 1 in colorectal cancer cell lines decreased invasiveness and cell proliferation[
36]. While the precise process of tumor inhibition remains undefined,
NEAT1 shows promise as a tumor suppressor in individuals with Alzheimer's disease, potentially playing a role in the reduced susceptibility of Alzheimer's patients to cancer.
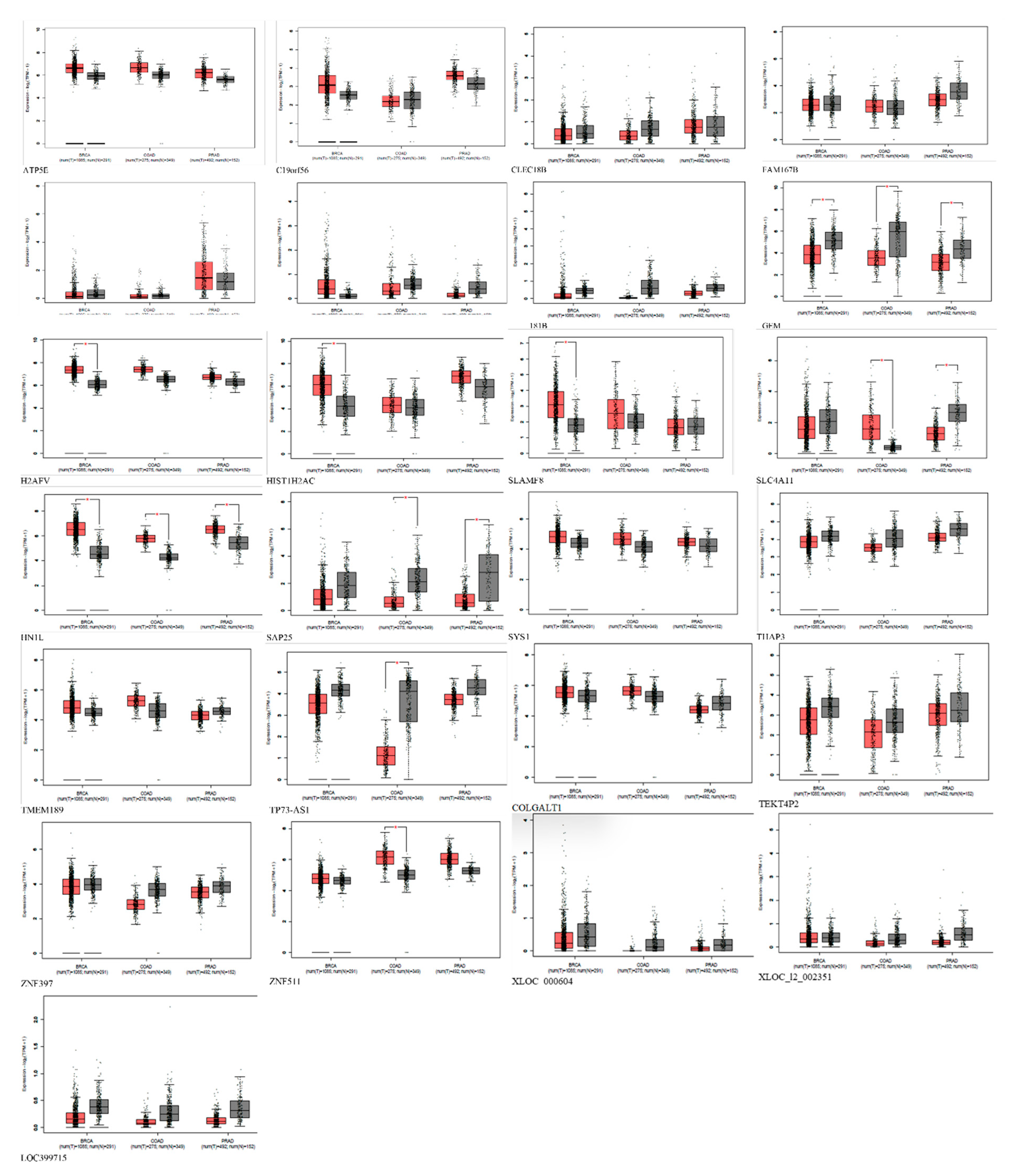
Furthermore, the comparative analysis also showed that 28 genes are exclusively upregulated in Alzheimer's disease and cannot be found in prostate, breast, and colorectal cancer. Therefore, we hypothesized that these genes could be responsible for the inverse comorbidity observed between cancers and Alzheimer's disease. The upregulation of genes exclusively related to Alzheimer's disease could be involved in inverse comorbidity. The fact that these 28 genes are upregulated in Alzheimer's pathology but not in the mentioned cancers suggests that Alzheimer's pathology and these cancers may involve distinct molecular and cellular pathways. The processes driving Alzheimer's pathology progression might differ from those driving these cancers' progression. If these genes influence the pathology of Alzheimer's, their absence or repression in certain cancers could contribute to the observed inverse association. The molecular processes they trigger may be involved in the protective effects against the onset or progression of these cancers. To draw more concrete conclusions, it is crucial to understand the biological functions of these 28 genes, particularly their involvement in cell growth, apoptosis, DNA repair, immune response, or neural functions.
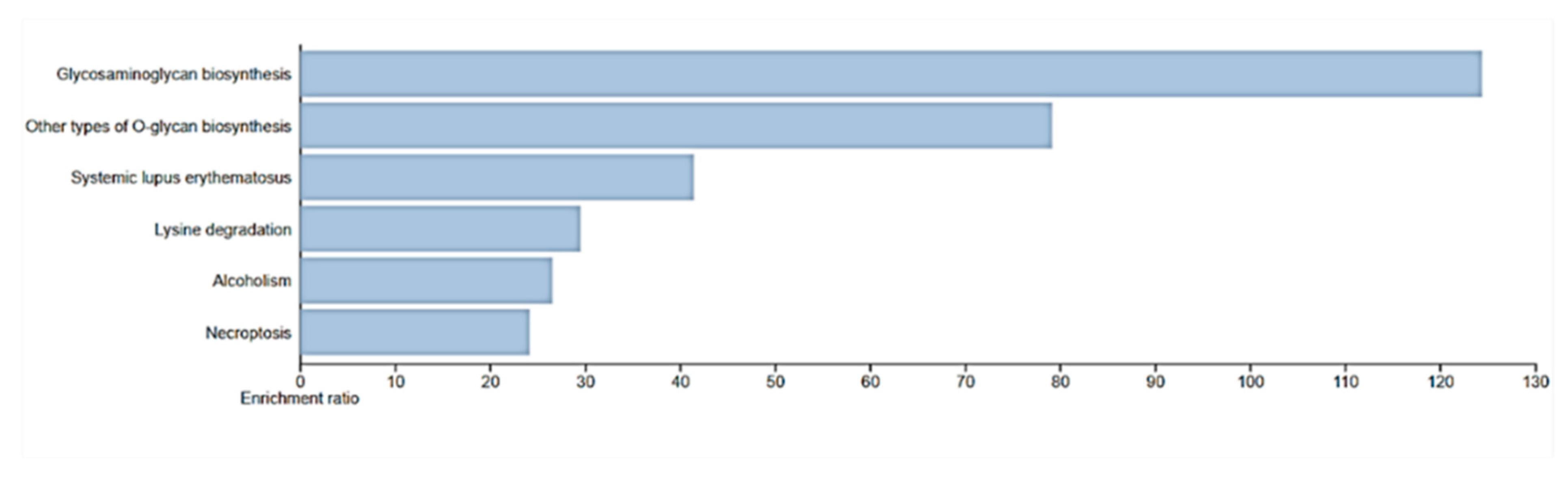
Functional analysis of the biological processes of these genes revealed that
HIST1H2AC is involved in necroptosis, a form of regulated cell death morphologically and mechanistically distinct from apoptosis. Unlike apoptosis, which is a form of programmed cell death generally considered non-inflammatory and involves the activation of caspases, necroptosis resembles necrotic cell death and is often associated with inflammation. As a member of the histone H2A family, the HIST1H2AC gene is essential to the structural arrangement of chromatin, which can affect a number of DNA-related functions, including transcription, replication, and repair. The expression levels of histone modifications have a significant impact on how genes and cellular pathways are regulated [
37]. In the context of uterine cervical preneoplastic lesions, studies have shown a trend in the expression of the
HIST1H2AC gene. Specifically, there has been a progressive downregulation of
HIST1H2AC in keratinocytes that are positive for Human papillomavirus (HPV). HPV is a well-established causative agent for cervical cancer, with certain high-risk strains being more associated with malignancy than others. Though the expression of
HIST1H2AC has not been investigated in various types of cancers, this observation suggests a potential correlation between decreased
HIST1H2AC expression and advancing cellular abnormalities. Therefore, we postulate that the upregulation of HIST1H2AC may be a factor for the inverse correlation between cancer and Alzheimer's pathology.
HIST1H2AC upregulation could influence chromatin structure, gene accessibility, and, consequently, cellular growth and differentiation in Alzheimer's pathologic patients. However, further studies on its exact role, mechanism of downregulation, and broader implications of
HIST1H2AC could provide valuable insights into cancer biology and potential therapeutic strategies.
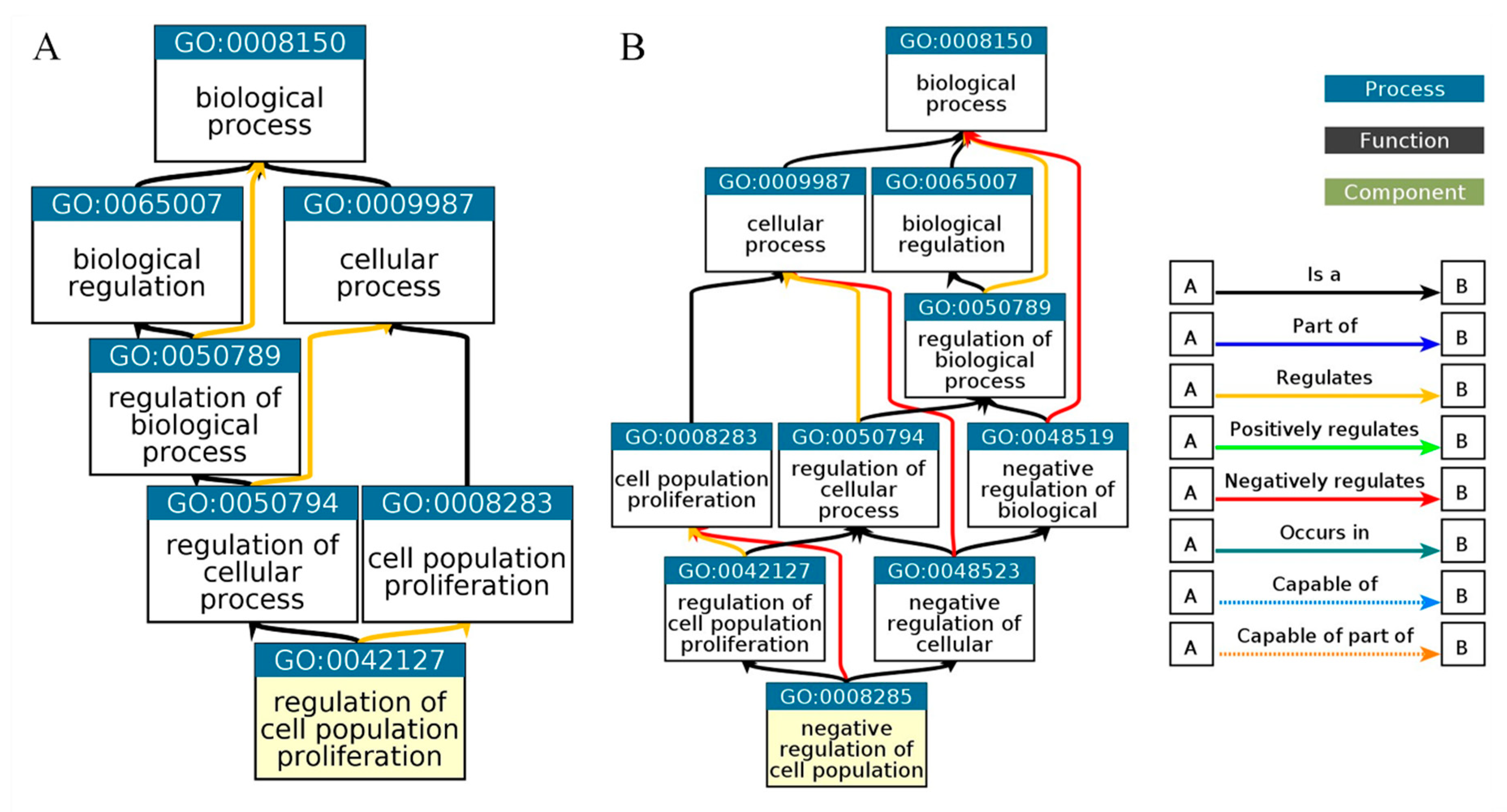
Biological process analysis reveals that
HIST1H2AC is involved in negative cell proliferation and chromatin assembling, this further confirms the involvement of
HIST1H2AC in necroptosis. Given the role of
HIST1H2AC regulation of cell proliferation and its upregulation in Alzheimer's disease, there is a need to further investigate
HIST1H2AC as a potential tumor suppressor responsible for Alzheimer's disease and cancer inverse association. Furthermore, the biological process also revealed that C1orf56 is involved in the regulation of cell proliferation. C1orf56 was involved in the control of cellular proliferation and served as an oncogenic modifier contributing to the tumor suppressor function of DNMT3B. Dnmt3b affects tumor progression [
38]. This shows that genes involved in Alzheimer's disease complexities continue to be explored. Characterizing these genes may also provide insight into their role in the inverse association between cancer and Alzheimer's disease.
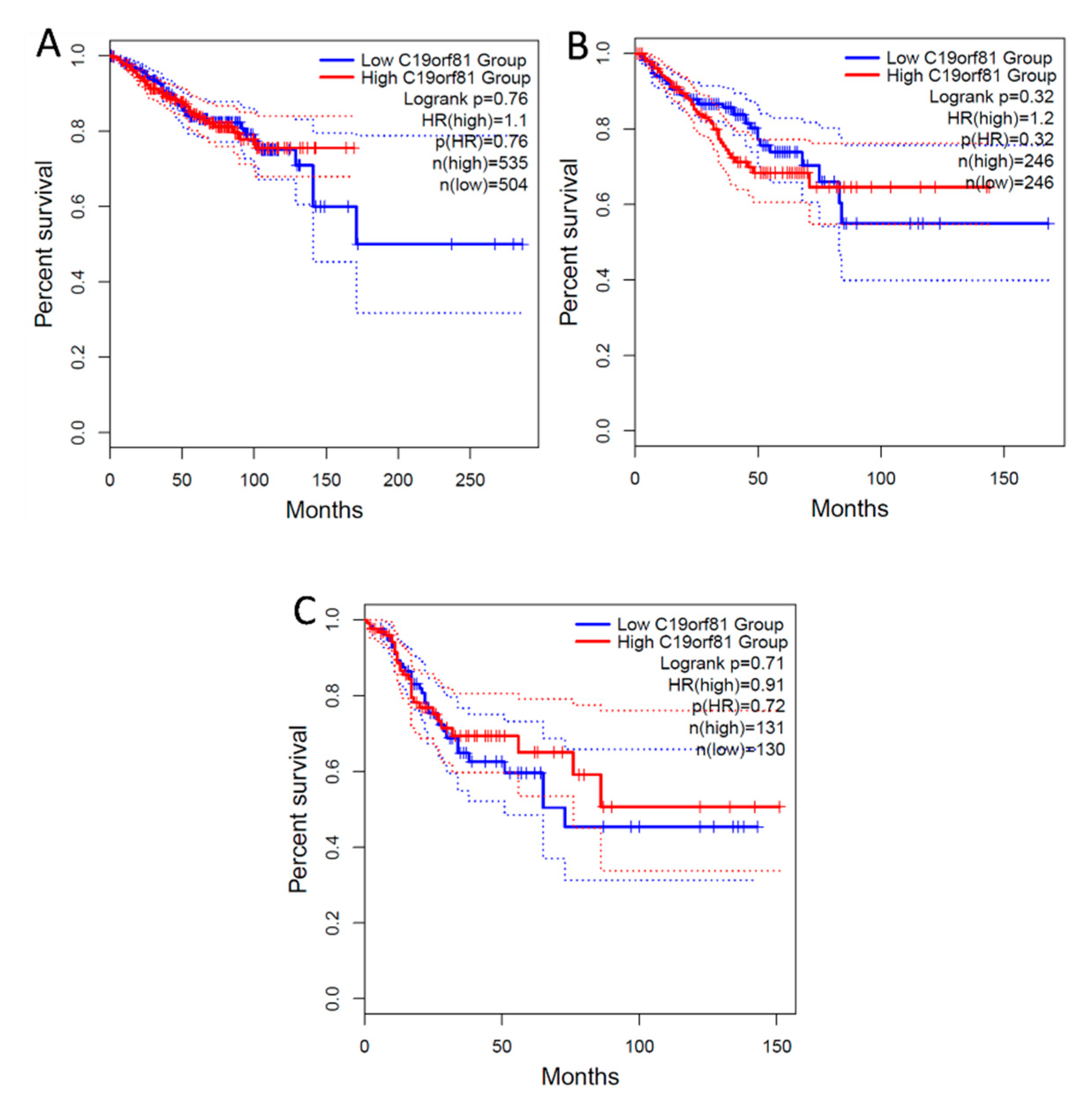
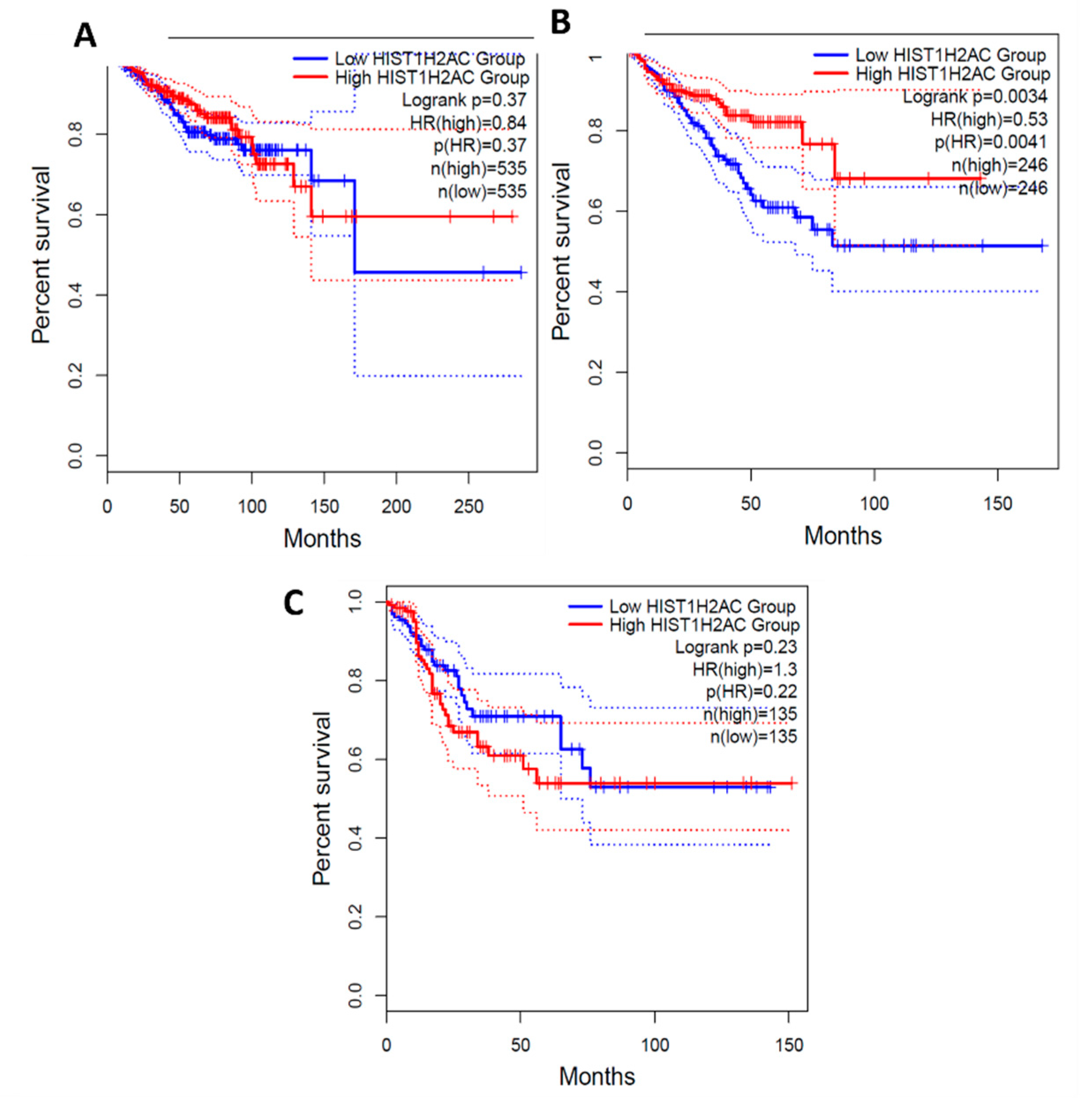
From the survival analysis, high expression of C1orf56 in the breast and prostate did show a significant improvement in survival with a hazard ratio of 1.1 and 1.2 however there was an observable difference in colorectal cancer patient survival with high expression of C1orf56. This indicates that C1orf56 could potentially suppress colorectal cancer progression and improve survival. Though the survival analysis did not give a consistent pattern in all cells, there is an indication that the role of these genes needs to be further elucidated. For HIST1H2AC survival analysis, high expression of HIST1H2AC was associated with improvement in breast and prostate cancer, however contrary to C1orf56, HIST1H2AC did not have an observable effect on improving patient survival. This indicates that different factors may work together to reduce the risk of cancer development in AD patients.
In conclusion, there is a need to further investigate HIST1H2AC and C1orf56 as potential tumor suppressors responsible for the inverse interaction betwixt Alzheimer's disease and cancer given the role of HIST1H2AC and C1orf56 in negative regulation of cell proliferation and their upregulation in Alzheimer's pathology. The inverse interaction betwixt Alzheimer's disease and cancer may depend on various factors, such as the upregulation of potential tumor suppressors whose roles are not clearly defined and the upregulation of genes involved in cell proliferation. From this study, it is also apparent that there is limited information on the role of most of the upregulated Alzheimer's disease genes in cancer; hence, further research is needed into some of these genes and their interacting pathways.
Figure 1.
A flowchart of the approach used in this study.
Figure 1.
A flowchart of the approach used in this study.
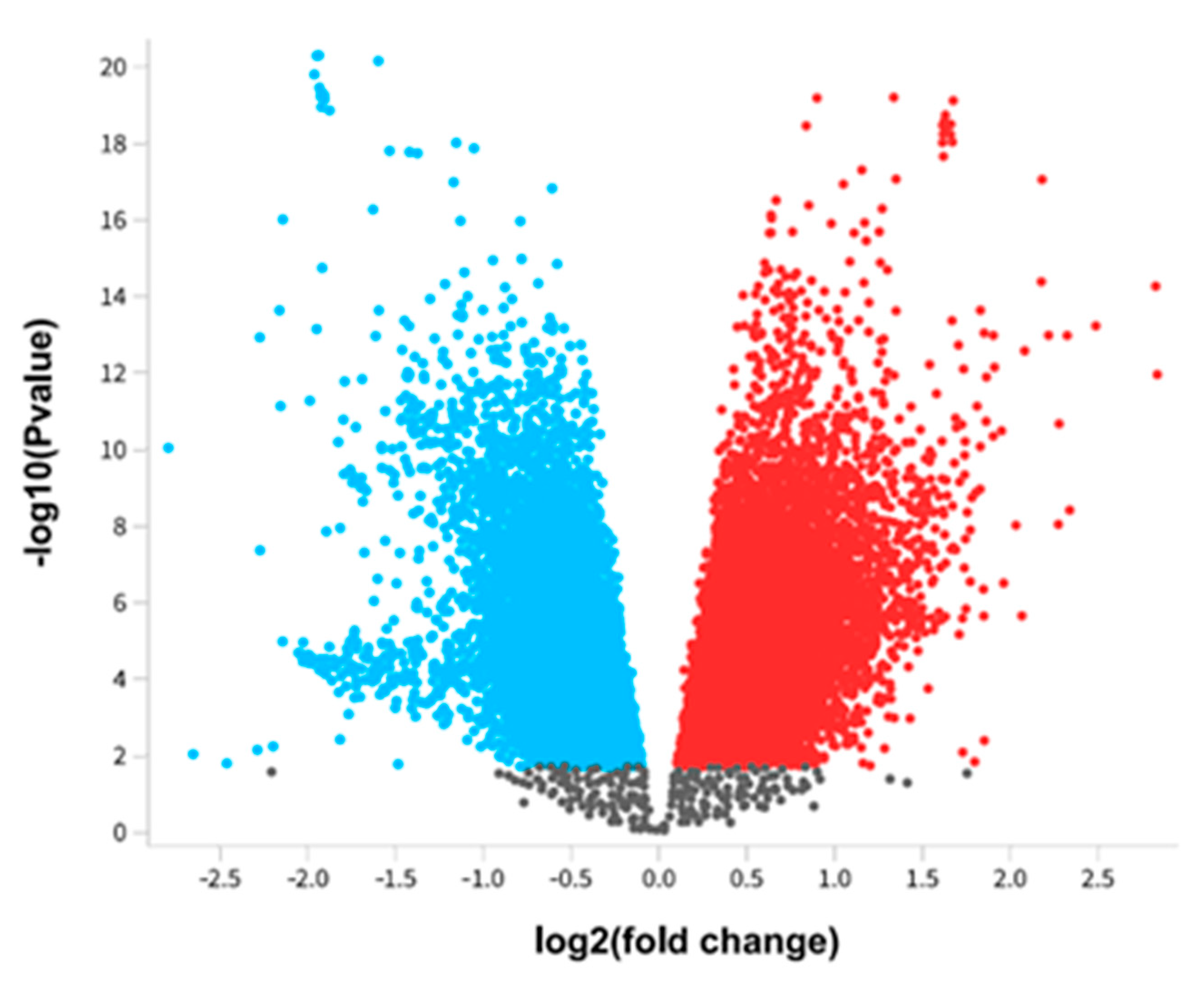
Figure 2.
Differentially expressed genes of Alzheimer's disease. The differential expressed genes of Alzheimer's pathology were presented using a Volcano plot. The red color represents upregulated genes, whereas the blue color represents downregulated genes.
Figure 2.
Differentially expressed genes of Alzheimer's disease. The differential expressed genes of Alzheimer's pathology were presented using a Volcano plot. The red color represents upregulated genes, whereas the blue color represents downregulated genes.
Figure 3.
Upregulated Alzheimer's disease-responsive genes among the top 250 DEG.
Figure 3.
Upregulated Alzheimer's disease-responsive genes among the top 250 DEG.
Figure 4.
Alzheimer's disease gene distribution in prostate, breast, and colon cancer.
Figure 4.
Alzheimer's disease gene distribution in prostate, breast, and colon cancer.
Figure 5.
Expression of Shared Alzheimer's disease upregulated genes in breast, prostate, and colon cancer using GEPIA. Red represents expression in tumor samples; black represents expression in normal samples, BRCA: breast invasive carcinoma, COAD: Colon adenocarcinoma, PRAD: Prostate adenocarcinoma: tumor, N: normal, * p-value < 0.01, |Log2FC| > 1.
Figure 5.
Expression of Shared Alzheimer's disease upregulated genes in breast, prostate, and colon cancer using GEPIA. Red represents expression in tumor samples; black represents expression in normal samples, BRCA: breast invasive carcinoma, COAD: Colon adenocarcinoma, PRAD: Prostate adenocarcinoma: tumor, N: normal, * p-value < 0.01, |Log2FC| > 1.
Figure 6.
Expression of AD-exclusive upregulated genes in breast, prostate, and colon cancer using GEPIA. Red represents expression in tumor samples; black represents expression in normal samples, BRCA: breast invasive carcinoma, COAD: Colon adenocarcinoma, PRAD: Prostate adenocarcinoma: tumor, N: normal, * p-value < 0.01, |Log2FC| > 1.
Figure 6.
Expression of AD-exclusive upregulated genes in breast, prostate, and colon cancer using GEPIA. Red represents expression in tumor samples; black represents expression in normal samples, BRCA: breast invasive carcinoma, COAD: Colon adenocarcinoma, PRAD: Prostate adenocarcinoma: tumor, N: normal, * p-value < 0.01, |Log2FC| > 1.
Figure 7.
Gene Ontology Biological Process. Biological process of C1orf56 (
Figure 4A), biological process of HIST1H2AC (
Figure 4B).
Figure 7.
Gene Ontology Biological Process. Biological process of C1orf56 (
Figure 4A), biological process of HIST1H2AC (
Figure 4B).
Figure 8.
Enrichment pathway for WebGestalt KEGG. Necroptosis was identified as a pathway for HIST1H2AC.
Figure 8.
Enrichment pathway for WebGestalt KEGG. Necroptosis was identified as a pathway for HIST1H2AC.
Figure 9.
Survival Analysis of C1orf56 Expression in Breast, prostate, and Colorectal cancer. 1.1, 1.2 and 0.91 hazard ratio (HR) was determined for breast, prostate, and colorectal cancer respectively. A: BRCA- Breast cancer, B: PRAD- Prostate Cancer, C: COAD- Colorectal Cancer.
Figure 9.
Survival Analysis of C1orf56 Expression in Breast, prostate, and Colorectal cancer. 1.1, 1.2 and 0.91 hazard ratio (HR) was determined for breast, prostate, and colorectal cancer respectively. A: BRCA- Breast cancer, B: PRAD- Prostate Cancer, C: COAD- Colorectal Cancer.
Figure 10.
Survival Analysis of HIST1H2AC Expression in Breast, prostate and Colorectal cancer.0.84, 0.53 and 1.3 Hazard ratio was determined for breast, prostate and colorectal cancer respectively. A: BRCA- Breast cancer, B: PRAD- Prostate Cancer, C: COAD- Colorectal Cancer.
Figure 10.
Survival Analysis of HIST1H2AC Expression in Breast, prostate and Colorectal cancer.0.84, 0.53 and 1.3 Hazard ratio was determined for breast, prostate and colorectal cancer respectively. A: BRCA- Breast cancer, B: PRAD- Prostate Cancer, C: COAD- Colorectal Cancer.