Submitted:
05 December 2023
Posted:
06 December 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and Methods
- -
- Ras-related protein Rab-5A (PDB Code: 1n6k,RAB5A_HUMAN); Grid box Coordinates of binding Center X ( 9,9289), Y( -2,608), Z(42,2021)
- -
- Ras-related C3 botulinum toxin substrate 3(PDB Code: qme AC3_HUMAN); Grid box Coordinates of binding Center X ( -10,6359), Y( 7,5954), Z(23,1647)
- -
- Ras-related protein Rab-28 (PDB Code: 2hxs,RAB28_HUMAN); Grid box Coordinates of binding Center X ( 20,2841), Y( 20,0731), Z(11,1145)
- -
- Ras-related GTP-binding protein C(PDB Code: 3llu,RRAGC_HUMAN); Grid box Coordinates of binding Center X ( 37,8726), Y( 15,9775), Z(25,5165)
- -
- Ras-related protein Rab-9A (PDB Code: 1wmsRAB9A_HUMAN); Grid box Coordinates of binding Center X ( -1,0272), Y( 21,5378), Z(7,3575)
- -
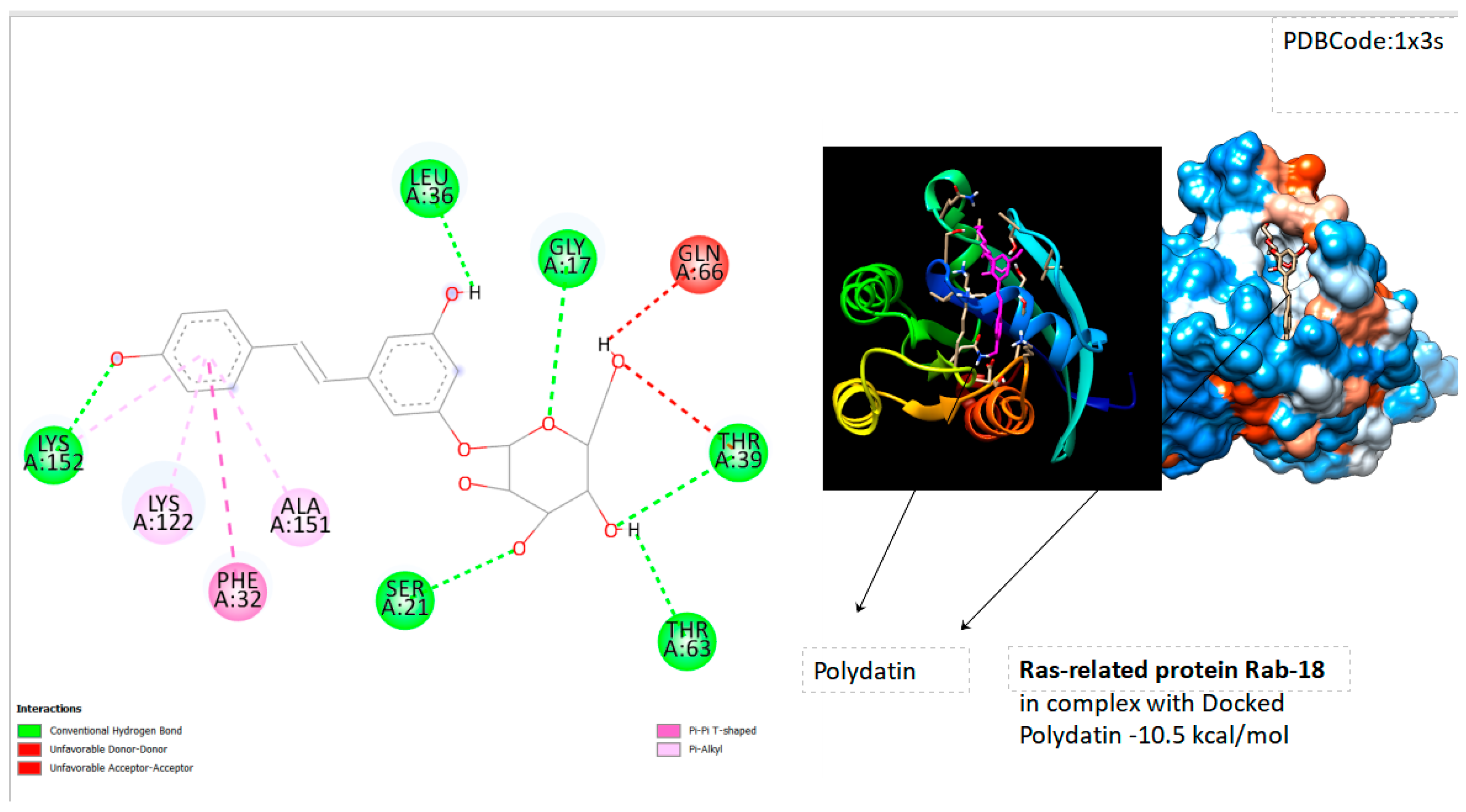
- Ras-related protein Rab-18 (PDB Code: 1x3s RAB18_HUMAN); Grid box Coordinates of binding Center X ( 29,773), Y( 32,4934), Z(17,9592)
- -
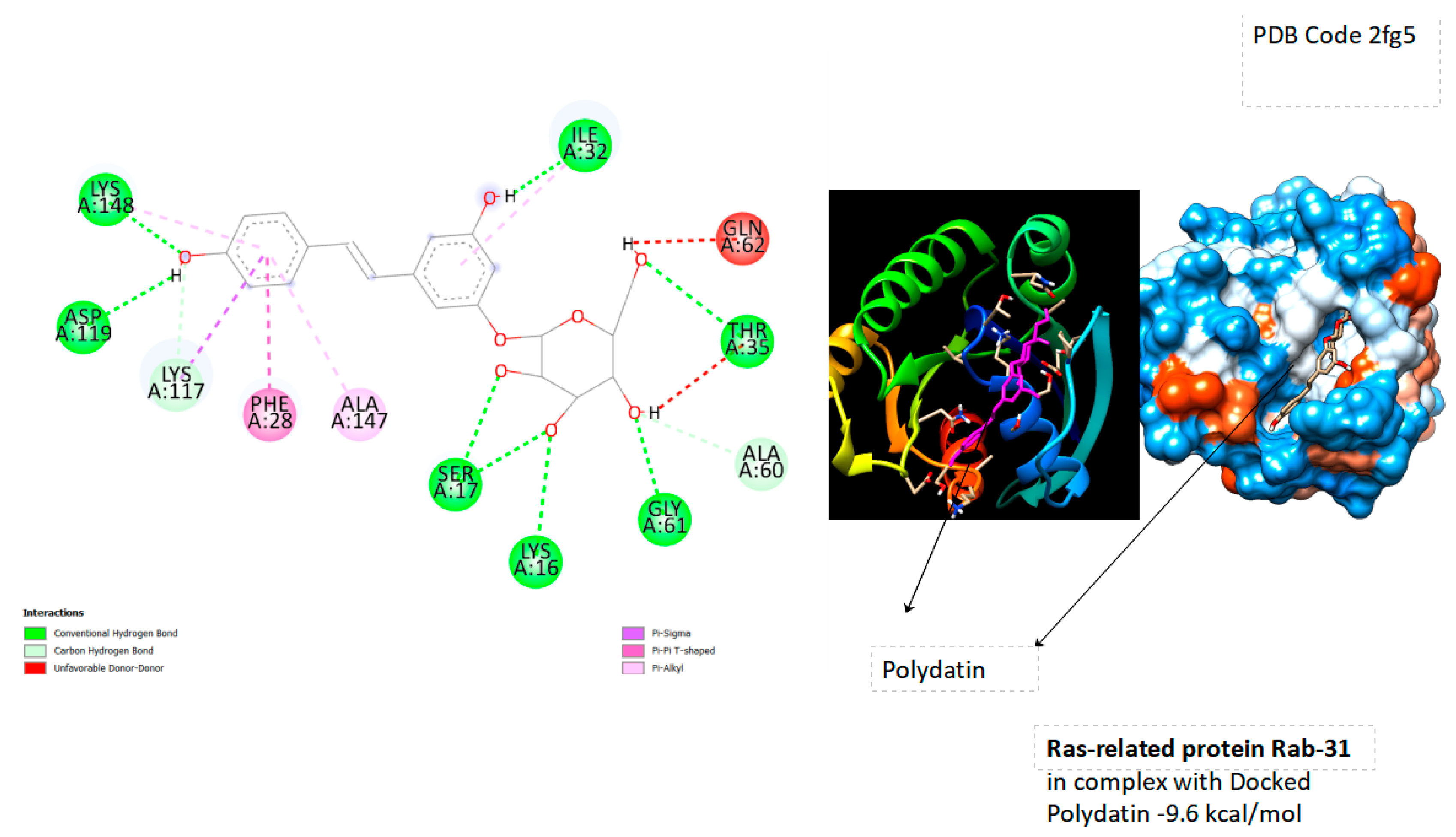
- Ras-related protein Rab-31 (PDB Code: 2fg5 RAB31_HUMAN); Grid box Coordinates of binding Center X ( 19,2312), Y( 6,1773 ), Z((2,0914)
- -
- Ras-related protein Rap-2A(PDB Code: 3rap RAP2A_HUMAN); Grid box Coordinates of binding Center X ( 37,5957), Y( 10,1612), Z(65,4559)
- -
- Ras-related protein Rab-11 B (PDB Code: 2f9l RB11B_HUMAN); Grid box Coordinates of binding Center X ( 5,0846), Y( 2,0916 ), Z(6,1611)
- -
- Ras-related protein Rab-3D(PDB Code: 2gf9 RAB3D_HUMAN); Grid box Coordinates of binding Center X ( 3,0868), Y( 9,6966 ), Z(6,4)
- -
- Ras-related protein Rab-4A(PDB Code: 2bme RAB3D_HUMAN); Grid box Coordinates of binding Center X ( 73,0035), Y( 13,8161), Z(0,889)
- -
- GTPase HRas(PDB Code: 1qra RAB3D_HUMAN); Grid box Coordinates of binding Center X ( 12,0003), Y( 32,9114), Z(19,526)
- -
- Ras-related protein R-Ras(PDB Code: 2fn4 RRAS_HUMAN); Grid box Coordinates of binding Center X ( -6,1087), Y( 25,4637 ), Z(14,5476)
- -
- Ras-related protein Rab-1B(PDB Code: 3nkv RAB1B_HUMAN); Grid box Coordinates of binding Center X (-15,1987), Y( 19,8262), Z(-7,1299)
- -
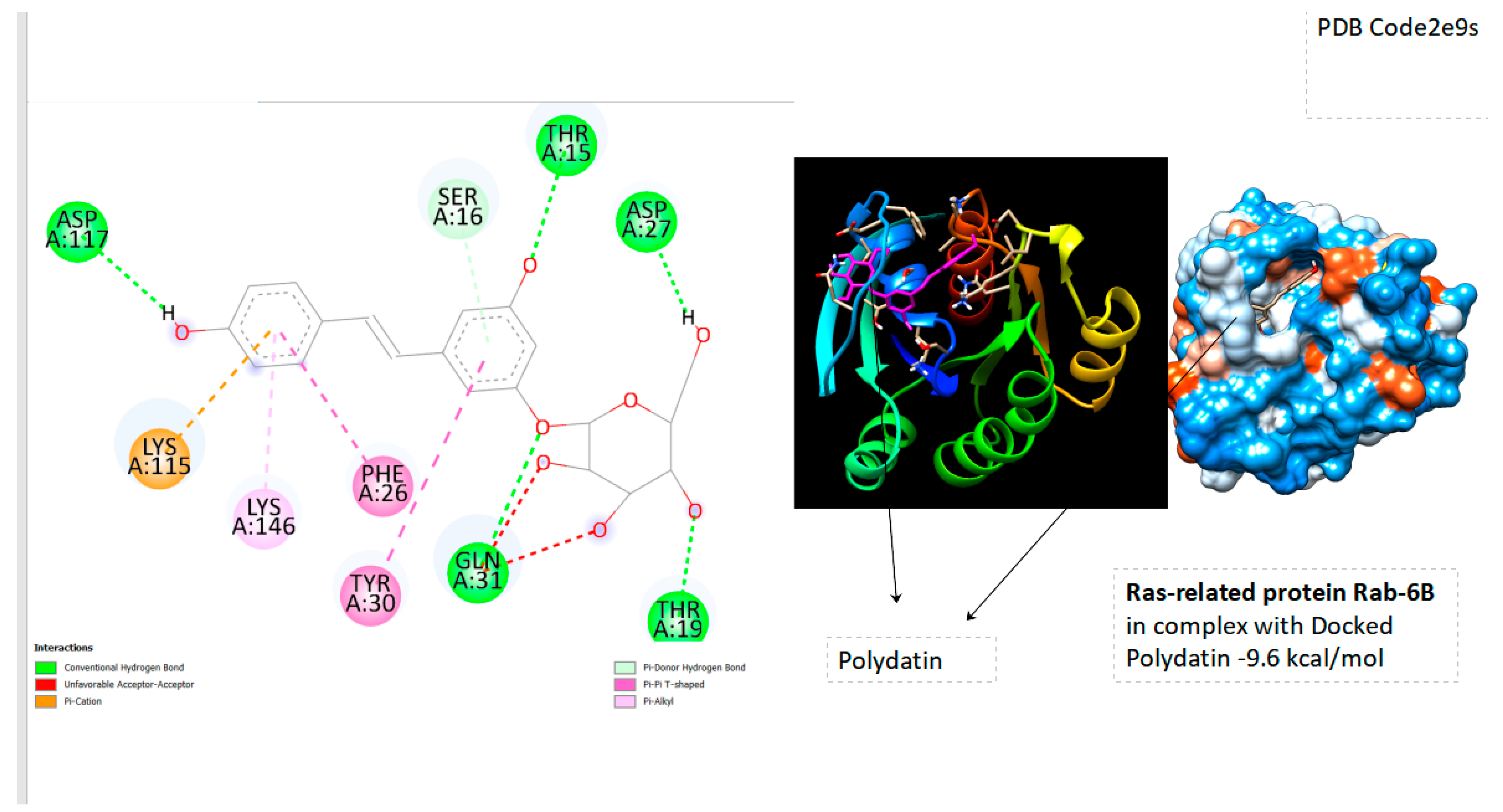
- Ras-related protein Rab-6B(PDB Code: 2e9s RAB6B_HUMAN); Grid box Coordinates of binding Center X (3,6002), Y( 33,0027), Z(3,0894)
- -
- Ras-related protein Rab-6A(PDB Code: 1yzq RAB6A_HUMAN); Grid box Coordinates of binding Center X (25,1083), Y( 17,85 ), Z(13,8221)
3. Results and Discussion
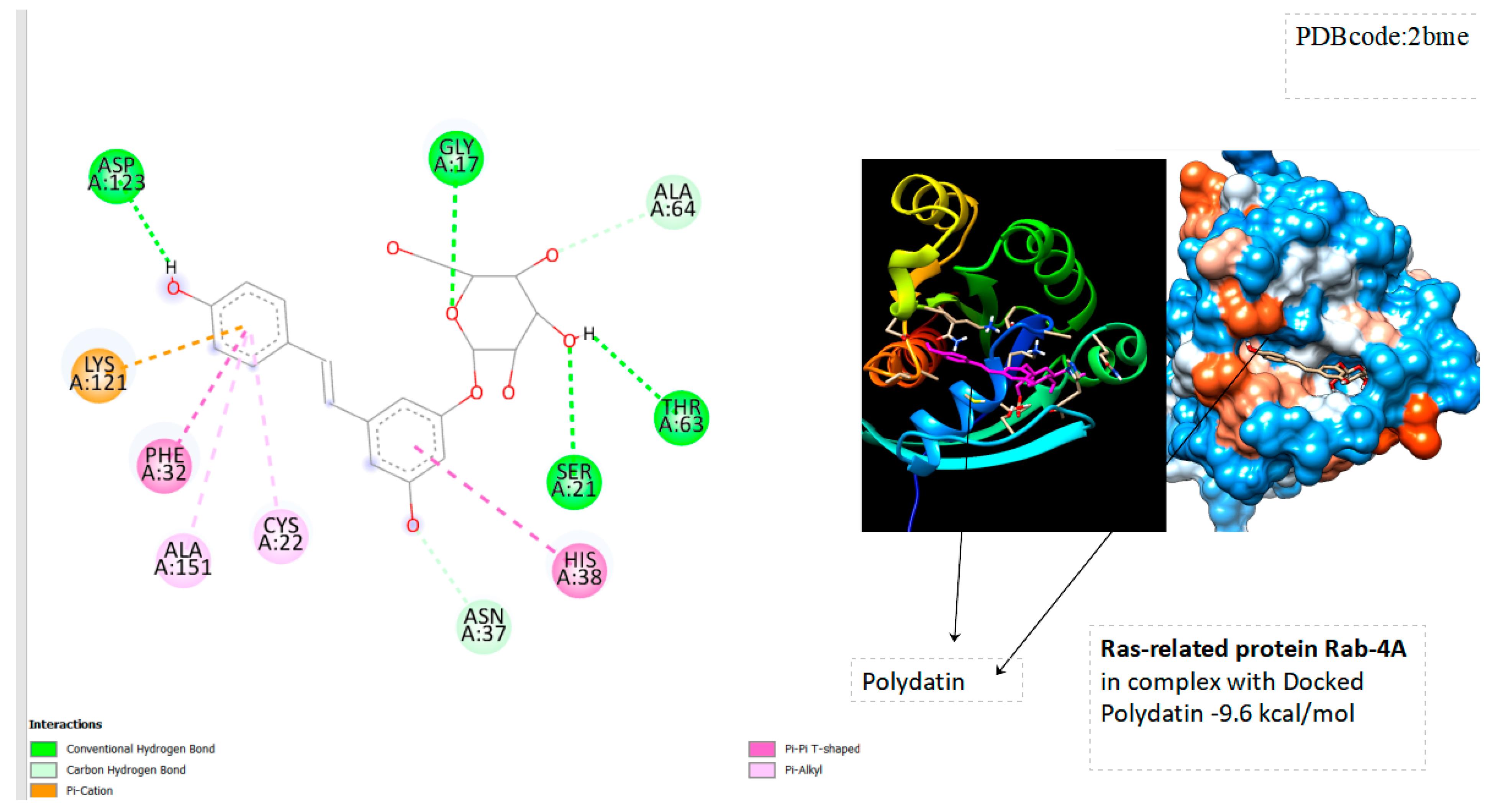
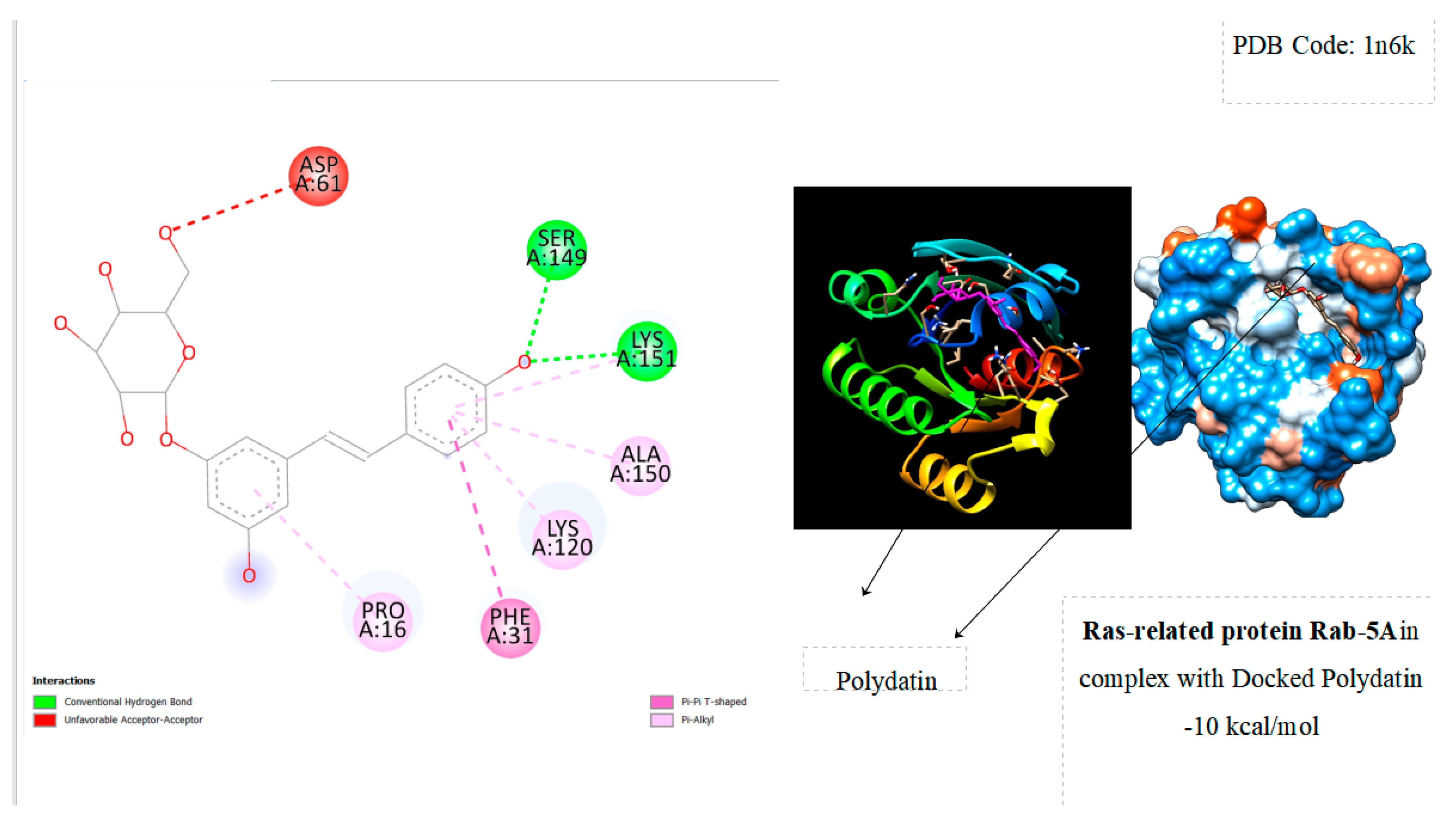
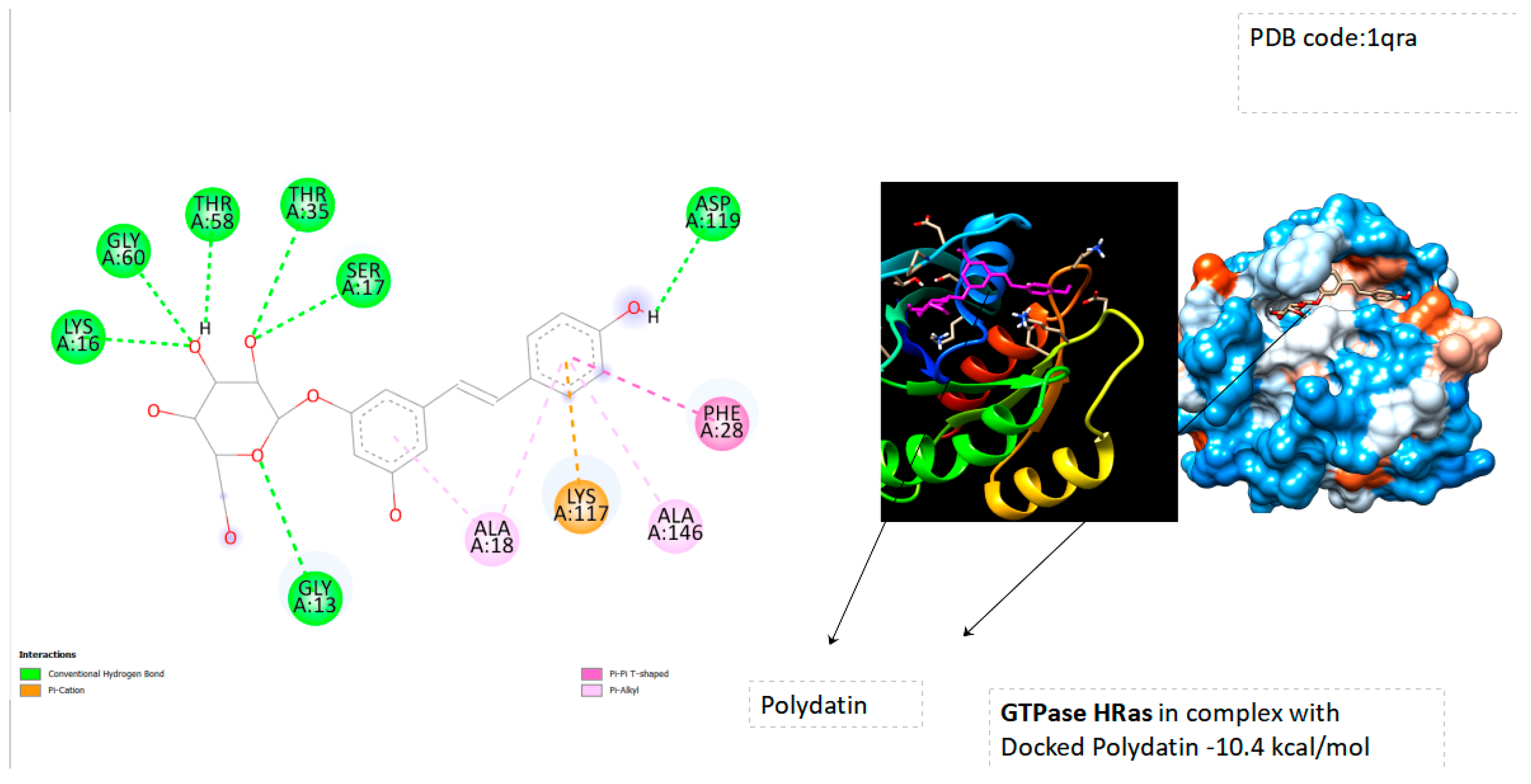
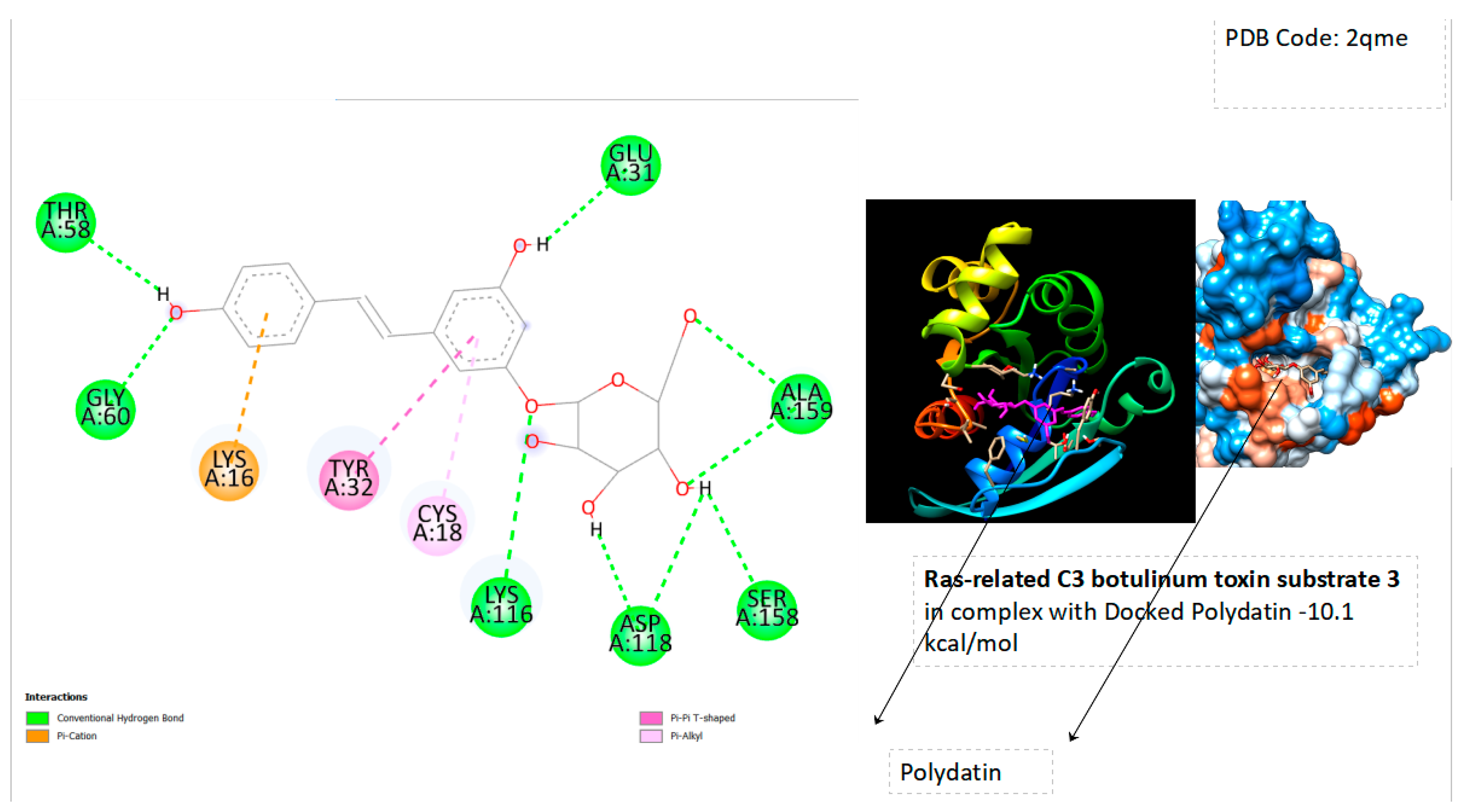
| Ras-Related Protein Rab-Proteins | Binding Energy (kcal/mol) |
|---|---|
| Ras-related protein Rab-28 | -7.0 |
| Ras-related GTP-binding protein C | -9.0 |
| Ras-related protein Rab-9A | -8.1 |
| Ras-related protein Rab-18 | -10.5 |
| Ras-related protein Rab-31 | -9.6 |
| Ras-related protein Rap-2a | -8.1 |
| Ras-related protein Rab-11B | -9.1 |
| Ras-related protein Rab-5A | -10 |
| Ras-related protein Rab-3D | -9.2 |
| Ras-related protein Rab-4A | -9.6 |
| GTPase HRas | -10.4 |
| Ras-related protein R-Ras | -9.1 |
| Ras-related protein Rap-2A | -8.4 |
| Ras-related protein Rab-1B | -8.9 |
| Ras-related C3 botulinum toxin substrate 3 | -10.1 |
| Ras-related protein Rab-6B | -9.6 |
| Ras-related protein Rab-6A | -9.3 |
4. Conclusion
References
- Bae, J. W., Kim, S. H., Kim, D. H., Ha, J. J., Yi, J. K., Hwang, S., ... & Kwon, W. S. (2019). Ras-related proteins (Rab) are key proteins related to male fertility following a unique activation mechanism. Reproductive biology, 19(4), 356-362.
- Martinez, O., & Goud, B. (1998). Rab proteins. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1404(1-2), 101-112.
- Barnekow, A., Thyrock, A., & Kessler, D. (2009). Rab proteins and their interaction partners. International review of cell and molecular biology, 274, 235-274.
- Bae, J. W., Yi, J. K., Jeong, E. J., Lee, W. J., Hwang, J. M., Kim, D. H., ... & Kwon, W. S. (2022). Ras-related proteins (Rab) play significant roles in sperm motility and capacitation status. Reproductive Biology, 22(2), 100617.
- Du, Q. H., Peng, C., & Zhang, H. (2013). Polydatin: a review of pharmacology and pharmacokinetics. Pharmaceutical biology, 51(11), 1347-1354. [CrossRef]
- Meng, X. Y., Zhang, H. X., Mezei, M., & Cui, M. (2011). Molecular docking: a powerful approach for structure-based drug discovery. Current computer-aided drug design, 7(2), 146-157. [CrossRef]
- Agarwal, S., & Mehrotra, R. J. J. C. (2016). An overview of molecular docking. JSM chem, 4(2), 1024-1028.
- Odhar, H. A., Rayshan, A. M., Ahjel, S. W., Hashim, A. A., & Albeer, A. A. M. A. (2019). Molecular docking enabled updated screening of the matrix protein VP40 from Ebola virus with millions of compounds in the MCULE database for potential inhibitors. Bioinformation, 15(9), 627.
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry, 31(2), 455-461. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





