1. Introduction
Pim-1 protein kinase is an enzyme encoded by the PIM1 gene in humans [
1,
2,
3]. Originally identified as a proto-oncogene in murine T-cell lymphomas, it has been frequently activated by Moloney murine leukemia virus [
4]. Subsequently, it has been associated with various human cancers, including prostate cancer, acute myeloid leukemia, and other hematopoietic malignancies. Pim-1 is expressed in several tissues, including spleen, thymus, bone marrow, prostate, oral epithelium, hippocampus, and fetal liver, and is particularly elevated in cell cultures isolated from human tumors [
1,
2,
3,
4].
Its main participation occurs in the regulation of cell cycle progression, apoptosis and transcriptional activation, as well as being involved in more general signal transduction pathways. Given its implications in oncogenic signaling, Pim-1 has become a significant target in cancer research. Numerous drug candidates that aim to inhibit this protein are currently being studied, indicating its potential as a therapeutic target in the treatment of tumors[
1,
2,
3,
4]. Due to its association with cancer and its involvement in oncogenic signaling pathways, Pim-1 has become a subject of intense investigation for potential therapeutic interventions.
The primary objective of this brief theoretical study is to simulate the interaction between the natural substance Hypericin with the enzyme Pim-1. The focus is on comprehending the extent of their interaction, identifying the types of chemical bonds formed during this process, and assessing the energetic affinity between Hypericin and the Pim-1 enzyme. This simulation aims to provide insights into the molecular dynamics of their association, shedding light on potential mechanisms of action and contributing to a deeper understanding of the biochemical processes involved.
Hypericin is a natural compound that belongs to a class of organic compounds known as polycyclic polyprenylated acylphloroglucinols (PPAPs). It is found in the genus Hypericum, commonly known as St. John's wort. Hypericin is known for its pharmacological properties, and it has been studied for various potential medical applications.
There have been several studies suggesting potential anticancer properties of hypericin, the natural compound found in St. John's wort. The anticancer effects of hypericin have been explored in various in vitro and in vivo experiments[
5,
6].
2. Material and Methods
Proto-oncogene serine/threonine-protein kinase pim-1 target was taken from Protein Data Bank (PDB Code: 3bgz) and was prepared from Mcule Database [
7], before to perform docking analysis with Autodock Vina[
8]. Molecular Docking investigation was evaluated in the Binding site center of this target with the following coordinated: X(-19,2277), Y( 37,3128) Z( -1,4445).
3. Results and Discussion
Detailed understanding of Pim-1 and its regulatory mechanisms may provide crucial information for the development of new therapeutic strategies in the treatment of pathological conditions, particularly those related to cancer [
1,
2,
3,
4]. The protein kinase Pim-1 is a crucial enzyme in the regulation of cellular processes such as growth, survival and differentiation. Deriving from the PIM1 gene, it is a proto-oncogene involved in the transformation of normal cells into cancerous cells. Initially associated with murine T-cell lymphomas, it is frequently activated by Moloney murine leukemia virus[
1,
2,
3,
4]. Expressed in different tissues and in human tumor cultures, Pim-1 regulates the cell cycle, apoptosis and transcriptional activation, also participating in cellular signaling pathways. Involved in several neoplasms, such as prostate cancer and acute myeloid leukemia, Pim-1 is the subject of intense oncological research[
1,
2,
3,
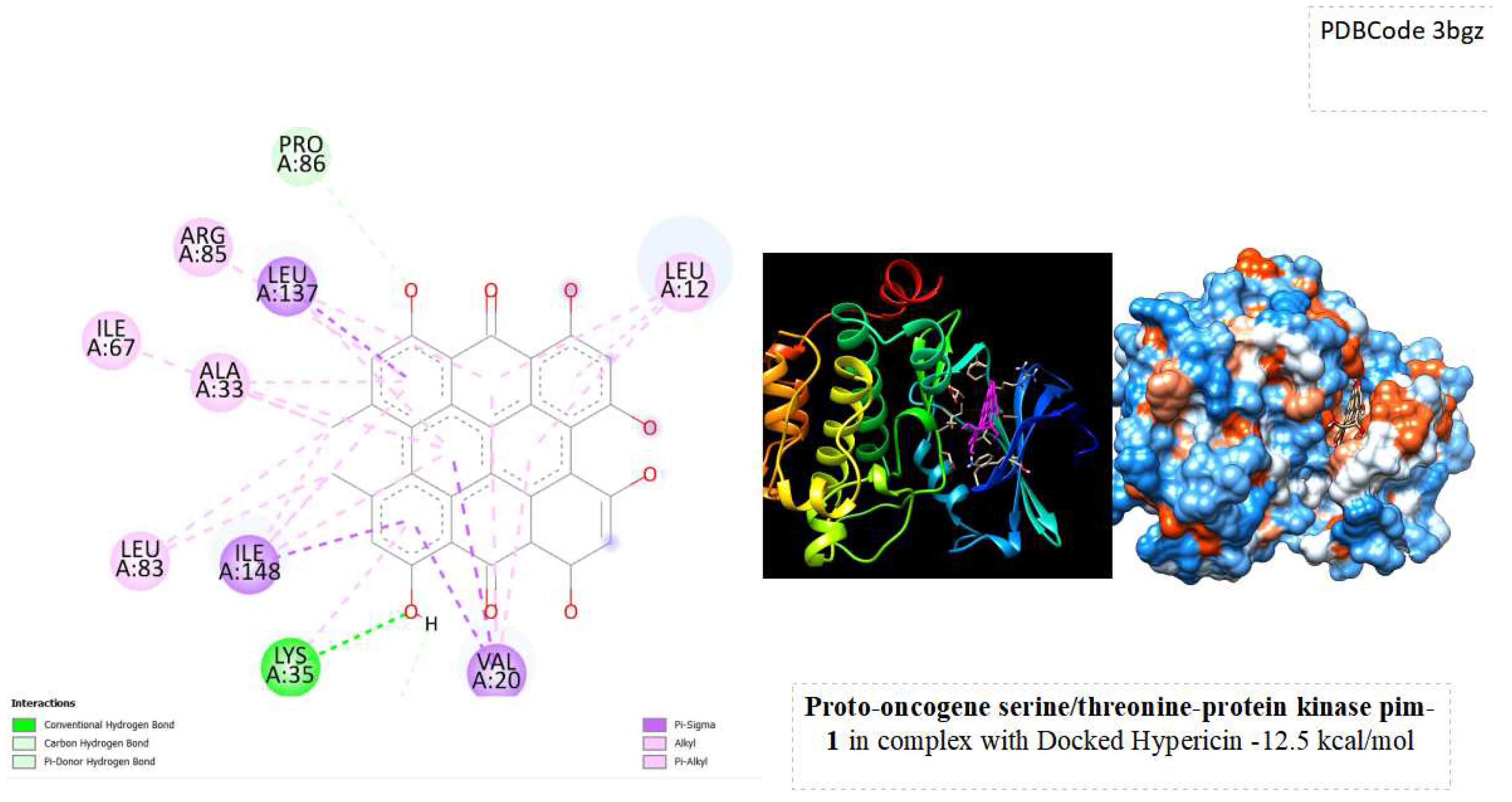
4].Considered a potential therapeutic target, studies are underway on drugs that aim to inhibit Pim-1, suggesting new prospects for cancer treatment. From docking results Hypericin showed a high score of binding energy of about -12.5 kcal/mol with this target, indicating a relatively high binding affinity, suggesting that hypericin may form stable interactions with the Pim-1 enzyme ( See below
Figure 1). Lower scores, in this context, are generally associated with stronger interactions.This strong binding could have implications for potential therapeutic applications or further research exploring the interaction's biological significance.
4. Conclusion
From docking simulations Hypericin with Pim-1 enzyme in the Ligand Binding Site obtained a Vina score of -12.5 kcal/mol , indicating a robust binding affinity. The negative docking score suggests that hypericin is energetically favorable in its interaction with the Pim-1 enzyme. This could be indicative of a potential for hypericin to modulate the activity of Pim-1.
While docking studies provide valuable insights into potential interactions, experimental validation is crucial to confirm these predictions. Further studies, such as molecular dynamics simulations or binding assays, would help validate and characterize the nature of the Pim-1 and hypericin interaction.
References
- Cen, B., Xiong, Y., Song, J. H., Mahajan, S., DuPont, R., McEachern, K.,... & Kraft, A. S. The Pim-1 protein kinase is an important regulator of MET receptor tyrosine kinase levels and signaling. Molecular and cellular biology 2014, 34, 2517–2532. [Google Scholar] [CrossRef] [PubMed]
- Wang, K., Deng, X., Shen, Z., Jia, Y., Ding, R., Li, R., ... & Jie, W. High glucose promotes vascular smooth muscle cell proliferation by upregulating proto-oncogene serine/threonine-protein kinase Pim-1 expression. Oncotarget, 2017, 8, 88320. [Google Scholar] [CrossRef]
- Domen, J., Von Lindern, M., Hermans, A., Breuer, M., Grosveld, G., & Berns, A. Comparison of the human and mouse PIM-1 cDNAs: nucleotide sequence and immunological identification of the in vitro synthesized PIM-1 protein. Oncogene research 1987, 1, 103–112. [Google Scholar]
- Bachmann, M., & Möröy, T. The serine/threonine kinase Pim-1. The international journal of biochemistry & cell biology, 2005, 37, 726–730. [Google Scholar]
- Agostinis, P., Vantieghem, A., Merlevede, W., & de Witte, P. A. Hypericin in cancer treatment: more light on the way. The international journal of biochemistry & cell biology, 2002, 34, 221–241. [Google Scholar]
- Kubin, A., Wierrani, F., Burner, U., Alth, G., & Grunberger, W. Hypericin-the facts about a controversial agent. Current pharmaceutical design, 2005, 11, 233–253. [Google Scholar] [CrossRef]
- Odhar, H. A., Rayshan, A. M., Ahjel, S. W., Hashim, A. A., & Albeer, A. A. M. A. Molecular docking enabled updated screening of the matrix protein VP40 from Ebola virus with millions of compounds in the MCULE database for potential inhibitors. Bioinformation, 2019, 15, 627. [Google Scholar]
- Trott, O., & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry, 2010, 31, 455–461. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).