Submitted:
26 November 2023
Posted:
28 November 2023
You are already at the latest version
Abstract
Keywords:
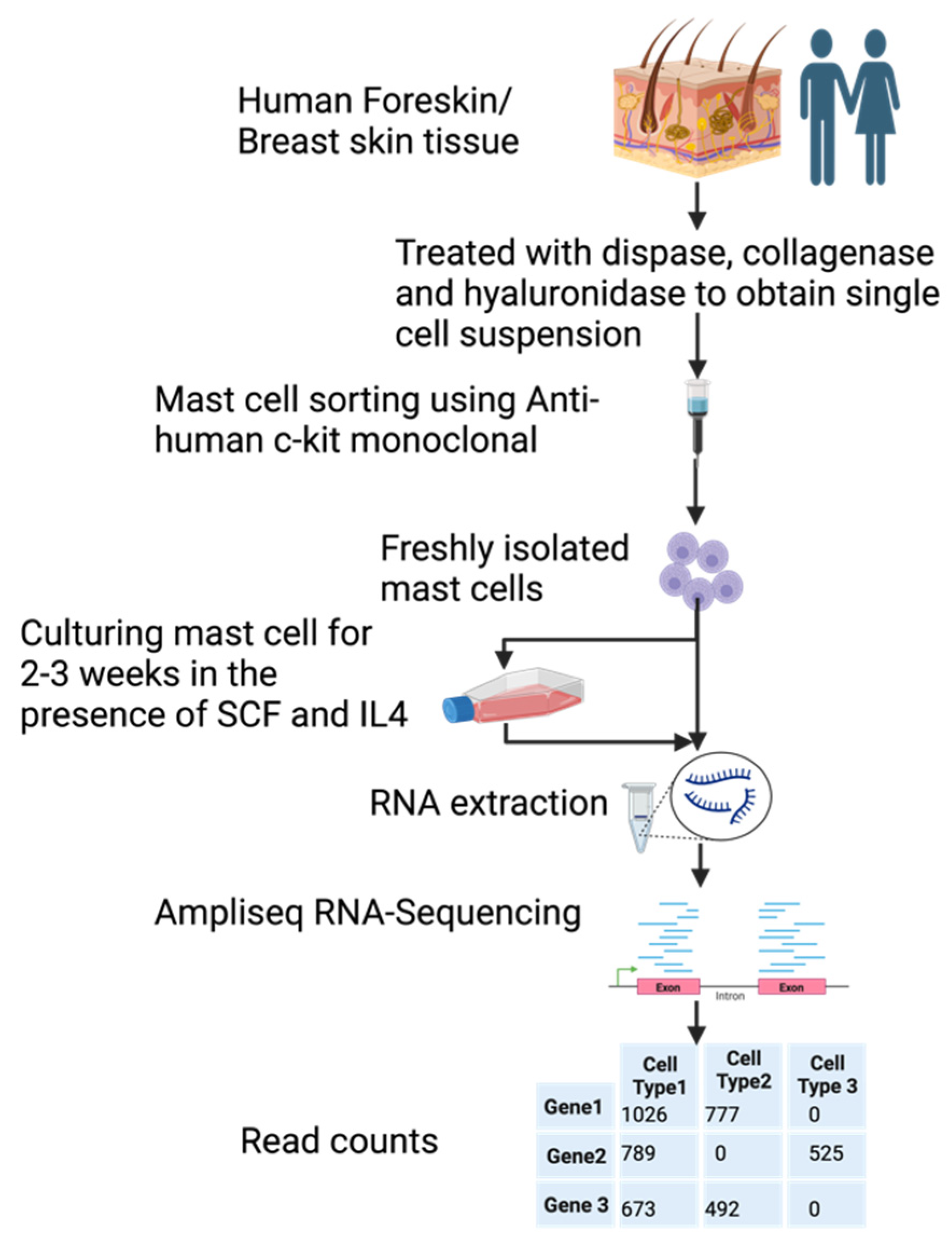
1. Introduction
2. Materials and Methods
2.1. Purification of human skin MCs
2.2. In vitro culture of human skin MCs
2.3. RNA isolation and heparinase treatment
2.4. Ampliseq analysis of the total transcriptome
2.5. Validations and RT-qPCR
| Gene | Forward Primer (5′->3′) | Reverse Primer (5′->3′) |
| FOS | AGTGACCGTGGGAATGAAGT | GCTTCAACAGACTACGAG |
| FOSB | CTACGGAGCCTGCACTTTCA | AGCGAGTCCTCAAAGTACGC |
| HSPA1A | AGCTGGAGCAGGTGTGTAAC | CAGCAATCTTGGAAAGGCCC |
| HSPA1B | TGTAACCCCATCATCAGCGG | TCCCAACAGTCCACCTCAAAG |
| ACTB | CTGGAACGGTGAAGGTGACA | AAGGGACTTCCTGTAACAATGCA |
| GAPDH | ACATCGCTCAGACACCATG | TGTAGTTGAGGTCAATGAAGGG |
| HPRT | GCCTCCCATCTCCTTCATCA | CCTGGCGTCGTGATTAGTGA |
| PPIB | AAGATGTCCCTGTGCCCTAC | ATGGCAAGCATGTGGTGTTT |
3. Results
3.1. Samples used for analysis
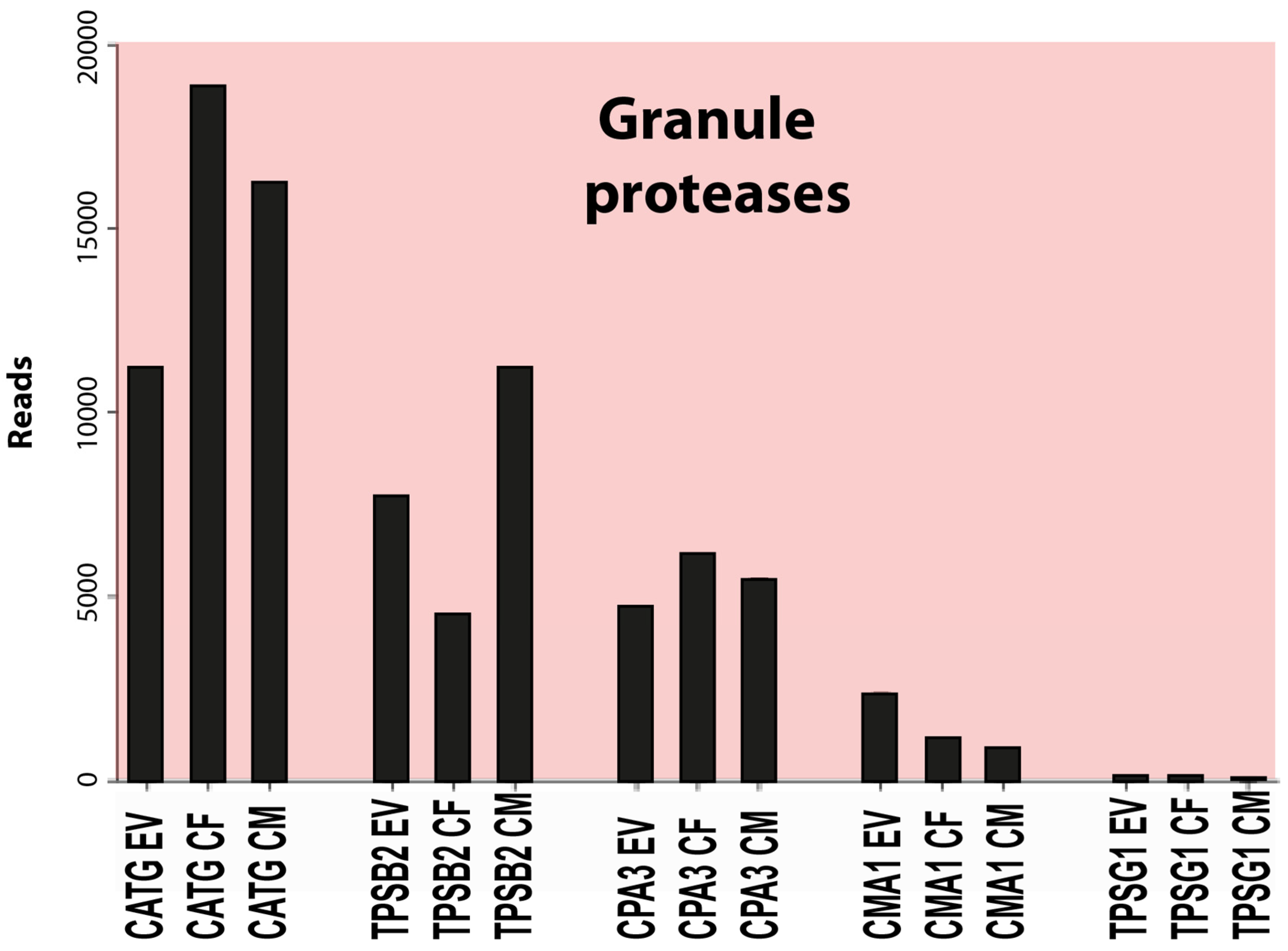
3.2. Transcript levels for the major granule-stored proteases and other proteases
3.3. Transcript levels for protease inhibitors.
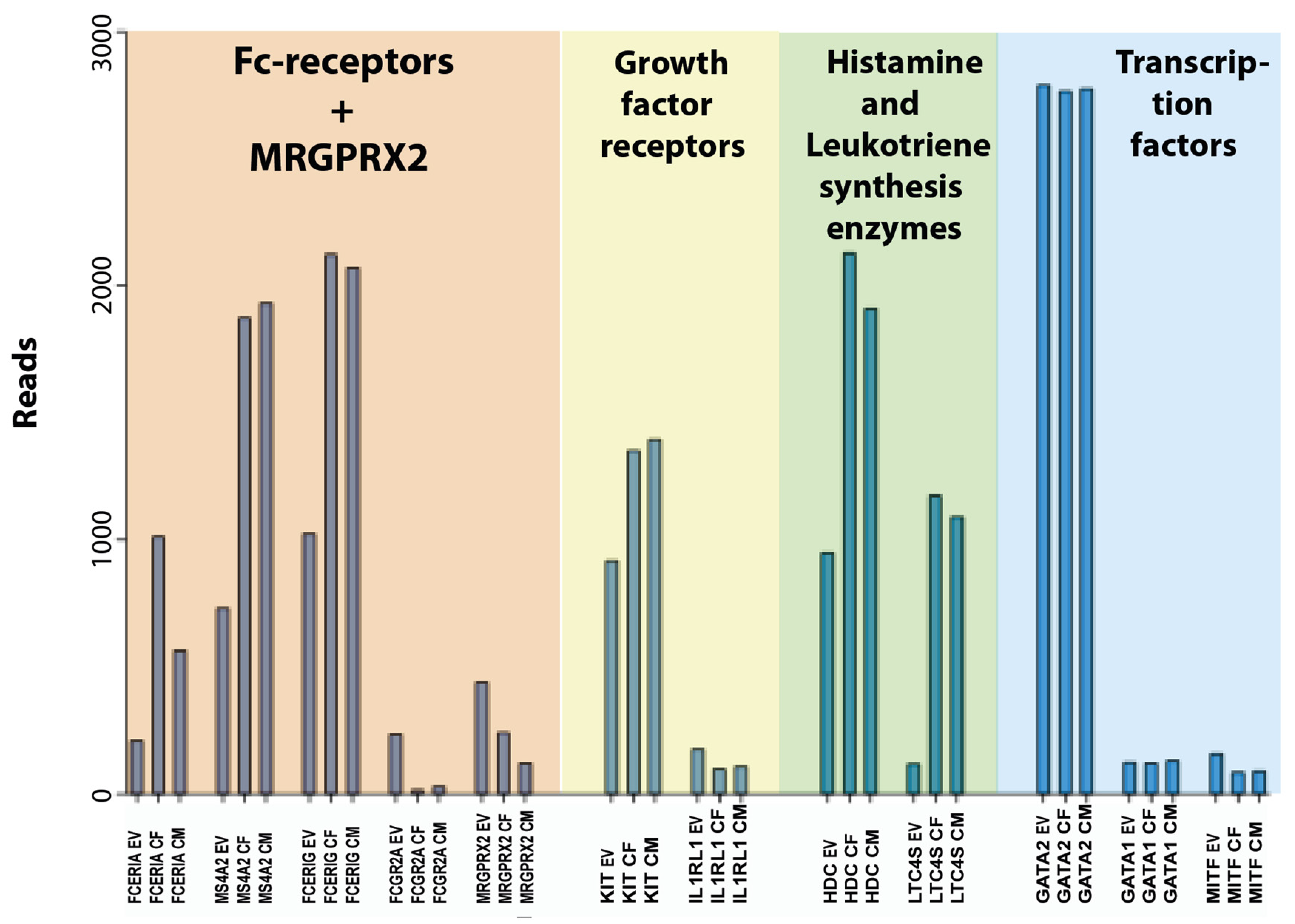
3.4. Transcript levels for Fc receptors and other cell surface receptors.
3.5. Transcript levels for MRGPRX2, purinergic, cannabinoid and anti-Mullerian hormone receptors.
3.6. Transcript levels for growth factor receptors.
3.7. Transcript levels for MHC class I and class II genes
3.8. Transcript levels for enzymes involved in proteoglycan, histamine, prostaglandin and leukotriene synthesis
3.9. Transcript levels for cell adhesion molecules
3.10. Transcript levels for transcription factors
3.11. Transcript levels for growth-related genes
3.12. Transcript levels for cytokine and growth factor genes
3.13. Transcript levels for several cluster of differentiation (CD) cell surface expressed proteins
3.14. Transcript levels for circadian clock related genes
3.15. Transcript levels for other proteins of potential interest
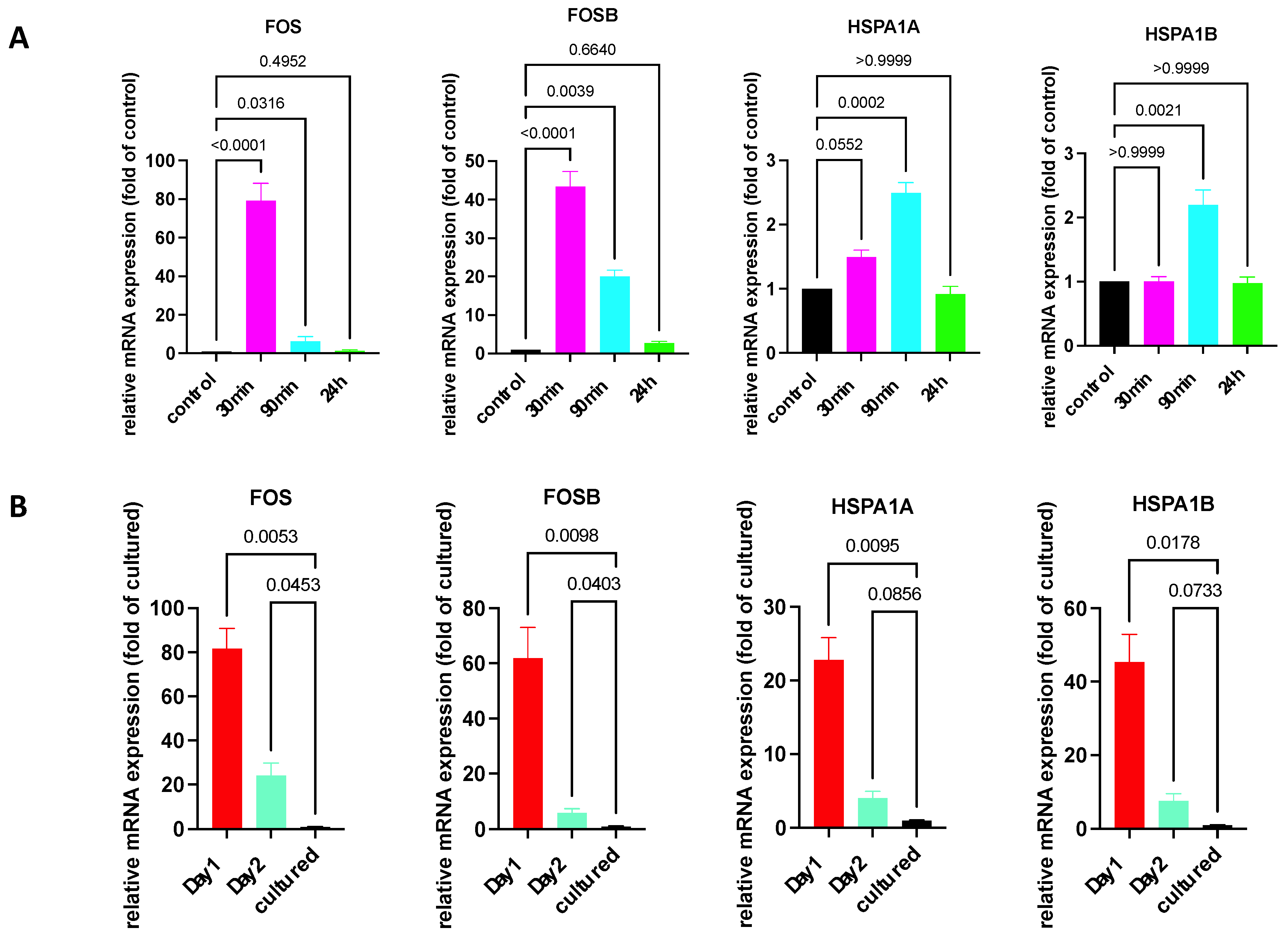
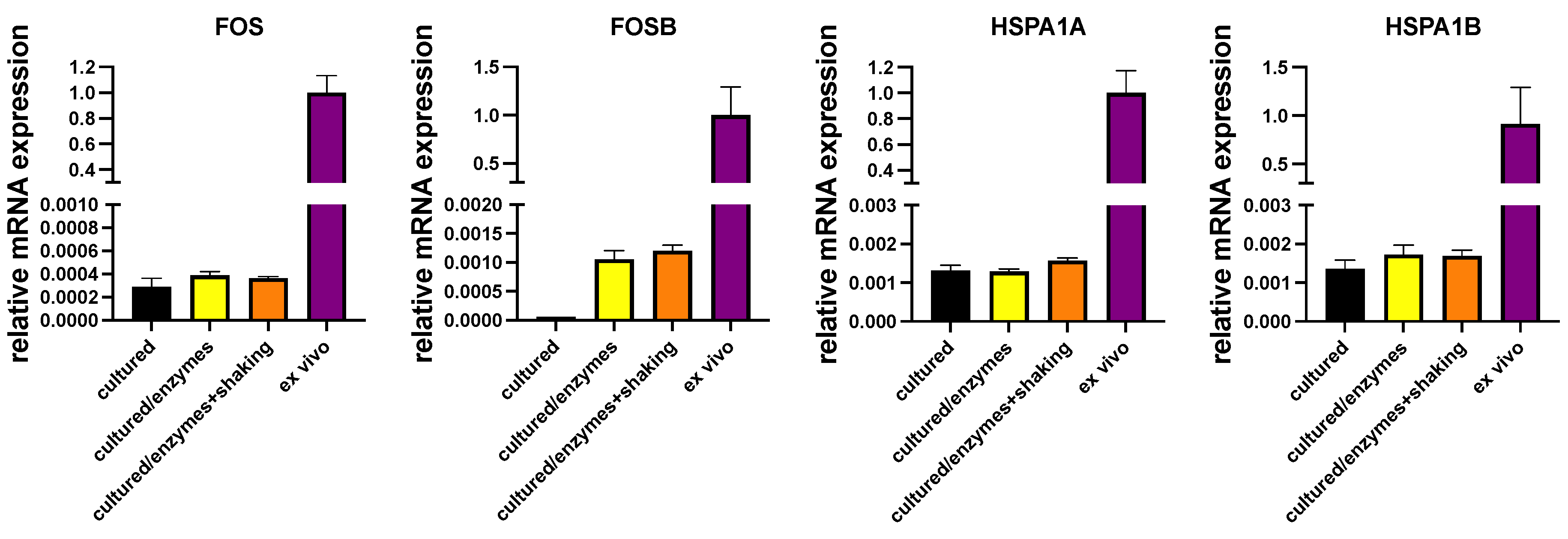
3.16. Transcript levels for heat shock proteins (HSPs) and immediate early genes (IEGs)
4. Discussion
Supplementary Materials
Acknowledgments
Abbreviations
References
- Akula, S., et al., Quantitative In-Depth Analysis of the Mouse Mast Cell Transcriptome Reveals Organ-Specific Mast Cell Heterogeneity. Cells, 2020. 9(1). [CrossRef]
- Schwartz, L.B., et al., Quantitation of histamine, tryptase, and chymase in dispersed human T and TC mast cells. J Immunol, 1987. 138(8): p. 2611-5. [CrossRef]
- Lutzelschwab, C., et al., Secretory granule proteases in rat mast cells. Cloning of 10 different serine proteases and a carboxypeptidase A from various rat mast cell populations. J Exp Med, 1997. 185(1): p. 13-29. [CrossRef]
- Tatemoto, K., et al., Immunoglobulin E-independent activation of mast cell is mediated by Mrg receptors. Biochem Biophys Res Commun, 2006. 349(4): p. 1322-8. [CrossRef]
- Motakis, E., et al., Redefinition of the human mast cell transcriptome by deep-CAGE sequencing. Blood, 2014. 123(17): p. e58-67. [CrossRef]
- McNeil, B.D., et al., Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature, 2015. 519(7542): p. 237-41. [CrossRef]
- Valent, P., et al., Induction of differentiation of human mast cells from bone marrow and peripheral blood mononuclear cells by recombinant human stem cell factor/kit-ligand in long-term culture. Blood, 1992. 80(9): p. 2237-45. [CrossRef]
- Irani, A.M., et al., Recombinant human stem cell factor stimulates differentiation of mast cells from dispersed human fetal liver cells. Blood, 1992. 80(12): p. 3009-21. [CrossRef]
- Mitsui, H., et al., Development of human mast cells from umbilical cord blood cells by recombinant human and murine c-kit ligand. Proc Natl Acad Sci U S A, 1993. 90(2): p. 735-9. [CrossRef]
- Franke, K., et al., The SCF/KIT axis in human mast cells: Capicua acts as potent KIT repressor and ERK predominates PI3K. Allergy, 2022. 77(11): p. 3337-3349. [CrossRef]
- Okayama, Y. and T. Kawakami, Development, migration, and survival of mast cells. Immunol Res, 2006. 34(2): p. 97-115. [CrossRef]
- Cruse, G., D.D. Metcalfe, and A. Olivera, Functional deregulation of KIT: link to mast cell proliferative diseases and other neoplasms. Immunol Allergy Clin North Am, 2014. 34(2): p. 219-37. [CrossRef]
- Toru, H., et al., Interleukin-4 promotes the development of tryptase and chymase double-positive human mast cells accompanied by cell maturation. Blood, 1998. 91(1): p. 187-95. [CrossRef]
- Bischoff, S.C., et al., IL-4 enhances proliferation and mediator release in mature human mast cells. Proc Natl Acad Sci U S A, 1999. 96(14): p. 8080-5. [CrossRef]
- Thienemann, F., B.M. Henz, and M. Babina, Regulation of mast cell characteristics by cytokines: divergent effects of interleukin-4 on immature mast cell lines versus mature human skin mast cells. Arch Dermatol Res, 2004. 296(3): p. 134-8. [CrossRef]
- Kulka, M. and D.D. Metcalfe, High-resolution tracking of cell division demonstrates differential effects of TH1 and TH2 cytokines on SCF-dependent human mast cell production in vitro: correlation with apoptosis and Kit expression. Blood, 2005. 105(2): p. 592-9. [CrossRef]
- Guhl, S., et al., Long-term cultured human skin mast cells are suitable for pharmacological studies of anti-allergic drugs due to high responsiveness to FcepsilonRI cross-linking. Biosci Biotechnol Biochem, 2011. 75(2): p. 382-4. [CrossRef]
- Babina, M., et al., IL-4 and human skin mast cells revisited: reinforcement of a pro-allergic phenotype upon prolonged exposure. Arch Dermatol Res, 2016. 308(9): p. 665-670. [CrossRef]
- Allakhverdi, Z., et al., Cutting edge: The ST2 ligand IL-33 potently activates and drives maturation of human mast cells. J Immunol, 2007. 179(4): p. 2051-4. [CrossRef]
- Iikura, M., et al., IL-33 can promote survival, adhesion and cytokine production in human mast cells. Lab Invest, 2007. 87(10): p. 971-8. [CrossRef]
- Babina, M., et al., Yin-Yang of IL-33 in Human Skin Mast Cells: Reduced Degranulation, but Augmented Histamine Synthesis through p38 Activation. J Invest Dermatol, 2019. 139(7): p. 1516-1525 e3. [CrossRef]
- Wang, Z., et al., IL-33 and MRGPRX2-Triggered Activation of Human Skin Mast Cells-Elimination of Receptor Expression on Chronic Exposure, but Reinforced Degranulation on Acute Priming. Cells, 2019. 8(4). [CrossRef]
- Kitamura, Y., et al., Effect of MITF on mast cell differentiation. Mol Immunol, 2002. 38(16-18): p. 1173-6. [CrossRef]
- Ohneda, K., S. Ohmori, and M. Yamamoto, Mouse Tryptase Gene Expression is Coordinately Regulated by GATA1 and GATA2 in Bone Marrow-Derived Mast Cells. Int J Mol Sci, 2019. 20(18). [CrossRef]
- Babina, M., et al., The transcription factor profile of human mast cells in comparison with monocytes and granulocytes. Cell Mol Life Sci, 2005. 62(2): p. 214-26. [CrossRef]
- Akula, S., et al., How Relevant Are Bone Marrow-Derived Mast Cells (BMMCs) as Models for Tissue Mast Cells? A Comparative Transcriptome Analysis of BMMCs and Peritoneal Mast Cells. Cells, 2020. 9(9). [CrossRef]
- Li, Z., et al., Adult Connective Tissue-Resident Mast Cells Originate from Late Erythro-Myeloid Progenitors. Immunity, 2018. 49(4): p. 640-653 e5. [CrossRef]
- Gentek, R., et al., Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity, 2018. 48(6): p. 1160-1171 e5. [CrossRef]
- Hallgren, J. and M.F. Gurish, Mast cell progenitor trafficking and maturation. Adv Exp Med Biol, 2011. 716: p. 14-28. [CrossRef]
- Sieweke, M.H. and J.E. Allen, Beyond stem cells: self-renewal of differentiated macrophages. Science, 2013. 342(6161): p. 1242974. [CrossRef]
- Amit, I., D.R. Winter, and S. Jung, The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol, 2016. 17(1): p. 18-25. [CrossRef]
- Perdiguero, E.G., et al., The Origin of Tissue-Resident Macrophages: When an Erythro-myeloid Progenitor Is an Erythro-myeloid Progenitor. Immunity, 2015. 43(6): p. 1023-4. [CrossRef]
- Hoeffel, G., et al., C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity, 2015. 42(4): p. 665-78. [CrossRef]
- Ginhoux, F., et al., Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science, 2010. 330(6005): p. 841-5. [CrossRef]
- Kierdorf, K., et al., Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci, 2013. 16(3): p. 273-80. [CrossRef]
- Bain, C.C., et al., Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol, 2014. 15(10): p. 929-937. [CrossRef]
- Babina, M., et al., Comparative cytokine profile of human skin mast cells from two compartments--strong resemblance with monocytes at baseline but induction of IL-5 by IL-4 priming. J Leukoc Biol, 2004. 75(2): p. 244-52. [CrossRef]
- Guhl, S., et al., Skin mast cells develop non-synchronized changes in typical lineage characteristics upon culture. Exp Dermatol, 2014. 23(12): p. 933-5. [CrossRef]
- Franke, K., et al., CREB Is Activated by the SCF/KIT Axis in a Partially ERK-Dependent Manner and Orchestrates Survival and the Induction of Immediate Early Genes in Human Skin Mast Cells. Int J Mol Sci, 2023. 24(4). [CrossRef]
- Franke, K., et al., Clorfl86/RHEX Is a Negative Regulator of SCF/KIT Signaling in Human Skin Mast Cells. Cells, 2023. 12(9). [CrossRef]
- Inoue, K., et al., Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science, 2010. 330(6003): p. 496-9. [CrossRef]
- Korkmaz, B., et al., Neutrophil proteinase 3 and dipeptidyl peptidase I (cathepsin C) as pharmacological targets in granulomatosis with polyangiitis (Wegener granulomatosis). Semin Immunopathol, 2013. 35(4): p. 411-21. [CrossRef]
- Brown, J., et al., Lymphopain, a cytotoxic T and natural killer cell-associated cysteine proteinase. Leukemia, 1998. 12(11): p. 1771-81. [CrossRef]
- Hazzan, T., et al., Thymic Stromal Lymphopoietin Interferes with the Apoptosis of Human Skin Mast Cells by a Dual Strategy Involving STAT5/Mcl-1 and JNK/Bcl-xL. Cells, 2019. 8(8). [CrossRef]
- Paivandy, A., et al., Quantitative In-Depth Transcriptome Analysis Implicates Peritoneal Macrophages as Important Players in the Complement and Coagulation Systems. Int J Mol Sci, 2022. 23(3). [CrossRef]
- Wallner, B.P., et al., Cloning and expression of human lipocortin, a phospholipase A2 inhibitor with potential anti-inflammatory activity. Nature, 1986. 320(6057): p. 77-81. [CrossRef]
- Kuch, E.M., et al., Differentially localized acyl-CoA synthetase 4 isoenzymes mediate the metabolic channeling of fatty acids towards phosphatidylinositol. Biochim Biophys Acta, 2014. 1841(2): p. 227-39. [CrossRef]
- Morimoto, K., et al., Prostaglandin E2-EP3 signaling induces inflammatory swelling by mast cell activation. J Immunol, 2014. 192(3): p. 1130-7. [CrossRef]
- Babina, M., K. Franke, and G. Bal, How "Neuronal" Are Human Skin Mast Cells? Int J Mol Sci, 2022. 23(18). [CrossRef]
- Shelburne, C.P., et al., Stat5 expression is critical for mast cell development and survival. Blood, 2003. 102(4): p. 1290-7. [CrossRef]
- Li, Y., et al., The STAT5-GATA2 pathway is critical in basophil and mast cell differentiation and maintenance. J Immunol, 2015. 194(9): p. 4328-38. [CrossRef]
- Pearson, R., et al., Kruppel-like transcription factors: a functional family. Int J Biochem Cell Biol, 2008. 40(10): p. 1996-2001. [CrossRef]
- Noguchi, S., et al., FANTOM5 CAGE profiles of human and mouse samples. Sci Data, 2017. 4: p. 170112. [CrossRef]
- Chen, X., et al., Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem, 2001. 276(32): p. 30423-8. [CrossRef]
- Carballo, E., W.S. Lai, and P.J. Blackshear, Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science, 1998. 281(5379): p. 1001-5. [CrossRef]
- Ayyar, V.S., et al., Quantitative tissue-specific dynamics of in vivo GILZ mRNA expression and regulation by endogenous and exogenous glucocorticoids. Physiol Rep, 2015. 3(6). [CrossRef]
- Bullwinkel, J., et al., Ki-67 protein is associated with ribosomal RNA transcription in quiescent and proliferating cells. J Cell Physiol, 2006. 206(3): p. 624-35. [CrossRef]
- Iuliano, C., et al., Fetal Tissue-Derived Mast Cells (MC) as Experimental Surrogate for In Vivo Connective Tissue MC. Cells, 2022. 11(6). [CrossRef]
- Babina, M., et al., MRGPRX2 is negatively targeted by SCF and IL-4 to diminish pseudo-allergic stimulation of skin mast cells in culture. Exp Dermatol, 2018. 27(11): p. 1298-1303. [CrossRef]
- Babina, M., et al., Skin mast cell phenotypes between two highly divergent cohorts - more pronounced variability within than between groups. Exp Dermatol, 2017. 26(5): p. 446-449. [CrossRef]
- Hsieh, F.H., et al., T helper cell type 2 cytokines coordinately regulate immunoglobulin E-dependent cysteinyl leukotriene production by human cord blood-derived mast cells: profound induction of leukotriene C(4) synthase expression by interleukin 4. J Exp Med, 2001. 193(1): p. 123-33. [CrossRef]
- Ng, K., et al., Xist and the order of silencing. EMBO Rep, 2007. 8(1): p. 34-9. [CrossRef]
- Dwyer, D.F., et al., Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat Immunol, 2016. 17(7): p. 878-87. [CrossRef]
- Friedman, A.D., Transcriptional regulation of granulocyte and monocyte development. Oncogene, 2002. 21(21): p. 3377-90. [CrossRef]
| Gene | Mouse Peritoneal MCs | Mouse BMMCs |
| Proteases | ||
| Cma1 (Mcpt5) | 45221 | 15683 |
| Mcpt4 (mMCP-4) | 31290 | 7 |
| Tpsb2 (Mcpt6) | 67773 | 3119 |
| Cpa3 (CPA-3) | 45604 | 22478 |
| Tpsab1 (Mcpt7) | 96 | 54 |
| Mcpt9 (mMCP-9) | 0 | 0 |
| CtsG (CTS-G) | 512 | 18 |
| Mcpt8 (mMCP-8) | 12 | 46 |
| Gzm B | 236 | 386 |
| Gzm C | 0.8 | 0.1 |
| Gzm D | 0.4 | 0 |
| Gzm E | 0 | 0.1 |
| Receptors | ||
| FcεRI alpha | 252 | 1631 |
| Ms4a2 (IgE rec. beta) | 1297 | 4288 |
| c-kit | 720 | 833 |
| Mrgprb2 | 899 | 8 |
| Transcription factors | ||
| Gata1 | 74 | 296 |
| Gata2 | 2272 | 5205 |
| Gata3 | 27 | 14 |
| Mitf | 370 | 144 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| XIST | 1 | 54 | 0 | 389 | 311 | 0 | 0 |
| RPS4Y1 | 81 | 73 | 53 | 0.1 | 0.2 | 193 | 172 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| Proteases | |||||||
| CATG | 9445 | 6194 | 12156 | 16285 | 21210 | 18847 | 13470 |
| TPSB2 | 7096 | 6177 | 7970 | 3771 | 5143 | 6791 | 15604 |
| CPA3 | 4403 | 3086 | 4976 | 5717 | 6465 | 6134 | 4812 |
| CMA1 | 1713 | 1359 | 2958 | 1177 | 1160 | 954 | 655 |
| TPSD1 | 418 | 67 | 145 | 196 | 156 | 78 | 62 |
| TPSG1 | 86 | 49 | 107 | 73 | 88 | 39 | 53 |
| CPM | 140 | 81 | 19 | 87 | 91 | 48 | 52 |
| GZMA | 0 | 0 | 0.1 | 0.1 | 2 | 0.3 | 0.5 |
| GZMK | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GZMB | 0.4 | 0.2 | 0 | 0.6 | 0.2 | 11 | 2 |
| GZMH | 0 | 0 | 0 | 0 | 0.2 | 0 | 0 |
| GZMM | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CTSC | 23 | 28 | 36 | 83 | 72 | 74 | 83 |
| CTSW | 32 | 12 | 12 | 294 | 177 | 175 | 184 |
| CTSD | 2356 | 1723 | 2806 | 3354 | 5192 | 3513 | 4318 |
| CTSB | 241 | 315 | 334 | 495 | 517 | 499 | 608 |
| CTSL1 | 48 | 158 | 55 | 22 | 28 | 29 | 27 |
| PLAT (tPA) | 136 | 288 | 137 | 164 | 218 | 317 | 158 |
| PLAU (uPA) | 102 | 38 | 100 | 16 | 20 | 18 | 12 |
| ADAMTS7 | 80 | 58 | 56 | 68 | 53 | 41 | 45 |
| ADAMTS14 | 2 | 1 | 3 | 108 | 141 | 98 | 159 |
| ADAM12 | 9 | 6 | 4 | 158 | 146 | 144 | 144 |
| PRSS12 | 38 | 59 | 98 | 3 | 4 | 10 | 3 |
| CASP3 | 88 | 74 | 119 | 294 | 223 | 168 | 217 |
| BACE2 | 103 | 49 | 39 | 473 | 434 | 399 | 425 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| Protease inhibitors | |||||||
| TIMP1 | 407 | 357 | 502 | 569 | 982 | 692 | 630 |
| TIMP3 | 1270 | 995 | 939 | 434 | 521 | 414 | 529 |
| CST3 | 564 | 586 | 395 | 219 | 337 | 258 | 219 |
| CST7 | 111 | 91 | 44 | 760 | 1210 | 418 | 482 |
| LXN | 27 | 132 | 49 | 16 | 13 | 16 | 17 |
| SERPINB1 | 333 | 156 | 222 | 1669 | 1663 | 1652 | 1569 |
| SERPINE1 | 200 | 339 | 115 | 6 | 2 | 9 | 0.7 |
| SERPINH1 | 270 | 352 | 470 | 11 | 13 | 17 | 10 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| Fc receptors | |||||||
| FCER1A (α) | 214 | 127 | 207 | 987 | 1041 | 494 | 633 |
| MS4A2 (β) | 817 | 441 | 642 | 1966 | 1782 | 2129 | 1732 |
| FCER1G (γ) | 908 | 848 | 1140 | 1852 | 2391 | 2064 | 2074 |
| FCER2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FCGR1A | 0 | 0 | 0 | 0 | 0 | 0.3 | 0 |
| FCGR2A | 142 | 199 | 325 | 23 | 6 | 36 | 20 |
| FCGR2B | 2 | 0.4 | 0.2 | 0.1 | 0 | 0 | 0 |
| FCGR2C | 2 | 1 | 1 | 0.4 | 0 | 0 | 0 |
| FCGR3A | 0.3 | 0 | 0 | 0 | 0 | 0 | 0 |
| FCGR3B | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| MILR1 | 57 | 29 | 68 | 135 | 204 | 162 | 145 |
| CD200R1 | 24 | 6 | 5 | 109 | 88 | 84 | 85 |
| CD300A | 56 | 35 | 74 | 152 | 167 | 119 | 151 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| Multiligand pseudo-allergy receptor, Purinergic and Cannabinoid receptors, anti-Mullerian hormone receptors | |||||||
| MRGPRX2 | 365 | 253 | 511 | 143 | 338 | 134 | 109 |
| MAS1L | 119 | 26 | 344 | 76 | 34 | 30 | 105 |
| P2RX1 | 334 | 226 | 337 | 328 | 335 | 259 | 356 |
| P2RX6 | 4 | 3 | 4 | 12 | 12 | 8 | 11 |
| P2RY1 | 11 | 3 | 2 | 68 | 61 | 75 | 77 |
| CNRIP1 | 113 | 74 | 81 | 382 | 124 | 241 | 224 |
| DRD2 | 35 | 19 | 56 | 94 | 47 | 71 | 57 |
| PAQR5 | 142 | 71 | 126 | 171 | 187 | 171 | 192 |
| EDNRB | 101 | 63 | 65 | 21 | 69 | 54 | 68 |
| ADORA2B | 8 | 6 | 5 | 21 | 23 | 17 | 30 |
| ADORA3 | 16 | 3 | 5 | 10 | 12 | 14 | 20 |
| CNR1 | 1 | 4 | 0.3 | 0.1 | 0 | 0 | 0 |
| CNR2 | 0 | 0 | 0 | 13 | 8 | 10 | 11 |
| AMHR2 | 59 | 26 | 57 | 85 | 101 | 86 | 104 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||||||
| Growth Factor Receptors, Hormone receptors and Retinoid receptors | |||||||||||
| KIT | 1023 | 458 | 811 | 1347 | 1352 | 1381 | 1403 | ||||
| EPOR | 105 | 38 | 112 | 69 | 73 | 58 | 63 | ||||
| CSF2RA (GM-CSFR) | 14 | 6 | 6 | 1 | 1 | 3 | 0.8 | ||||
| CSF2RB (beta) | 512 | 347 | 789 | 448 | 326 | 177 | 312 | ||||
| CRLF2 (TSLP-R) | 194 | 427 | 257 | 85 | 77 | 87 | 76 | ||||
| IL1RL1 (IL33R) | 125 | 289 | 226 | 96 | 99 | 115 | 99 | ||||
| BMPR1A | 59 | 42 | 47 | 78 | 92 | 74 | 74 | ||||
| PTAFR | 53 | 25 | 46 | 90 | 128 | 64 | 144 | ||||
| IL2RA | 17 | 32 | 14 | 102 | 11 | 28 | 90 | ||||
| IL3RA | 10 | 11 | 0.6 | 4 | 0.7 | 4 | 3 | ||||
| IL5RA | 17 | 11 | 14 | 9 | 9 | 16 | 12 | ||||
| IL6R | 69 | 98 | 27 | 16 | 15 | 7 | 13 | ||||
| IL6ST | 124 | 252 | 115 | 65 | 110 | 61 | 62 | ||||
| IL9R | 4 | 11 | 2 | 110 | 87 | 155 | 122 | ||||
| IL18R1 | 280 | 216 | 510 | 62 | 47 | 88 | 56 | ||||
| GPR34 | 39 | 9 | 14 | 332 | 304 | 206 | 224 | ||||
| MC1R | 9 | 6 | 19 | 67 | 130 | 44 | 136 | ||||
| TNFRSF9 | 159 | 351 | 282 | 10 | 1 | 11 | 15 | ||||
| TNFRSF21 | 292 | 226 | 503 | 11 | 17 | 6 | 9 | ||||
| LTBR | 74 | 65 | 43 | 3 | 4 | 8 | 10 | ||||
| ACVR1B | 28 | 62 | 27 | 12 | 13 | 17 | 12 | ||||
| ADIPOR2 | 48 | 101 | 58 | 62 | 64 | 64 | 71 | ||||
| GABARAPL1 | 43 | 71 | 25 | 11 | 12 | 9 | 6 | ||||
| AGTRAP | 115 | 50 | 109 | 215 | 293 | 201 | 253 | ||||
| ADRB2 | 561 | 590 | 832 | 244 | 278 | 262 | 252 | ||||
| RXRA | 181 | 109 | 142 | 117 | 129 | 68 | 94 | ||||
| NRP1 | 58 | 71 | 26 | 171 | 213 | 150 | 149 | ||||
| NRP2 | 87 | 132 | 47 | 47 | 39 | 69 | 55 | ||||
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| MHC and related genes | |||||||
| HLA-A | 344 | 326 | 253 | 231 | 329 | 167 | 201 |
| HLA-B | 297 | 188 | 70 | 338 | 48 | 93 | 106 |
| HLA-C | 552 | 241 | 200 | 449 | 475 | 251 | 196 |
| HLA-DPA1 | 93 | 60 | 21 | 13 | 5 | 10 | 23 |
| HLA-DPB1 | 39 | 28 | 9 | 0.2 | 0.4 | 1 | 0.3 |
| HLA-DRA | 75 | 65 | 19 | 0.4 | 0.2 | 0.3 | 0 |
| HLA-DRB1 | 146 | 121 | 118 | 52 | 0 | 0.7 | 0.8 |
| HLA-DQA1 | 17 | 8 | 4 | 0.2 | 0 | 0.4 | 0 |
| HLA-DOA | 2 | 0.6 | 0.4 | 0 | 0.1 | 0 | 0 |
| HLA-DOB | 0.1 | 0.1 | 0 | 0 | 0 | 0 | 0 |
| CIITA | 7 | 3 | 0.5 | 0.1 | 0 | 0.4 | 0 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | |||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
||||||
| Histamine, prostaglandin and leukotriene synthesis and receptors | ||||||||
| HDC | 853 | 796 | 1044 | 1844 | 2409 | 1941 | 1880 | |
| HRH4 | 27 | 7 | 31 | 23 | 6 | 15 | 9 | |
| HPGD | 1296 | 1021 | 1491 | 8857 | 9230 | 7877 | 8074 | |
| HPGDS | 705 | 484 | 588 | 2728 | 2015 | 2133 | 2715 | |
| PTGS2 (Cox2) | 523 | 392 | 353 | 293 | 248 | 881 | 364 | |
| PTGS1 (Cox1) | 378 | 357 | 401 | 335 | 390 | 340 | 389 | |
| ALOX5 | 307 | 143 | 109 | 282 | 380 | 369 | 351 | |
| PLA2G2A | 272 | 579 | 606 | 0 | 0 | 7 | 0 | |
| PLA2G3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| PLA2G4A | 18 | 12 | 9 | 20 | 28 | 33 | 43 | |
| LTC4S | 129 | 60 | 105 | 1111 | 1236 | 923 | 1258 | |
| TBXAS1 | 26 | 18 | 26 | 128 | 108 | 104 | 129 | |
| ANXA1 | 3919 | 5423 | 6675 | 1907 | 1793 | 1938 | 1558 | |
| ACSL4 | 529 | 841 | 576 | 2027 | 1607 | 1961 | 1830 | |
| PTGER2 | 23 | 10 | 15 | 53 | 28 | 33 | 36 | |
| PTGER3 | 43 | 18 | 34 | 122 | 117 | 115 | 102 | |
| PTGER4 | 217 | 203 | 210 | 234 | 272 | 268 | 230 | |
| Complement and coagulation components | ||||||||
| C2 | 13 | 13 | 13 | 3 | 10 | 43 | 32 | |
| C3AR1 | 31 | 6 | 8 | 157 | 146 | 111 | 138 | |
| PROCR | 36 | 19 | 14 | 12 | 18 | 20 | 22 | |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| Cell adhesion molecules | |||||||
| ITGA2B | 4 | 3 | 6 | 649 | 654 | 547 | 479 |
| ITGA3 | 98 | 75 | 92 | 18 | 34 | 34 | 23 |
| ITGA5 | 162 | 358 | 176 | 25 | 22 | 25 | 21 |
| ITGA9 | 148 | 127 | 156 | 28 | 22 | 23 | 17 |
| ITGAV | 29 | 148 | 24 | 20 | 18 | 25 | 13 |
| ITGAX | 86 | 231 | 76 | 81 | 115 | 83 | 99 |
| PXN | 180 | 127 | 147 | 34 | 37 | 31 | 29 |
| SELPLG | 60 | 21 | 34 | 262 | 209 | 135 | 228 |
| ICAM1 | 146 | 53 | 15 | 52 | 37 | 38 | 42 |
| L1CAM | 304 | 232 | 233 | 24 | 20 | 52 | 58 |
| FAT1 | 34 | 36 | 24 | 60 | 54 | 85 | 71 |
| CEACAM1 | 11 | 6 | 14 | 301 | 254 | 261 | 309 |
| NINJ1 | 181 | 266 | 123 | 37 | 16 | 19 | 24 |
| PMP22 | 504 | 523 | 279 | 270 | 321 | 365 | 351 |
| NTM | 119 | 97 | 158 | 52 | 99 | 57 | 43 |
| JPH4 | 93 | 62 | 127 | 72 | 94 | 80 | 109 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| Transcription factors | |||||||
| GATA2 | 2859 | 1421 | 2719 | 2575 | 2955 | 2735 | 2815 |
| GATA1 | 105 | 108 | 141 | 119 | 123 | 116 | 147 |
| GATA3 | 1 | 2 | 1 | 10 | 15 | 29 | 12 |
| MITF | 195 | 64 | 118 | 80 | 87 | 94 | 83 |
| HEY1 | 86 | 206 | 145 | 46 | 40 | 67 | 23 |
| HES1 | 138 | 117 | 88 | 13 | 13 | 15 | 16 |
| CREB1 | 75 | 83 | 54 | 73 | 71 | 67 | 64 |
| STAT5A | 80 | 56 | 70 | 86 | 65 | 78 | 86 |
| STAT5B | 111 | 100 | 156 | 150 | 133 | 127 | 128 |
| BHLHE40 | 1449 | 969 | 715 | 1612 | 1267 | 976 | 1046 |
| NFE2L3 | 18 | 19 | 27 | 22 | 18 | 20 | 21 |
| PBX1 | 53 | 44 | 50 | 131 | 134 | 112 | 136 |
| PHTF2 | 37 | 26 | 32 | 100 | 99 | 87 | 89 |
| HOXB2 | 22 | 14 | 12 | 58 | 67 | 64 | 60 |
| HOXB4 | 31 | 12 | 24 | 56 | 52 | 46 | 55 |
| RUNX2 | 61 | 23 | 40 | 17 | 14 | 14 | 5 |
| NR4A1 | 997 | 147 | 476 | 148 | 80 | 136 | 61 |
| IKZF1 | 158 | 133 | 124 | 353 | 312 | 262 | 353 |
| KLF2 | 2008 | 820 | 2018 | 3 | 10 | 8 | 8 |
| KLF4 | 1751 | 1041 | 1184 | 0.6 | 0.1 | 0.7 | 0 |
| TSC22D3 | 2856 | 2648 | 2859 | 7 | 6 | 5 | 8 |
| ZFP36 | 15395 | 10607 | 11923 | 289 | 138 | 122 | 53 |
| ZNF618 | 54 | 26 | 74 | 67 | 73 | 61 | 69 |
| ZNF521 | 43 | 19 | 35 | 69 | 77 | 70 | 56 |
| ZCCHC24 | 53 | 65 | 46 | 70 | 77 | 60 | 77 |
| ZMIZ1 | 190 | 300 | 224 | 153 | 159 | 135 | 158 |
| GFI1 | 116 | 41 | 54 | 148 | 257 | 195 | 229 |
| EGR3 | 494 | 194 | 902 | 379 | 248 | 422 | 220 |
| MEIS2 | 275 | 193 | 320 | 272 | 285 | 287 | 260 |
| STAT3 | 222 | 199 | 87 | 243 | 261 | 248 | 223 |
| AFF2 | 95 | 48 | 90 | 89 | 104 | 91 | 90 |
| TAL1 | 87 | 41 | 72 | 114 | 112 | 92 | 86 |
| E2F8 | 2 | 1 | 1 | 31 | 20 | 39 | 32 |
| FOXM1 | 0.3 | 0.3 | 0.2 | 75 | 55 | 111 | 81 |
| GLI3 | 57 | 47 | 32 | 11 | 25 | 22 | 21 |
| EPAS1 | 436 | 835 | 592 | 206 | 129 | 321 | 286 |
| MAF | 23 | 59 | 54 | 28 | 36 | 19 | 25 |
| PTRF | 465 | 707 | 280 | 198 | 140 | 175 | 125 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| Cell growth related transcripts | |||||||
| HIST1H3G | 8 | 9 | 10 | 1145 | 645 | 1695 | 982 |
| HIST1H3J | 5 | 4 | 4 | 363 | 174 | 481 | 319 |
| HIST1H3F | 4 | 2 | 1 | 277 | 185 | 229 | 238 |
| RRM2 | 1 | 0.3 | 1 | 259 | 133 | 323 | 254 |
| TOP2A | 1 | 1 | 2 | 207 | 151 | 326 | 189 |
| CDK1 | 1 | 1 | 1 | 108 | 74 | 140 | 120 |
| MKI67 | 0.3 | 0.4 | 0.7 | 111 | 65 | 143 | 107 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| Growth factors | |||||||
| VEGFA | 1948 | 1323 | 2848 | 17 | 15 | 23 | 23 |
| VEGFB | 138 | 87 | 108 | 155 | 213 | 185 | 240 |
| VEGFC | 7 | 9 | 0.4 | 0 | 0.4 | 1 | 0.2 |
| PDGFA | 270 | 366 | 442 | 7 | 5 | 8 | 7 |
| PDGFB | 4 | 14 | 0.8 | 0 | 0 | 0 | 0 |
| PDGFC | 17 | 20 | 14 | 0.7 | 0.7 | 0.9 | 0.2 |
| CSF1 (M-CSF) | 1579 | 933 | 1736 | 561 | 274 | 466 | 232 |
| CSF2 (GM-CSF) | 9 | 61 | 6 | 627 | 799 | 276 | 360 |
| CCL2 | 1518 | 1644 | 807 | 1379 | 925 | 1789 | 925 |
| CCL4 | 34 | 117 | 47 | 1 | 98 | 29 | 34 |
| CXCL16 | 263 | 160 | 310 | 185 | 252 | 117 | 156 |
| LIF | 659 | 307 | 1402 | 1582 | 1085 | 1798 | 1210 |
| TGFA | 31 | 21 | 41 | 65 | 87 | 58 | 59 |
| TGFB1I1 | 137 | 149 | 75 | 8 | 11 | 27 | 13 |
| TNF | 138 | 124 | 213 | 5 | 8 | 19 | 43 |
| TNFSF10 | 89 | 93 | 96 | 149 | 201 | 264 | 163 |
| IL13 | 29 | 47 | 28 | 0.5 | 0.4 | 1 | 0.3 |
| IL7 | 1 | 2 | 0.1 | 25 | 11 | 10 | 12 |
| IL5 | 0.1 | 0 | 0 | 3 | 1 | 5 | 8 |
| POSTN | 19 | 24 | 1 | 32 | 25 | 40 | 4 |
| GDF15 | 136 | 27 | 78 | 77 | 97 | 52 | 54 |
| EMR2 | 154 | 207 | 168 | 148 | 118 | 127 | 109 |
| OPTN | 37 | 27 | 10 | 63 | 56 | 80 | 58 |
| Growth factor induced proteins | |||||||
| TNFAIP3 | 5148 | 1318 | 588 | 24 | 11 | 23 | 18 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| Surface Markers | |||||||
| CD4 | 238 | 157 | 188 | 365 | 382 | 201 | 342 |
| CD9 | 931 | 1078 | 997 | 896 | 770 | 914 | 833 |
| CD14 | 22 | 26 | 16 | 9 | 36 | 21 | 27 |
| CD22 | 146 | 110 | 118 | 730 | 738 | 411 | 585 |
| CD33 | 42 | 28 | 70 | 199 | 179 | 140 | 100 |
| CD34 | 4 | 2 | 3 | 0 | 0 | 0.1 | 0 |
| CD52 | 29 | 20 | 9 | 833 | 697 | 954 | 776 |
| CD63 | 265 | 306 | 270 | 1274 | 1264 | 1255 | 1092 |
| CD68 | 355 | 305 | 275 | 686 | 999 | 1124 | 1240 |
| CD274 (PD-L1) | 106 | 158 | 130 | 15 | 10 | 17 | 17 |
| CD276 | 17 | 32 | 28 | 34 | 41 | 32 | 44 |
| CD109 | 16 | 13 | 7 | 30 | 31 | 31 | 32 |
| PROS1 | 67 | 55 | 45 | 297 | 301 | 220 | 243 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| Circadian proteins | |||||||
| PER1 | 2028 | 1197 | 2104 | 26 | 23 | 26 | 18 |
| CLOCK | 20 | 17 | 13 | 41 | 43 | 46 | 39 |
| PER2 | 45 | 54 | 29 | 11 | 17 | 10 | 12 |
| TIMELESS | 4 | 2 | 1 | 98 | 62 | 97 | 71 |
| PER3 | 39 | 21 | 15 | 25 | 39 | 40 | 36 |
| ARNTL | 5 | 3 | 1 | 15 | 14 | 13 | 15 |
| NR1D1 | 45 | 36 | 25 | 10 | 11 | 13 | 13 |
| NR1D2 | 37 | 33 | 12 | 34 | 32 | 31 | 31 |
| Freshly isolated cells | Cells cultured for 2-3 weeks | ||||||
| Foreskin (Male) |
Breast skin (Female) |
Foreskin (Male) |
|||||
| Stress and Growth response | |||||||
| FOSB | 13651 | 8861 | 14157 | 39 | 6 | 49 | 5 |
| FOS | 6395 | 3043 | 2026 | 158 | 60 | 140 | 51 |
| JUN | 30 | 17 | 16 | 0.2 | 0.1 | 0.3 | 0.2 |
| JUNB | 5676 | 3118 | 3982 | 331 | 315 | 231 | 250 |
| HSPA1A | 33988 | 35501 | 75250 | 259 | 293 | 211 | 293 |
| HSPA1B | 11715 | 5094 | 23085 | 57 | 45 | 65 | 85 |
| HSPA6 | 417 | 396 | 950 | 0.4 | 0 | 0 | 0 |
| HSPH1 | 885 | 423 | 702 | 94 | 108 | 115 | 111 |
| HSP90AB1 | 2350 | 2500 | 4641 | 432 | 479 | 391 | 430 |
| PLAUR | 888 | 415 | 1012 | 70 | 47 | 65 | 41 |
| CLU | 958 | 799 | 797 | 2460 | 2376 | 1523 | 1963 |
| CREB3L2 | 338 | 307 | 323 | 401 | 350 | 375 | 471 |
| IER3 | 1186 | 1693 | 836 | 1175 | 1143 | 1059 | 749 |
| EGR3 | 494 | 194 | 902 | 379 | 248 | 422 | 220 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





