Submitted:
23 February 2024
Posted:
26 February 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Objectives of this Work
1.2. Virological Context
1.3. Medicinal Chemistry Context
2. Results and Discussion
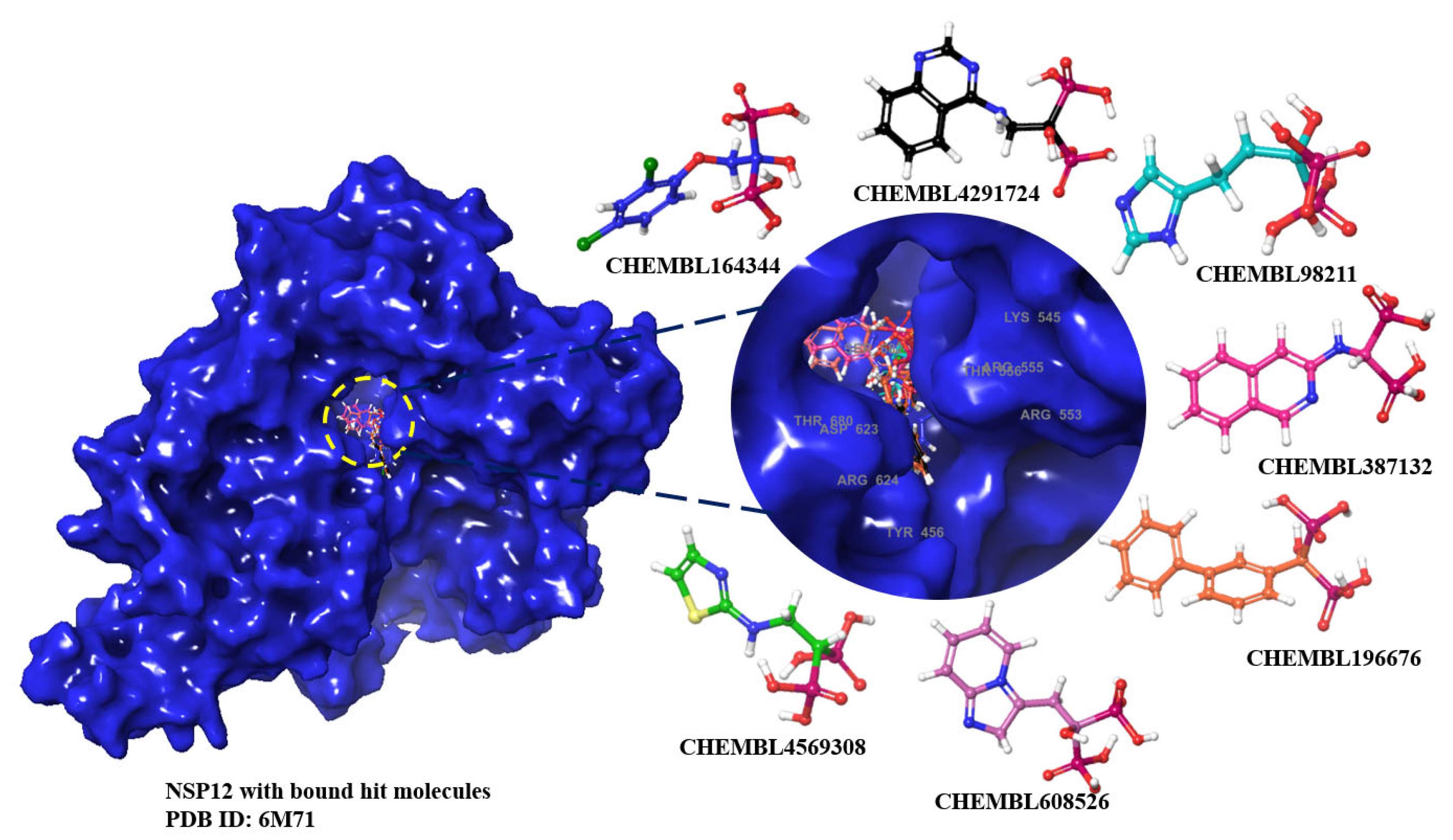
2.1. MM-GBSA Studies
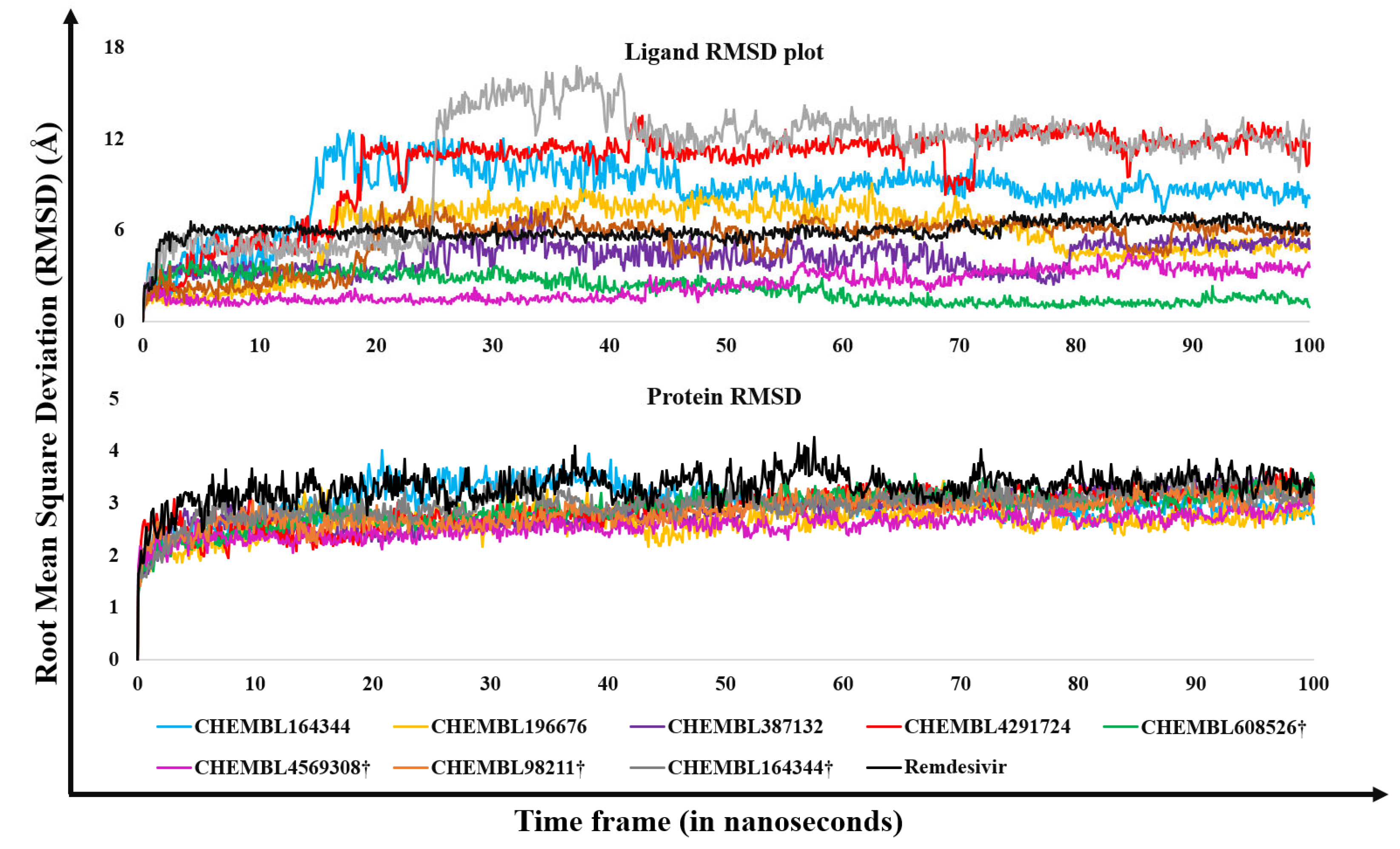
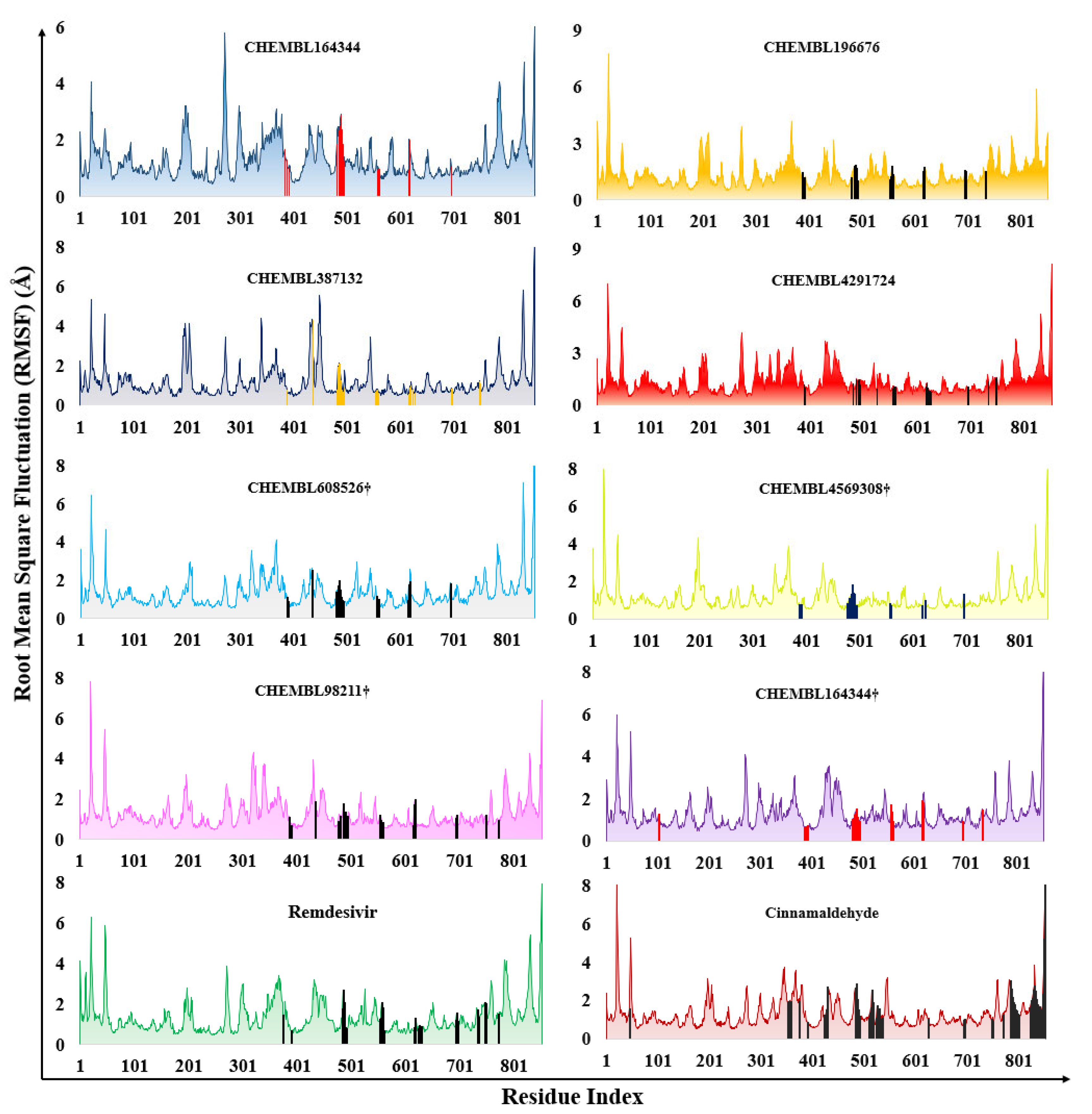
2.2. Molecular Dynamics Results
2.3. Estimation of Entropic Contribution by gmx_MMPBSA
2.4. Results of in Silico Predicted ADMET Profiles
3. Materials and Methods
3.1. Protein Preparation
3.2. Ligand Preparation
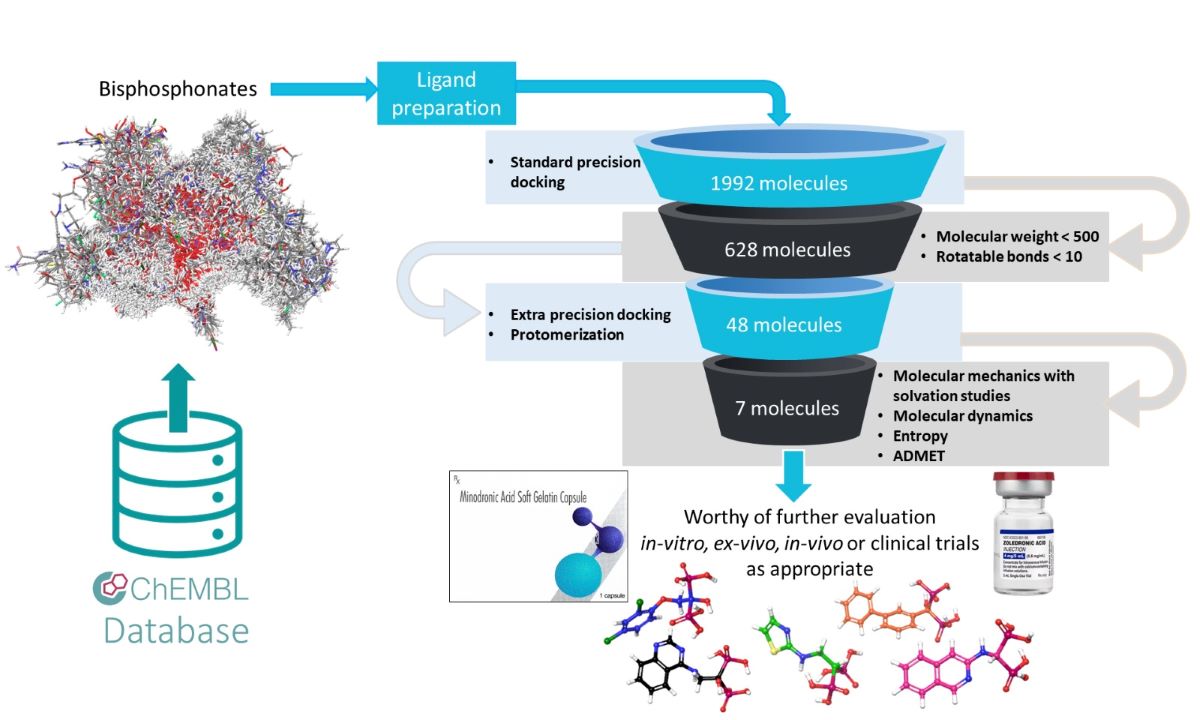
3.3. Molecular Docking
3.4. MM-GBSA Calculations
3.5. Molecular Dynamics Studies
3.6. Entropy Calculation for Molecular Dynamics Trajectories
3.7. ADMET (Absorption, Distribution, Metabolism, Excretion, Toxicity) Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mesel-Lemoine, M.; Millet, J.; Vidalain, P.-O.; Law, H.; Vabret, A.; Lorin, V.; Escriou, N.; Albert, M.L.; Nal, B.; Tangy, F. A Human Coronavirus Responsible for the Common Cold Massively Kills Dendritic Cells but Not Monocytes. J. Virol. 2012, 86, 7577–7587. [Google Scholar] [CrossRef]
- Zhu, Z.; Lian, X.; Su, X.; Wu, W.; Marraro, G.A.; Zeng, Y. From SARS and MERS to COVID-19: A Brief Summary and Comparison of Severe Acute Respiratory Infections Caused by Three Highly Pathogenic Human Coronaviruses. Respir. Res. 2020, 21, 224. [Google Scholar] [CrossRef]
- WHO Coronavirus (COVID-19) Dashboard. WHO Coronavirus (COVID-19) Dashboard With Vaccination Data. Available online: https://covid19.who.int/ (accessed on 2 November 2023).
- Thaweethai, T.; Jolley, S.E.; Karlson, E.W.; Levitan, E.B.; Levy, B.; Mccomsey, G.A.; Mccorkell, L.; Nadkarni, G.N.; Parthasarathy, S.; Singh, U.; et al. Development of a Definition of Postacute Sequelae of SARS-CoV-2 Infection. JAMA 2023, 329, 1934–1946. [Google Scholar] [CrossRef] [PubMed]
- Proal, A.D.; VanElzakker, M.B. Long COVID or Post-Acute Sequelae of COVID-19 (PASC): An Overview of Biological Factors That May Contribute to Persistent Symptoms. Front. Microbiol. 2021, 12, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Office for National Statistics (ONS) Prevalence of Ongoing Symptoms Following Coronavirus (COVID-19) Infection in the UK: 2 February 2023. Available online: https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/prevalenceofongoingsymptomsfollowingcoronaviruscovid19infectionintheuk/2february2023 (accessed on 6 November 2023).
- McDonald, E.G.; Lee, T.C. Nirmatrelvir-Ritonavir for COVID-19. CMAJ 2022, 194, E218. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration (FDA) Revocation of Emergency Use of a Drug During the COVID-19 Pandemic; Availability. Available online: https://www.federalregister.gov/documents/2022/07/26/2022-15956/revocation-of-emergency-use-of-a-drug-during-the-covid-19-pandemic-availability#citation-1-p44407 (accessed on 9 February 2024).
- Gilead. FDA Approves Veklury® (Remdesivir) to Treat COVID-19 in People With Mild to Severe Hepatic Impairment With No Dose Adjustment. Available online: https://www.gilead.com/news-and-press/press-room/press-releases/2023/8/fda-approves-veklury-remdesivir-to-treat-covid19-in-people-with-mild-to-severe-hepatic-impairment-with-no-dose-adjustment (accessed on 9 February 2024).
- Consortium, W.S.T. Remdesivir and Three Other Drugs for Hospitalised Patients with COVID-19: Final Results of the WHO Solidarity Randomised Trial and Updated Meta-Analyses. Lancet 2022, 399, 1941–1953. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration (FDA) FDA Approves First Oral Antiviral for Treatment of COVID-19 in Adults. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-oral-antiviral-treatment-covid-19-adults (accessed on 6 November 2023).
- Jayk Bernal, A.; Gomes da Silva, M.M.; Musungaie, D.B.; Kovalchuk, E.; Gonzalez, A.; Delos Reyes, V.; Martín-Quirós, A.; Caraco, Y.; Williams-Diaz, A.; Brown, M.L.; et al. Molnupiravir for Oral Treatment of Covid-19 in Nonhospitalized Patients. N. Engl. J. Med. 2022, 386, 509–520. [Google Scholar] [CrossRef]
- Manabe, T.; Kambayashi, D.; Akatsu, H.; Kudo, K. Favipiravir for the Treatment of Patients with COVID-19: A Systematic Review and Meta-Analysis. BMC Infect. Dis. 2021, 21, 489. [Google Scholar] [CrossRef] [PubMed]
- McAuley, A.J.; Vuren, P.J. van; Mohammed, M.-U.-R.; Faheem, F.; Goldie, S.; Riddell, S.; Gödde, N.J.; Styles, I.K.; Bruce, M.P.; Chahal, S.; et al. Use of Human Lung Tissue Models for Screening of Drugs against SARS-CoV-2 Infection. Viruses 2022, 14, 2417. [Google Scholar] [CrossRef]
- Reis, G.; dos Santos Moreira-Silva, E.A.; Silva, D.C.M.; Thabane, L.; Milagres, A.C.; Ferreira, T.S.; dos Santos, C.V.Q.; de Souza Campos, V.H.; Nogueira, A.M.R.; de Almeida, A.P.F.G.; et al. Effect of Early Treatment with Fluvoxamine on Risk of Emergency Care and Hospitalisation among Patients with COVID-19: The TOGETHER Randomised, Platform Clinical Trial. Lancet Glob. Heal. 2022, 10, e42–e51. [Google Scholar] [CrossRef]
- Butler, C.C.; Hobbs, F.D.R.; Gbinigie, O.A.; Rahman, N.M.; Hayward, G.; Richards, D.B.; Dorward, J.; Lowe, D.M.; Standing, J.F.; Breuer, J.; et al. Molnupiravir plus Usual Care versus Usual Care Alone as Early Treatment for Adults with COVID-19 at Increased Risk of Adverse Outcomes (PANORAMIC): An Open-Label, Platform-Adaptive Randomised Controlled Trial. Lancet 2023, 401, 281–293. [Google Scholar] [CrossRef]
- Wise, J. Covid-19: Molnupiravir Does Not Cut Hospital Admissions or Deaths in Vaccinated People at High Risk, Trial Finds. BMJ 2022, o3055. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, M. Merck’s COVID Pill Loses Its Lustre: What That Means for the Pandemic. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
- Batool, S.; Vuthaluru, K.; Hassan, A.; Bseiso, O.; Tehseen, Z.; Pizzorno, G.; Rodriguez Reyes, Y.; Saleem, F. Efficacy and Safety of Favipiravir in Treating COVID-19 Patients: A Meta-Analysis of Randomized Control Trials. Cureus 2023, 15, e33676. [Google Scholar] [CrossRef]
- Bosaeed, M.; Alharbi, A.; Mahmoud, E.; Alrehily, S.; Bahlaq, M.; Gaifer, Z.; Alturkistani, H.; Alhagan, K.; Alshahrani, S.; Tolbah, A.; et al. Efficacy of Favipiravir in Adults with Mild COVID-19: A Randomized, Double-Blind, Multicentre, Placebo-Controlled Clinical Trial. Clin. Microbiol. Infect. 2022, 28, 602–608. [Google Scholar] [CrossRef]
- Shah, P.L.; Orton, C.M.; Grinsztejn, B.; Donaldson, G.C.; Crabtree Ramírez, B.; Tonkin, J.; Santos, B.R.; Cardoso, S.W.; Ritchie, A.I.; Conway, F.; et al. Favipiravir in Patients Hospitalised with COVID-19 (PIONEER Trial): A Multicentre, Open-Label, Phase 3, Randomised Controlled Trial of Early Intervention versus Standard Care. Lancet Respir. Med. 2023, 11, 415–424. [Google Scholar] [CrossRef]
- Siripongboonsitti, T.; Ungtrakul, T.; Tawinprai, K.; Nimmol, T.; Buttakosa, M.; Sornsamdang, G.; Jarrusrojwuttikul, T.; Silapant, P.; Mahanonda, N. Efficacy of Combination Therapy of Fluvoxamine and Favipiravir vs Favipiravir Monotherapy to Prevent Severe COVID-19 among Mild to Moderate COVID-19 Patients: Open-Label Randomized Controlled Trial (EFFaCo Study). Int. J. Infect. Dis. 2023, 134, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Lenze, E.J.; Mattar, C.; Zorumski, C.F.; Stevens, A.; Schweiger, J.; Nicol, G.E.; Miller, J.P.; Yang, L.; Yingling, M.; Avidan, M.S.; et al. Fluvoxamine vs Placebo and Clinical Deterioration in Outpatients With Symptomatic COVID-19. JAMA 2020, 324, 2292. [Google Scholar] [CrossRef]
- Calusic, M.; Marcec, R.; Luksa, L.; Jurkovic, I.; Kovac, N.; Mihaljevic, S.; Likic, R. Safety and Efficacy of Fluvoxamine in COVID-19 ICU Patients: An Open Label, Prospective Cohort Trial with Matched Controls. Br. J. Clin. Pharmacol. 2022, 88, 2065–2073. [Google Scholar] [CrossRef]
- Jain, H.A.; Agarwal, V.; Bansal, C.; Kumar, A.; Faheem, F.; Mohammed, M.-U.-R.; Murugesan, S.; Simpson, M.M.; Karpe, A.V.; Chandra, R.; et al. CoviRx: A User-Friendly Interface for Systematic Down-Selection of Repurposed Drug Candidates for COVID-19. Data 2022, 7, 164. [Google Scholar] [CrossRef]
- MacRaild, C.A.; Mohammed, M.U.R.; Faheem, *!!! REPLACE !!!*; Murugesan, S.; Styles, I.K.; Peterson, A.L.; Kirkpatrick, C.M.J.; Cooper, M.A.; Palombo, E.A.; Simpson, M.M.; et al. Systematic Down-Selection of Repurposed Drug Candidates for COVID-19. Int. J. Mol. Sci. 2022, 23, 11851. [Google Scholar] [CrossRef]
- Muzaffar-Ur-Rehman, M.; Suryakant, C.K.; Chandu, A.; Kumar, B.K.; Joshi, R.P.; Jadav, S.R.; Sankaranarayanan, M.; Vasan, S.S. Molecular Docking and Dynamics Identify Potential Drugs to Be Repurposed as SARS-CoV-2 Inhibitors. J. Comput. Biophys. Chem. 2023, 1–23. [Google Scholar] [CrossRef]
- Thompson, J.; Wang, Y.; Dreischulte, T.; Barreiro, O.; Gonzalez, R.J.; Hanč, P.; Matysiak, C.; Neely, H.R.; Rottenkolber, M.; Haskell, T.; et al. Association between Bisphosphonate Use and COVID-19 Related Outcomes. Elife 2023, 12, e79548. [Google Scholar] [CrossRef]
- Huang, B. Use Of A Nitrogen-Containing Bisphosphonate In Combination With A Glucocorticoid In Preventing Or Treating Viral Pneumonia. 2023, 1–26, US20230149429A1. [Google Scholar]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 Genome, Structure, Evolution, Pathogenesis and Therapies: Structural Genomics Approach. Biochim. Biophys. acta. Mol. basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-Dependent RNA Polymerase from COVID-19 Virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Malone, B.; Urakova, N.; Snijder, E.J.; Campbell, E.A. Structures and Functions of Coronavirus Replication–Transcription Complexes and Their Relevance for SARS-CoV-2 Drug Design. Nat. Rev. Mol. Cell Biol. 2021, 23, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Gangadharan, S.; Ambrose, J.M.; Rajajagadeesan, A.; Kullappan, M.; Patil, S.; Gandhamaneni, S.H.; Veeraraghavan, V.P.; Nakkella, A.K.; Agarwal, A.; Jayaraman, S.; et al. Repurposing of Potential Antiviral Drugs against RNA-Dependent RNA Polymerase of SARS-CoV-2 by Computational Approach. J. Infect. Public Health 2022, 15, 1180–1191. [Google Scholar] [CrossRef] [PubMed]
- Sivaraman, H.; Er, S.Y.; Choong, Y.K.; Gavor, E.; Sivaraman, J. Structural Basis of SARS-CoV-2- and SARS-CoV-Receptor Binding and Small-Molecule Blockers as Potential Therapeutics. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 465–493. [Google Scholar] [CrossRef]
- Bertolin, A.P.; Weissmann, F.; Zeng, J.; Posse, V.; Milligan, J.C.; Canal, B.; Ulferts, R.; Wu, M.; Drury, L.S.; Howell, M.; et al. Identifying SARS-CoV-2 Antiviral Compounds by Screening for Small Molecule Inhibitors of Nsp12/7/8 RNA-Dependent RNA Polymerase. Biochem. J. 2021, 478, 2425–2443. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural Basis for Inhibition of the RNA-Dependent RNA Polymerase from SARS-CoV-2 by Remdesivir. Science (80-.) 2020, 368, 1499–1504. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Ward, A.B. Structure of the SARS-CoV Nsp12 Polymerase Bound to Nsp7 and Nsp8 Co-Factors. Nat. Commun. 2019, 10, 2342. [Google Scholar] [CrossRef]
- Ahmad, J.; Ikram, S.; Ahmad, F.; Rehman, I.U.; Mushtaq, M. SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp)–A Drug Repurposing Study. Heliyon 2020, 6, e04502. [Google Scholar] [CrossRef]
- El Sohaimy, S.; Abdo, N.; Shehata, M.; Moheyeldin, O. Inhibition of COVID-19 RNA-Dependent RNA Polymerase by Natural Bioactive Compounds: Molecular Docking Analysis. Egypt. J. Chem. 2021, 64, 1989–2001. [Google Scholar] [CrossRef]
- Zamzami, M.A. Molecular Docking, Molecular Dynamics Simulation and MM-GBSA Studies of the Activity of Glycyrrhizin Relevant Substructures on SARS-CoV-2 RNA-Dependent-RNA Polymerase. J. Biomol. Struct. Dyn. 2023, 41, 1846–1858. [Google Scholar] [CrossRef] [PubMed]
- Brunt, D.; Lakernick, P.M.; Wu, C. Discovering New Potential Inhibitors to SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp) Using High Throughput Virtual Screening and Molecular Dynamics Simulations. Sci. Rep. 2022, 12, 19986. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Lu, W.; Jiang, H.; Yang, C.; Dong, X. Molecular Docking and Dynamics of Phytochemicals From Chinese Herbs With SARS-CoV-2 RdRp. Nat. Prod. Commun. 2022, 17. [Google Scholar] [CrossRef]
- Askari, F.S.; Ebrahimi, M.; Parhiz, J.; Hassanpour, M.; Mohebbi, A.; Mirshafiey, A. Digging for the Discovery of SARS-CoV-2 Nsp12 Inhibitors: A Pharmacophore-Based and Molecular Dynamics Simulation Study. Future Virol. 2022, 17, 743–759. [Google Scholar] [CrossRef] [PubMed]
- Uengwetwanit, T.; Chutiwitoonchai, N.; Wichapong, K.; Karoonuthaisiri, N. Identification of Novel SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp) Inhibitors: From in Silico Screening to Experimentally Validated Inhibitory Activity. Comput. Struct. Biotechnol. J. 2022, 20, 882–890. [Google Scholar] [CrossRef]
- Chinnamadhu, A.; Ramakrishnan, J.; Suresh, S.; Ramadurai, P.; Poomani, K. Dynamics and Binding Affinity of Nucleoside and Non-Nucleoside Inhibitors with RdRp of SARS-CoV-2: A Molecular Screening, Docking, and Molecular Dynamics Simulation Study. J. Biomol. Struct. Dyn. 2022, 41, 10396–10410. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, F.A.; Alkarim, S.A.; Hawsawi, Y.M.; Abdulaal, W.H.; Albiheyri, R.; Kurdi, B.; Alguridi, H.; El-Magd, M.A. 25 (S)-Hydroxycholesterol Acts as a Possible Dual Enzymatic Inhibitor of SARS-CoV-2 M pro and RdRp–: An Insight from Molecular Docking and Dynamics Simulation Approaches. J. Biomol. Struct. Dyn. 2023, 41, 4744–4755. [Google Scholar] [CrossRef] [PubMed]
- Alexpandi, R.; De Mesquita, J.F.; Pandian, S.K.; Ravi, A.V. Quinolines-Based SARS-CoV-2 3CLpro and RdRp Inhibitors and Spike-RBD-ACE2 Inhibitor for Drug-Repurposing Against COVID-19: An in Silico Analysis. Front. Microbiol. 2020, 11, 1796. [Google Scholar] [CrossRef]
- Kushwaha, P.P.; Singh, A.K.; Bansal, T.; Yadav, A.; Prajapati, K.S.; Shuaib, M.; Kumar, S. Identification of Natural Inhibitors Against SARS-CoV-2 Drugable Targets Using Molecular Docking, Molecular Dynamics Simulation, and MM-PBSA Approach. Front. Cell. Infect. Microbiol. 2021, 11, 730288. [Google Scholar] [CrossRef]
- Shady, N.H.; Hayallah, A.M.; Mohamed, M.F.A.; Ghoneim, M.M.; Chilingaryan, G.; Al-Sanea, M.M.; Fouad, M.A.; Kamel, M.S.; Abdelmohsen, U.R. Targeting 3CLpro and SARS-CoV-2 RdRp by Amphimedon Sp. Metabolites: A Computational Study. Molecules 2021, 26, 3775. [Google Scholar] [CrossRef]
- Gajjar, N.D.; Dhameliya, T.M.; Shah, G.B. In Search of RdRp and Mpro Inhibitors against SARS CoV-2: Molecular Docking, Molecular Dynamic Simulations and ADMET Analysis. J. Mol. Struct. 2021, 1239, 130488. [Google Scholar] [CrossRef]
- Parihar, A.; Sonia, Z.F.; Akter, F.; Ali, M.A.; Hakim, F.T.; Hossain, M.S. Phytochemicals-Based Targeting RdRp and Main Protease of SARS-CoV-2 Using Docking and Steered Molecular Dynamic Simulation: A Promising Therapeutic Approach for Tackling COVID-19. Comput. Biol. Med. 2022, 145, 105468. [Google Scholar] [CrossRef]
- M A Kawsar, S.; Hosen, M.A.; Ahmad, S.; El Bakri, Y.; Laaroussi, H.; Ben Hadda, T.; Almalki, F.A.; Ozeki, Y.; Goumri-Said, S. Potential SARS-CoV-2 RdRp Inhibitors of Cytidine Derivatives: Molecular Docking, Molecular Dynamic Simulations, ADMET, and POM Analyses for the Identification of Pharmacophore Sites. PLoS ONE 2022, 17, e0273256. [Google Scholar] [CrossRef]
- Veerasamy, R.; Karunakaran, R. Molecular Docking Unveils the Potential of Andrographolide Derivatives against COVID-19: An in Silico Approach. J. Genet. Eng. Biotechnol. 2022, 20, 58. [Google Scholar] [CrossRef]
- Mohammed, A.O.; Abo-Idrees, M.I.; Makki, A.A.; Ibraheem, W.; Alzain, A.A. Drug Repurposing against Main Protease and RNA-Dependent RNA Polymerase of SARS-CoV-2 Using Molecular Docking, MM-GBSA Calculations and Molecular Dynamics. Struct. Chem. 2022, 33, 1553–1567. [Google Scholar] [CrossRef] [PubMed]
- Ribaudo, G.; Ongaro, A.; Oselladore, E.; Zagotto, G.; Memo, M.; Gianoncelli, A. A Computational Approach to Drug Repurposing against SARS-CoV-2 RNA Dependent RNA Polymerase (RdRp). J. Biomol. Struct. Dyn. 2022, 40, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Baby, K.; Maity, S.; Mehta, C.H.; Suresh, A.; Nayak, U.Y.; Nayak, Y. Targeting SARS-CoV-2 RNA-Dependent RNA Polymerase: An in Silico Drug Repurposing for COVID-19. F1000Research 2020, 9, 1166. [Google Scholar] [CrossRef]
- Hosseini, M.; Chen, W.; Xiao, D.; Wang, C. Computational Molecular Docking and Virtual Screening Revealed Promising SARS-CoV-2 Drugs. Precis. Clin. Med. 2021, 4, 1–16. [Google Scholar] [CrossRef]
- El Hassab, M.A.; Hemeda, L.R.; Elsayed, Z.M.; Al-Rashood, S.T.; Abdel-Hamid Amin, M.K.; Abdel-Aziz, H.A.; Eldehna, W.M. Computational Prediction of the Potential Target of SARS-CoV-2 Inhibitor Plitidepsin via Molecular Docking, Dynamic Simulations and MM-PBSA Calculations. Chem. Biodivers. 2022, 19, e202100719. [Google Scholar] [CrossRef]
- Vesga, L.C.; Ruiz-Hernández, C.A.; Alvarez-Jacome, J.J.; Duque, J.E.; Rincon-Orozco, B.; Mendez-Sanchez, S.C. Repurposing of Four Drugs as Anti-SARS-CoV-2 Agents and Their Interactions with Protein Targets. Sci. Pharm. 2022, 90, 24. [Google Scholar] [CrossRef]
- Elfiky, A.A. Dual Targeting of RdRps of SARS-CoV-2 and the Mucormycosis-Causing Fungus: An in Silico Perspective. Future Microbiol. 2022, 17, 755–762. [Google Scholar] [CrossRef]
- Wang, B.; Svetlov, D.; Artsimovitch, I. NMPylation and De-NMPylation of SARS-CoV-2 Nsp9 by the NiRAN Domain. Nucleic Acids Res. 2021, 49, 8822–8835. [Google Scholar] [CrossRef] [PubMed]
- McClung, M.; Harris, S.T.; Miller, P.D.; Bauer, D.C.; Davison, K.S.; Dian, L.; Hanley, D.A.; Kendler, D.L.; Yuen, C.K.; Lewiecki, E.M. Bisphosphonate Therapy for Osteoporosis: Benefits, Risks, and Drug Holiday. Am. J. Med. 2013, 126, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Shen, Q.; Guo, W.; He, W.; Li, J.; Zhang, Y.; Wang, Y.; Zhou, Z.; Deng, D.; Ouyang, X.; et al. Clinical Characteristics of Elderly Patients with COVID-19 in Hunan Province, China: A Multicenter, Retrospective Study. Gerontology 2020, 66, 467–475. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, M.; Wang, Y.; Zheng, F.; Huang, Y.; Huang, K.; Yu, Q.; Cai, C.; Chen, D.; Tian, Y.; et al. Clinical Characteristics of Older and Younger Patients Infected with SARS-CoV-2. Aging (Albany. NY). 2020, 12, 11296–11305. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, V.; Iannacone, M. The Interplay of Drug Therapeutics and Immune Responses to SARS-CoV-2. Cell. Mol. Immunol. 2023, 21, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Drake, M.T.; Clarke, B.L.; Khosla, S. Bisphosphonates: Mechanism of Action and Role in Clinical Practice. Mayo Clin. Proc. 2008, 83, 1032–1045. [Google Scholar] [CrossRef]
- Kunzmann, V.; Bauer, E.; Feurle, J.; Tony Florian Weißinger, H.-P.; Wilhelm, M.; Florian Weißinger, H.-P.; Wilhelm, M. Stimulation of Γδ T Cells by Aminobisphosphonates and Induction of Antiplasma Cell Activity in Multiple Myeloma. Blood 2000, 96, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.; Dou, H.; Zhang, X.; Uhagaze, D.S.; Ding, X.; Dong, Y. Determination of PKa Values of Alendronate Sodium in Aqueous Solution by Piecewise Linear Regression Based on Acid-Base Potentiometric Titration. J. Pharm. Anal. 2016, 6, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Homeyer, N.; Gohlke, H. Free Energy Calculations by the Molecular Mechanics Poisson−Boltzmann Surface Area Method. Mol. Inform. 2012, 31, 114–122. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Liu, X.; Zhang, J.Z.H. Interaction Entropy: A New Paradigm for Highly Efficient and Reliable Computation of Protein-Ligand Binding Free Energy. J. Am. Chem. Soc. 2016, 138, 5722–5728. [Google Scholar] [CrossRef]
- MacRaild, C.A.; Daranas, A.H.; Bronowska, A.; Homans, S.W. Global Changes in Local Protein Dynamics Reduce the Entropic Cost of Carbohydrate Binding in the Arabinose-Binding Protein. J. Mol. Biol. 2007, 368, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, S.C.J.; Kebriaei, R.; Dresser, L.D. Remdesivir: Review of Pharmacology, Pre-clinical Data, and Emerging Clinical Experience for COVID-19. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2020, 40, 659–671. [Google Scholar] [CrossRef]
- Cervelli, M.J.; Russ, G.R. Principles of Drug Therapy, Dosing, and Prescribing in Chronic Kidney Disease and Renal Replacement Therapy. In Comprehensive Clinical Nephrology; Elsevier, 2010; pp. 871–893. ISBN 9780323077668. [Google Scholar]
- Davies, M.; Nowotka, M.; Papadatos, G.; Dedman, N.; Gaulton, A.; Atkinson, F.; Bellis, L.; Overington, J.P. ChEMBL Web Services: Streamlining Access to Drug Discovery Data and Utilities. Nucleic Acids Res. 2015, 43, W612–W620. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A Software Program for PK(a) Prediction and Protonation State Generation for Drug-like Molecules. J. Comput. Aided. Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Tanimoto, T.T. An Elementary Mathematical Theory of Classification and Prediction. In Proceedings of the Proc. IBM Internal Report; International Business Machines Corp; 1958; pp. 1–11. [Google Scholar]
- Koulgi, S.; Jani, V.; Uppuladinne, M.V.N.; Sonavane, U.; Joshi, R. Remdesivir-Bound and Ligand-Free Simulations Reveal the Probable Mechanism of Inhibiting the RNA Dependent RNA Polymerase of Severe Acute Respiratory Syndrome Coronavirus 2. RSC Adv. 2020, 10, 26792–26803. [Google Scholar] [CrossRef]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 2. Enrichment Factors in Database Screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M.; McCammon, J.A. Molecular Dynamics Simulations of Biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Nosé, S. A Unified Formulation of the Constant Temperature Molecular Dynamics Methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tuckerman, M.E.; Tobias, D.J.; Klein, M.L. Explicit Reversible Integrators for Extended Systems Dynamics. Mol. Phys. 1996, 87, 1117–1157. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Shirts, M.R.; Klein, C.; Swails, J.M.; Yin, J.; Gilson, M.K.; Mobley, D.L.; Case, D.A.; Zhong, E.D. Lessons Learned from Comparing Molecular Dynamics Engines on the SAMPL5 Dataset. J. Comput. Aided. Mol. Des. 2017, 31, 147–161. [Google Scholar] [CrossRef]
- Ekberg, V.; Ryde, U. On the Use of Interaction Entropy and Related Methods to Estimate Binding Entropies. J. Chem. Theory Comput. 2021, 17, 5379–5391. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Lee, P.; Ng, C.; Slattery, A.; Nair, P.; Eisman, J.A.; Center, J.R. Preadmission Bisphosphonate and Mortality in Critically Ill Patients. J. Clin. Endocrinol. Metab. 2016, 101, 1945–1953. [Google Scholar] [CrossRef] [PubMed]
- Sing, C.; Kiel, D.P.; Hubbard, R.B.; Lau, W.C.; Li, G.H.; Kung, A.W.; Wong, I.C.; Cheung, C. Nitrogen-Containing Bisphosphonates Are Associated With Reduced Risk of Pneumonia in Patients With Hip Fracture. J. Bone Miner. Res. 2020, 35, 1676–1684. [Google Scholar] [CrossRef] [PubMed]

| S. No | ChEMBL ID | Score* | S. No | ChEMBL ID | Score* | S. No | ChEMBL ID | Score* | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CHEMBL1213265
|
-10.235 -7.111 † |
2 | CHEMBL608526
|
-9.706 -8.235 † |
3 | CHEMBL319144
|
-9.657 -8.010 † |
|
| 4 | CHEMBL4802971
|
-9.355 -8.809 † |
5 | CHEMBL98211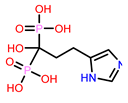
|
-9.347 -7.956 † |
6 | CHEMBL4291724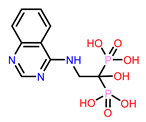
|
-9.308 -6.912 † |
|
| 7 | CHEMBL301247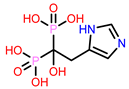
|
-9.219 -8.150 † |
8 | CHEMBL164344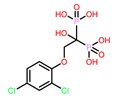
|
-9.213 -8.041 † |
9 | CHEMBL300361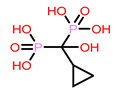
|
-9.151 -8.254 † |
|
| 10 | CHEMBL4289996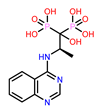
|
-9.119 -8.118 † |
11 | CHEMBL387132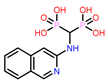
|
-9.11 -8.476 † |
12 | CHEMBL196676
|
-9.059 -7.558 † |
|
| 13 | CHEMBL4569308
|
-9.02 -8.208 † |
14 | CHEMBL338622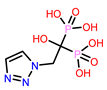
|
-9.014 -7.938 † |
15 | Cinnamaldehyde (Negative control) 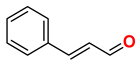
|
-1.707 | |
| 16 | Remdesivir (Reference drug) 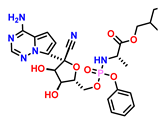
|
-3.270 |
17 | Favipiravir (Additional reference drug) 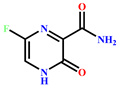
|
-3.443 | 18 | Molnupiravir (Additional reference drug) 
|
-4.927 | |
| S. no | Drugs | MM-GBSA dG Bind (kcal/mol) | |
|---|---|---|---|
| Uncharged state | at pH 7.0 ± 2.0 | ||
| 1. | CHEMBL1213265 | -7.74 | -33.25 |
| 2. | CHEMBL338622 | -24.14 | -26.52 |
| 3. | CHEMBL301247 | -24.22 | -27.73 |
| 4. | CHEMBL4289996 | -24.81 | -26.97 |
| 5. | CHEMBL98211 | -26.04 | -40.50† |
| 6. | CHEMBL300361 | -26.68 | -23.47 |
| 7. | CHEMBL608526 | -33.42 | -40.88† |
| 8. | CHEMBL319144 | -35.2 | -26.39 |
| 9. | CHEMBL4802971 | -36.77 | -39.13 |
| 10. | CHEMBL4569308 | -40.94† | -43.06† |
| 11. | CHEMBL4291724 | -41.51† | -34.06 |
| 12. | CHEMBL387132 | -43.28† | -38.34 |
| 13. | CHEMBL196676 | -44.14† | -37.10 |
| 14. | CHEMBL164344 | -46.65† | -46.73 |
| 15. | Favipiravir | -19.13 | -- |
| 16. | Molnupiravir | -34.13 | -- |
| 17. | Remdesivir | -40.32 | -- |
| 18. | Cinnamaldehyde | -30.05 | -- |
| S. No | ChEMBL ID | S. No | ChEMBL ID | S. No | ChEMBL ID | |
|---|---|---|---|---|---|---|
| 1 | CHEMBL4291724 Score: -9.308 kcal/mol 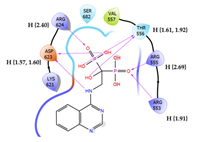
|
5 | CHEMBL608526 † Score: -8.235 kcal/mol 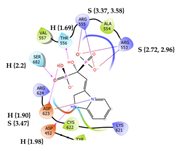
|
9 | Favipiravir Score: -3.443 kcal/mol 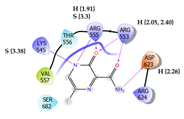
|
|
| 2 | CHEMBL164344 Score: -9.213 kcal/mol 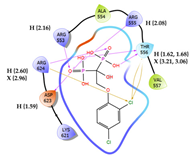
|
6. | CHEMBL98211† Score: -7.956 kcal/mol 
|
10 | Molnupiravir Score = -4.927 kcal/mol 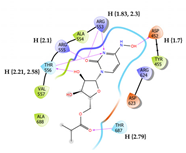
|
|
| 3 | CHEMBL387132 Score: -9.110 kcal/mol 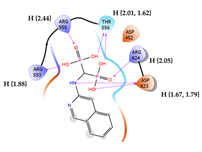
|
7 | CHEMBL4569308 † Score: -8.208 kcal/mol 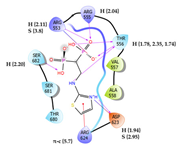
|
11 |
Cinnamaldehyde Score: -1.707 kcal/mol 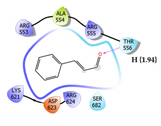
|
|
| 4 | CHEMBL196676 Score: -9.059 kcal/mol 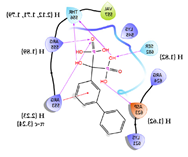
|
8 | Remdesivir Score: -3.270 kcal/mol 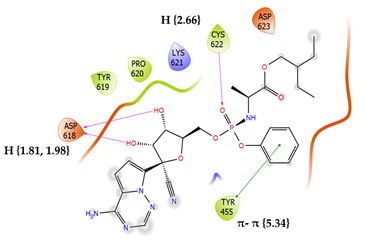
|
|||
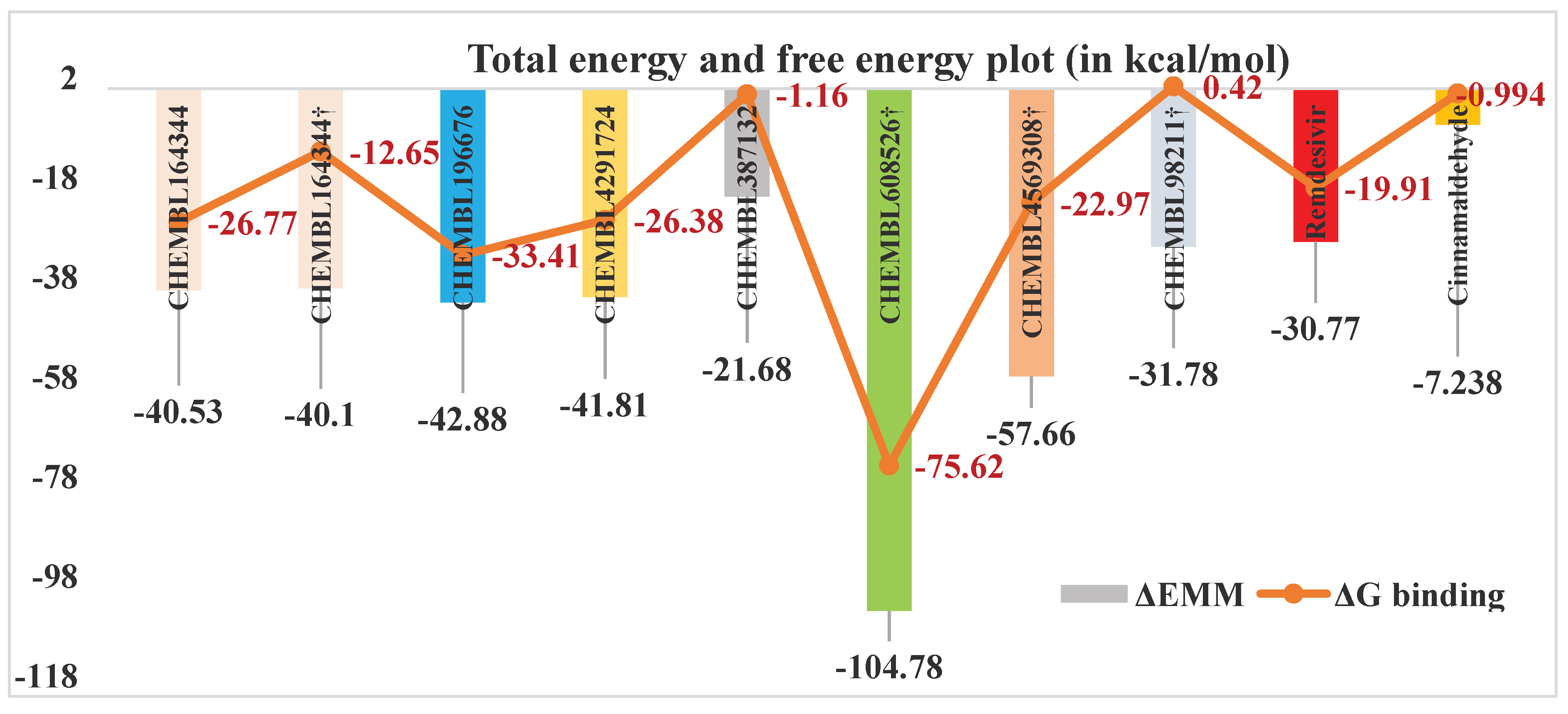
| Compounds | I.E. = -TΔS | Total energy contributions (ΔEMM) | ΔGbinding | ||||
|---|---|---|---|---|---|---|---|
| ΔEvdW | ΔEEL | ΔEPB | ΔENP | ΔEMM = ∑ ΔE | |||
| CHEMBL196676 | 9.48 | -13.06 | -90.55 | 63.70 | -2.98 | -42.88 | -33.41 |
| CHEMBL164344 | 13.76 | -22.77 | -93.71 | 79.14 | -3.19 | -40.53 | -26.77 |
| CHEMBL4291724 | 15.43 | -13.47 | -123.47 | 97.86 | -2.73 | -41.81 | -26.38 |
| CHEMBL387132 | 20.52 | -20.15 | -87.58 | 88.93 | -2.88 | -21.68 | -1.16 |
| CHEMBL608526† | 29.16 | 6.03 | -620.27 | 512.53 | -3.13 | -104.78 | -75.62 |
| CHEMBL4569308† | 34.69 | -3.58 | -373.84 | 322.48 | -2.71 | -57.66 | -22.97 |
| CHEMBL164344† | 27.45 | -11.38 | -133.86 | 107.72 | -2.57 | -40.10 | -12.65 |
| CHEMBL98211† | 32.20 | 1.81 | -253.57 | 222.44 | -2.46 | -31.78 | 0.42 |
| Remdesivir | 10.86 | -52.27 | -64.85 | 92.31 | -5.95 | -30.77 | -19.91 |
| Cinnamaldehyde | 6.24 | -11.16 | -3.24 | 8.20 | -1.03 | -7.24 | -0.99 |
| ADMET parameters | Hit molecules | |||||||||||||
| CHEMBL → | 4291724 | 164344 | 387132 | 196676 | 98211 | 608526 | 4569308 | Remdesivir | ||||||
| Absorption | WS (log mol/L) | -2.47 | -2.152 | -2.046 | -3.677 | -2.896 | -1.444 | -1.641 | -3.07 | |||||
| CP (log Papp in 10-6 cm/s) | -0.438 | -0.461 | 0.093 | 1.245 | -0.295 | 0.334 | -0.527 | 0.635 | ||||||
| IA (% Absorbed) | 42.008 | 16.774 | 71.627 | 38.34 | 24.799 | 34.854 | 43.351 | 71.109 | ||||||
| S.P. (log Kp) | -2.735 | -2.743 | -2.759 | -2.735 | -2.735 | -2.736 | -2.882 | -2.735 | ||||||
| P-glyco protein | Substrate | No | No | No | Yes | Yes | No | No | Yes | |||||
| Inhibitor | I | No | No | No | Yes | No | No | No | Yes | |||||
| II | No | No | No | No | No | No | No | No | ||||||
| Distribution | V.D. ss (log L/kg) | -0.768 | -0.558 | -0.814 | 0.578 | -0.84 | -0.439 | -0.641 | 0.307 | |||||
| F.U. (Fu) | 0.331 | 0.469 | 0.516 | 0.028 | 0.655 | 0.811 | 0.6 | 0.005 | ||||||
| BBB (log BB) | -2.302 | -2.284 | -1.86 | -1.908 | -2.489 | -1.522 | -2.541 | -2.056 | ||||||
| CNS (log P.S.) | -4.756 | -3.881 | -4.034 | -3.879 | -6.045 | -4.881 | -4.528 | -4.675 | ||||||
| Metabolism | CYP action | Substrate | 2D6 | No | No | No | No | No | No | No | No | |||
| 3A4 | No | No | No | No | No | No | No | Yes | ||||||
| Inhibition against 1A2, 2C19, 2C9, 2D6, 3A4 | No | No | No | No | No | No | No | No | ||||||
| Excretion | T.C. (log ml/min/kg) | 0.146 | 0.032 | -0.025 | -0.119 | 0.664 | 0.374 | 0.143 | 0.198 | |||||
| ROC | No | No | No | No | No | No | No | No | ||||||
| Toxicity | Ames assay | No | No | No | Yes | No | No | No | No | |||||
| MTD (log mg/kg/day) | 0.841 | 0.445 | 0.689 | 0.574 | -0.312 | 0.444 | 1.142 | 0.15 | ||||||
| hERG I inhibitor | No | No | No | No | No | No | No | No | ||||||
| hERG II inhibitor | No | No | No | Yes | No | No | No | Yes | ||||||
| Rat oral toxicity | Acute (LD50) (mol/kg) | 2.67 | 2.613 | 1.886 | 3.117 | 2.564 | 2.374 | 2.738 | 2.043 | |||||
| Chronic (LOAEL) (Log mg/kg_bw/day) |
3.195 | 3.597 | 3.052 | 3.404 | 4.773 | 3.253 | 3.826 | 1.639 | ||||||
| HT | Yes | No | Yes | No | No | Yes | No | Yes | ||||||
| SS | No | No | No | No | No | No | No | No | ||||||
| TT (log ug/L) | 0.285 | 0.285 | 0.288 | 0.285 | 0.285 | 0.285 | 0.285 | 0.285 | ||||||
| MT (log mM) | 2.992 | 1.686 | 2.938 | 0.427 | 2.709 | 2.355 | 2.875 | 0.291 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





