Submitted:
24 November 2023
Posted:
27 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Seed selection and accelerated aging treatment
2.2. Acquisition of hyperspectral images of samples
2.3. Standard germination test
2.4. Preprocessing of Vis/NIR reflection spectra
2.5. Architecture of CNN for classification of HSI images
2.5.1. AlexNet architecture
2.5.2. VGG architecture
2.6. Machine learning techniques for analyzing reflection spectra
2.6.1. Principal component analysis (PCA)
2.6.2. Linear discriminant analysis (LDA)
2.6.3. Support vector machine (SVM)
2.7. Evaluation criteria of models for peanut seed classification
3. Results and Discussion
3.1. Indices of seed viability
3.2. Modeling of seed viability classification based on hyperspectral images using CNN
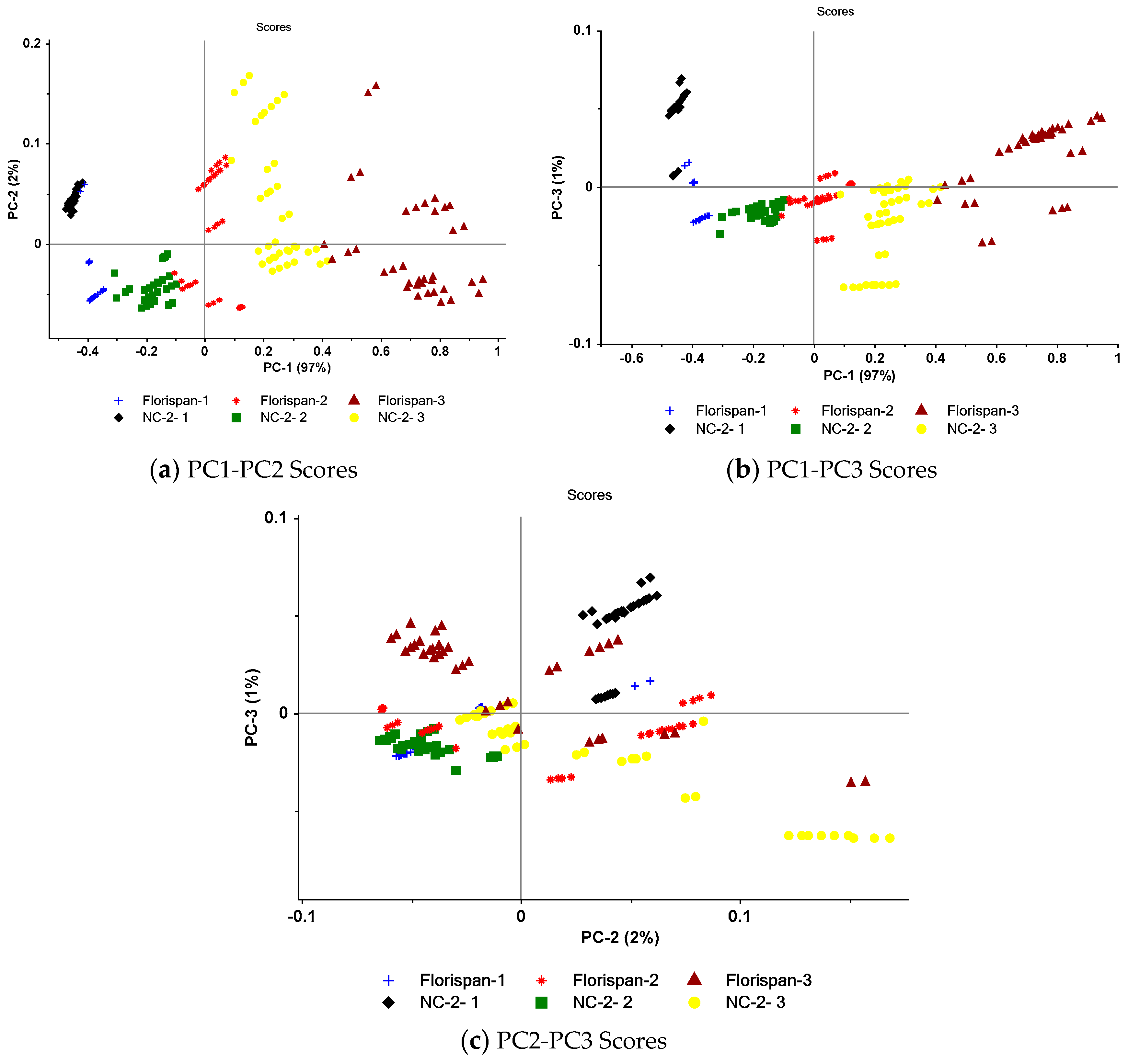
3.3. PC results
3.4. Modeling the seed viability classification based on reflection spectra using LDA and SVM
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, H.; Ni, J.; Gao, J.; Han, Z.; Luan, T. A novel method for peanut variety identification and classification by Improved VGG16. Scientific Reports 2021, 11, 15756. [CrossRef]
- Zhao, X.; Pang, L.; Wang, L.; Men, S.; Yan, L. Deep Convolutional Neural Network for Detection and Prediction of Waxy Corn Seed Viability Using Hyperspectral Reflectance Imaging. Mathematical and Computational Applications 2022, 27, 109. [CrossRef]
- Kandpal, L.M.; Lohumi, S.; Kim, M.S.; Kang, J.-S.; Cho, B.-K. Near-infrared hyperspectral imaging system coupled with multivariate methods to predict viability and vigor in muskmelon seeds. Sensors and Actuators B: Chemical 2016, 229, 534-544. [CrossRef]
- Pang, L.; Wang, L.; Yuan, P.; Yan, L.; Yang, Q.; Xiao, J. Feasibility study on identifying seed viability of Sophora japonica with optimized deep neural network and hyperspectral imaging. Computers and Electronics in Agriculture 2021, 190, 106426. [CrossRef]
- Kusumaningrum, D.; Lee, H.; Lohumi, S.; Mo, C.; Kim, M.S.; Cho, B.K. Non-destructive technique for determining the viability of soybean (Glycine max) seeds using FT-NIR spectroscopy. Journal of the Science of Food and Agriculture 2018, 98, 1734-1742. [CrossRef]
- Shrestha, S.; Knapič, M.; Žibrat, U.; Deleuran, L.C.; Gislum, R. Single seed near-infrared hyperspectral imaging in determining tomato (Solanum lycopersicum L.) seed quality in association with multivariate data analysis. Sensors and Actuators B: Chemical 2016, 237, 1027-1034. [CrossRef]
- Wang, X.; Zhang, H.; Song, R.; He, X.; Mao, P.; Jia, S. Non-destructive identification of naturally aged alfalfa seeds via multispectral imaging analysis. Sensors 2021, 21, 5804. [CrossRef]
- Marcos Filho, J. Seed vigor testing: an overview of the past, present and future perspective. Scientia agricola 2015, 72, 363-374. [CrossRef]
- Song, P.; Kim, G.; Song, P.; Yang, T.; Yue, X.; Gu, Y. Rapid and non-destructive detection method for water status and water distribution of rice seeds with different vigor. International Journal of Agricultural and Biological Engineering 2021, 14, 231-238. [CrossRef]
- ElMasry, G.; Mandour, N.; Al-Rejaie, S.; Belin, E.; Rousseau, D. Recent applications of multispectral imaging in seed phenotyping and quality monitoring—An overview. Sensors 2019, 19, 1090. [CrossRef]
- de Medeiros, A.D.; Bernardes, R.C.; da Silva, L.J.; de Freitas, B.A.L.; dos Santos Dias, D.C.F.; da Silva, C.B. Deep learning-based approach using X-ray images for classifying Crambe abyssinica seed quality. Industrial Crops and Products 2021, 164, 113378. [CrossRef]
- de Medeiros, A.D.; Pinheiro, D.T.; Xavier, W.A.; da Silva, L.J.; dos Santos Dias, D.C.F. Quality classification of Jatropha curcas seeds using radiographic images and machine learning. Industrial Crops and Products 2020, 146, 112162. [CrossRef]
- Li, L.; Chen, S.; Deng, M.; Gao, Z. Optical techniques in non-destructive detection of wheat quality: A review. Grain & Oil Science and Technology 2022, 5, 44-57. [CrossRef]
- Jia, B.; Wang, W.; Ni, X.; Lawrence, K.C.; Zhuang, H.; Yoon, S.-C.; Gao, Z. Essential processing methods of hyperspectral images of agricultural and food products. Chemometrics and Intelligent Laboratory Systems 2020, 198, 103936. [CrossRef]
- Saha, D.; Manickavasagan, A. Machine learning techniques for analysis of hyperspectral images to determine quality of food products: A review. Current Research in Food Science 2021, 4, 28-44. [CrossRef]
- Yang, Y.; Zhang, X.; Yin, J.; Yu, X. Rapid and nondestructive on-site classification method for consumer-grade plastics based on portable NIR spectrometer and machine learning. Journal of Spectroscopy 2020, 2020, 1-8. [CrossRef]
- Vega Diaz, J.J.; Sandoval Aldana, A.P.; Reina Zuluaga, D.V. Prediction of dry matter content of recently harvested ‘Hass’ avocado fruits using hyperspectral imaging. Journal of the Science of Food and Agriculture 2021, 101, 897-906. [CrossRef]
- Ravikanth, L.; Singh, C.B.; Jayas, D.S.; White, N.D. Classification of contaminants from wheat using near-infrared hyperspectral imaging. Biosystems Engineering 2015, 135, 73-86. [CrossRef]
- Zhou, S.; Sun, L.; Xing, W.; Feng, G.; Ji, Y.; Yang, J.; Liu, S. Hyperspectral imaging of beet seed germination prediction. Infrared Physics & Technology 2020, 108, 103363. [CrossRef]
- Caporaso, N.; Whitworth, M.B.; Fisk, I.D. Near-Infrared spectroscopy and hyperspectral imaging for non-destructive quality assessment of cereal grains. Applied spectroscopy reviews 2018, 53, 667-687. [CrossRef]
- Yu, Z.; Fang, H.; Zhangjin, Q.; Mi, C.; Feng, X.; He, Y. Hyperspectral imaging technology combined with deep learning for hybrid okra seed identification. Biosystems Engineering 2021, 212, 46-61. [CrossRef]
- Han, S.S.; Kim, M.S.; Lim, W.; Park, G.H.; Park, I.; Chang, S.E. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. Journal of Investigative Dermatology 2018, 138, 1529-1538. [CrossRef]
- Huang, Q.; Li, W.; Zhang, B.; Li, Q.; Tao, R.; Lovell, N.H. Blood cell classification based on hyperspectral imaging with modulated Gabor and CNN. IEEE journal of biomedical and health informatics 2019, 24, 160-170. [CrossRef]
- Zhao, W.; Du, S. Spectral–spatial feature extraction for hyperspectral image classification: A dimension reduction and deep learning approach. IEEE Transactions on Geoscience and Remote Sensing 2016, 54, 4544-4554. [CrossRef]
- Sony, S.; Dunphy, K.; Sadhu, A.; Capretz, M. A systematic review of convolutional neural network-based structural condition assessment techniques. Engineering Structures 2021, 226, 111347. [CrossRef]
- Chen, L.; Li, S.; Bai, Q.; Yang, J.; Jiang, S.; Miao, Y. Review of image classification algorithms based on convolutional neural networks. Remote Sensing 2021, 13, 4712. [CrossRef]
- Ghosh, A.; Sufian, A.; Sultana, F.; Chakrabarti, A.; De, D. Fundamental concepts of convolutional neural network. Recent trends and advances in artificial intelligence and Internet of Things 2020, 519-567. [CrossRef]
- Kabir, M.H.; Guindo, M.L.; Chen, R.; Liu, F.; Luo, X.; Kong, W. Deep Learning Combined with Hyperspectral Imaging Technology for Variety Discrimination of Fritillaria thunbergii. Molecules 2022, 27, 6042. [CrossRef]
- Yang, Y.; Liao, J.; Li, H.; Tan, K.; Zhang, X. Identification of high-oil content soybean using hyperspectral reflectance and one-dimensional convolutional neural network. Spectroscopy Letters 2023, 56, 28-41. [CrossRef]
- Ma, X.; Li, Y.; Wan, L.; Xu, Z.; Song, J.; Huang, J. Classification of seed corn ears based on custom lightweight convolutional neural network and improved training strategies. Engineering Applications of Artificial Intelligence 2023, 120, 105936. [CrossRef]
- Wang, Y.; Song, S. Variety identification of sweet maize seeds based on hyperspectral imaging combined with deep learning. Infrared Physics & Technology 2023, 130, 104611. [CrossRef]
- Boulent, J.; Foucher, S.; Théau, J.; St-Charles, P.-L. Convolutional neural networks for the automatic identification of plant diseases. Frontiers in plant science 2019, 10, 941. [CrossRef]
- De Vitis, M.; Hay, F.R.; Dickie, J.B.; Trivedi, C.; Choi, J.; Fiegener, R. Seed storage: maintaining seed viability and vigor for restoration use. Restoration Ecology 2020, 28, S249-S255. [CrossRef]
- Zou, Z.; Chen, J.; Zhou, M.; Zhao, Y.; Long, T.; Wu, Q.; Xu, L. Prediction of peanut seed vigor based on hyperspectral images. Food Science and Technology 2022, 42, e32822. [CrossRef]
- Hampton, J.G.; TeKRONY, D.M. Handbook of vigour test methods; The International Seed Testing Association, Zurich (Switzerland). 1995.
- Moghaddam, M.; Moradi, A.; Salehi, A.; Rezaei, R. The effect of various biological treatments on germination and some seedling indices of fennel (Foeniculum vulgare L.) under drought stress. Iranian Journal of Seed Science and Technology 2018, 7.
- Panwar, P.; Bhardwaj, S. Handbook of practical forestry; Agrobios (India): 2005.
- Mohajeri, F.; Taghvaei, M.; Ramrudi, M.; Galavi, M. Effect of priming duration and concentration on germination behaviors of (Phaseolus vulgaris L.) seeds. Int. J. Ecol. Environ. Conserv 2016, 22, 603-609.
- Maguire, J.D. Speed of germination-aid in selection and evaluation for seedling emergence and vigor. Crop Sci. 1962, 2, 176-177. [CrossRef]
- Hunter, E.; Glasbey, C.; Naylor, R. The analysis of data from germination tests. The Journal of Agricultural Science 1984, 102, 207-213. [CrossRef]
- Jiao, Y.; Li, Z.; Chen, X.; Fei, S. Preprocessing methods for near-infrared spectrum calibration. Journal of Chemometrics 2020, 34, e3306. [CrossRef]
- Xu, Y.; Zhong, P.; Jiang, A.; Shen, X.; Li, X.; Xu, Z.; Shen, Y.; Sun, Y.; Lei, H. Raman spectroscopy coupled with chemometrics for food authentication: A review. TrAC Trends in Analytical Chemistry 2020, 131, 116017. [CrossRef]
- Roger, J.-M.; Mallet, A.; Marini, F. preprocessing NIR spectra for aquaphotomics. Molecules 2022, 27, 6795. [CrossRef]
- Suhandy, D.; Yulia, M. The use of UV spectroscopy and SIMCA for the authentication of Indonesian honeys according to botanical, entomological and geographical origins. Molecules 2021, 26, 915. [CrossRef]
- Cozzolino, D.; Williams, P.; Hoffman, L. An overview of pre-processing methods available for hyperspectral imaging applications. Microchemical Journal 2023, 109129. [CrossRef]
- Saha, D.; Senthilkumar, T.; Sharma, S.; Singh, C.B.; Manickavasagan, A. Application of near-infrared hyperspectral imaging coupled with chemometrics for rapid and non-destructive prediction of protein content in single chickpea seed. Journal of Food Composition and Analysis 2023, 115, 104938. [CrossRef]
- Ajit, A.; Acharya, K.; Samanta, A. A Review of Convolutional Neural Networks. In Proceedings of 2020 International Conference on Emerging Trends in Information Technology and Engineering (ic-ETITE), Vellore, India; pp. 1-9. [CrossRef]
- Lv, M.; Zhou, G.; He, M.; Chen, A.; Zhang, W.; Hu, Y. Maize leaf disease identification based on feature enhancement and DMS-robust alexnet. IEEE access 2020, 8, 57952-57966. [CrossRef]
- Tan, Y.; Jing, X. Cooperative spectrum sensing based on convolutional neural networks. Applied Sciences 2021, 11, 4440. [CrossRef]
- Sakib, S.; Ahmed, N.; Kabir, A.J.; Ahmed, H. An overview of convolutional neural network: Its architecture and applications. 2019. [CrossRef]
- Li, Z.; Liu, F.; Yang, W.; Peng, S.; Zhou, J. A survey of convolutional neural networks: analysis, applications, and prospects. IEEE transactions on neural networks and learning systems 2021. [CrossRef]
- Naranjo-Torres, J.; Mora, M.; Hernández-García, R.; Barrientos, R.J.; Fredes, C.; Valenzuela, A. A review of convolutional neural network applied to fruit image processing. Applied Sciences 2020, 10, 3443. [CrossRef]
- Gebrehiwot, A.; Hashemi-Beni, L.; Thompson, G.; Kordjamshidi, P.; Langan, T.E. Deep convolutional neural network for flood extent mapping using unmanned aerial vehicles data. Sensors 2019, 19, 1486. [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 2014. [CrossRef]
- Yu, W.; Yang, K.; Bai, Y.; Xiao, T.; Yao, H.; Rui, Y. Visualizing and comparing AlexNet and VGG using deconvolutional layers. In Proceedings of Proceedings of the 33 rd International Conference on Machine Learning.
- Alsaadi, Z.; Alshamani, E.; Alrehaili, M.; Alrashdi, A.A.D.; Albelwi, S.; Elfaki, A.O. A real time Arabic sign language alphabets (ArSLA) recognition model using deep learning architecture. Computers 2022, 11, 78. [CrossRef]
- Khalifa, N.E.M.; Taha, M.H.N.; Hassanien, A.E. Aquarium family fish species identification system using deep neural networks. In Proceedings of International Conference on Advanced Intelligent Systems and Informatics; pp. 347-356. [CrossRef]
- Lang, S.; Bravo-Marquez, F.; Beckham, C.; Hall, M.; Frank, E. Wekadeeplearning4j: A deep learning package for weka based on deeplearning4j. Knowledge-Based Systems 2019, 178, 48-50. [CrossRef]
- Neo, E.R.K.; Yeo, Z.; Low, J.S.C.; Goodship, V.; Debattista, K. A review on chemometric techniques with infrared, Raman and laser-induced breakdown spectroscopy for sorting plastic waste in the recycling industry. Resources, Conservation and Recycling 2022, 180, 106217. [CrossRef]
- Park, H.; Son, J.-H. Machine learning techniques for THz imaging and time-domain spectroscopy. Sensors 2021, 21, 1186. [CrossRef]
- Peris-Díaz, M.D.; Krężel, A. A guide to good practice in chemometric methods for vibrational spectroscopy, electrochemistry, and hyphenated mass spectrometry. TrAC Trends in Analytical Chemistry 2021, 135, 116157. [CrossRef]
- Raschka, S.; Mirjalili, V. Python machine learning Packt Publishing Ltd. 2015.
- Brereton, R.G.; Lloyd, G.R. Partial least squares discriminant analysis: taking the magic away. Journal of Chemometrics 2014, 28, 213-225. [CrossRef]
- Qin, J.; Vasefi, F.; Hellberg, R.S.; Akhbardeh, A.; Isaacs, R.B.; Yilmaz, A.G.; Hwang, C.; Baek, I.; Schmidt, W.F.; Kim, M.S. Detection of fish fillet substitution and mislabeling using multimode hyperspectral imaging techniques. Food Control 2020, 114, 107234. [CrossRef]
- Delwiche, S.R.; Rodriguez, I.T.; Rausch, S.; Graybosch, R. Estimating percentages of fusarium-damaged kernels in hard wheat by near-infrared hyperspectral imaging. Journal of Cereal Science 2019, 87, 18-24. [CrossRef]
- Meza Ramirez, C.A.; Greenop, M.; Ashton, L.; Rehman, I.U. Applications of machine learning in spectroscopy. Applied Spectroscopy Reviews 2021, 56, 733-763. [CrossRef]
- Zhang, D.; Zhang, H.; Zhao, Y.; Chen, Y.; Ke, C.; Xu, T.; He, Y. A brief review of new data analysis methods of laser-induced breakdown spectroscopy: machine learning. Applied Spectroscopy Reviews 2022, 57, 89-111. [CrossRef]
- Bonah, E.; Huang, X.; Yi, R.; Aheto, J.H.; Yu, S. Vis-NIR hyperspectral imaging for the classification of bacterial foodborne pathogens based on pixel-wise analysis and a novel CARS-PSO-SVM model. Infrared Physics & Technology 2020, 105, 103220. [CrossRef]
- Pan, T.-t.; Chyngyz, E.; Sun, D.-W.; Paliwal, J.; Pu, H. Pathogenetic process monitoring and early detection of pear black spot disease caused by Alternaria alternata using hyperspectral imaging. Postharvest Biology and Technology 2019, 154, 96-104. [CrossRef]
- Liu, Y.; Nie, F.; Gao, Q.; Gao, X.; Han, J.; Shao, L. Flexible unsupervised feature extraction for image classification. Neural Networks 2019, 115, 65-71. [CrossRef]
- Chu, X.; Wang, W.; Ni, X.; Li, C.; Li, Y. Classifying maize kernels naturally infected by fungi using near-infrared hyperspectral imaging. Infrared Physics & Technology 2020, 105, 103242. [CrossRef]
- Dhakate, P.P.; Patil, S.; Rajeswari, K.; Abin, D. Preprocessing and Classification in WEKA using different classifiers. Inter J Eng Res Appl 2014, 4, 91-93.
- Muhammad, U.; Wang, W.; Chattha, S.P.; Ali, S. Pre-trained VGGNet architecture for remote-sensing image scene classification. In Proceedings of 2018 24th International Conference on Pattern Recognition (ICPR); pp. 1622-1627. [CrossRef]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Plant disease detection and classification by deep learning. Plants 2019, 8, 468. [CrossRef]
- Sharma, S.; Guleria, K. Deep learning models for image classification: comparison and applications. In Proceedings of 2022 2nd International Conference on Advance Computing and Innovative Technologies in Engineering (ICACITE); pp. 1733-1738. [CrossRef]
- Ullah, A.; Anwar, S.M.; Bilal, M.; Mehmood, R.M. Classification of arrhythmia by using deep learning with 2-D ECG spectral image representation. Remote Sensing 2020, 12, 1685. [CrossRef]
- Kolla, M.; Venugopal, T. Efficient classification of diabetic retinopathy using binary cnn. In Proceedings of 2021 International conference on computational intelligence and knowledge economy (ICCIKE); pp. 244-247. [CrossRef]
- Garillos-Manliguez, C.A.; Chiang, J.Y. Multimodal deep learning and visible-light and hyperspectral imaging for fruit maturity estimation. Sensors 2021, 21, 1288. [CrossRef]
- Bu, Y.; Jiang, X.; Tian, J.; Hu, X.; Han, L.; Huang, D.; Luo, H. Rapid nondestructive detecting of sorghum varieties based on hyperspectral imaging and convolutional neural network. Journal of the Science of Food and Agriculture 2023, 103, 3970-3983. [CrossRef]
- Yang, J.; Sun, L.; Xing, W.; Feng, G.; Bai, H.; Wang, J. Hyperspectral prediction of sugarbeet seed germination based on gauss kernel SVM. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 2021, 253, 119585. [CrossRef]
- Hubert, M.; Reynkens, T.; Schmitt, E.; Verdonck, T. Sparse PCA for high-dimensional data with outliers. Technometrics 2016, 58, 424-434. [CrossRef]
- Mejia, A.F.; Nebel, M.B.; Eloyan, A.; Caffo, B.; Lindquist, M.A. PCA leverage: outlier detection for high-dimensional functional magnetic resonance imaging data. Biostatistics 2017, 18, 521-536. [CrossRef]
- Rafajłowicz, E.; Steland, A. The Hotelling—Like T^ 2 T 2 Control Chart Modified for Detecting Changes in Images having the Matrix Normal Distribution. In Proceedings of Stochastic Models, Statistics and Their Applications: Dresden, Germany, March 2019 14; pp. 193-206. [CrossRef]
- Camacho-Tamayo, J.H.; Forero-Cabrera, N.M.; Ramírez-López, L.; Rubiano, Y. Near-infrared spectroscopic assessment of soil texture in an oxisol of the eastern plains of Colombia. Colombia Forestal 2017, 20, 5-18.
- Chen, X.; Zhang, B.; Wang, T.; Bonni, A.; Zhao, G. Robust principal component analysis for accurate outlier sample detection in RNA-Seq data. Bmc Bioinformatics 2020, 21, 1-20. [CrossRef]
- Lörchner, C.; Horn, M.; Berger, F.; Fauhl-Hassek, C.; Glomb, M.A.; Esslinger, S. Quality control of spectroscopic data in non-targeted analysis–Development of a multivariate control chart. Food Control 2022, 133, 108601. [CrossRef]
- Pang, L.; Men, S.; Yan, L.; Xiao, J. Rapid vitality estimation and prediction of corn seeds based on spectra and images using deep learning and hyperspectral imaging techniques. Ieee Access 2020, 8, 123026-123036. [CrossRef]
- Lee, H.; Kim, M.S.; Lim, H.-S.; Park, E.; Lee, W.-H.; Cho, B.-K. Detection of cucumber green mottle mosaic virus-infected watermelon seeds using a near-infrared (NIR) hyperspectral imaging system: Application to seeds of the “Sambok Honey” cultivar. Biosystems Engineering 2016, 148, 138-147. [CrossRef]
- Ma, T.; Tsuchikawa, S.; Inagaki, T. Rapid and non-destructive seed viability prediction using near-infrared hyperspectral imaging coupled with a deep learning approach. Computers and Electronics in Agriculture 2020, 177, 105683. [CrossRef]
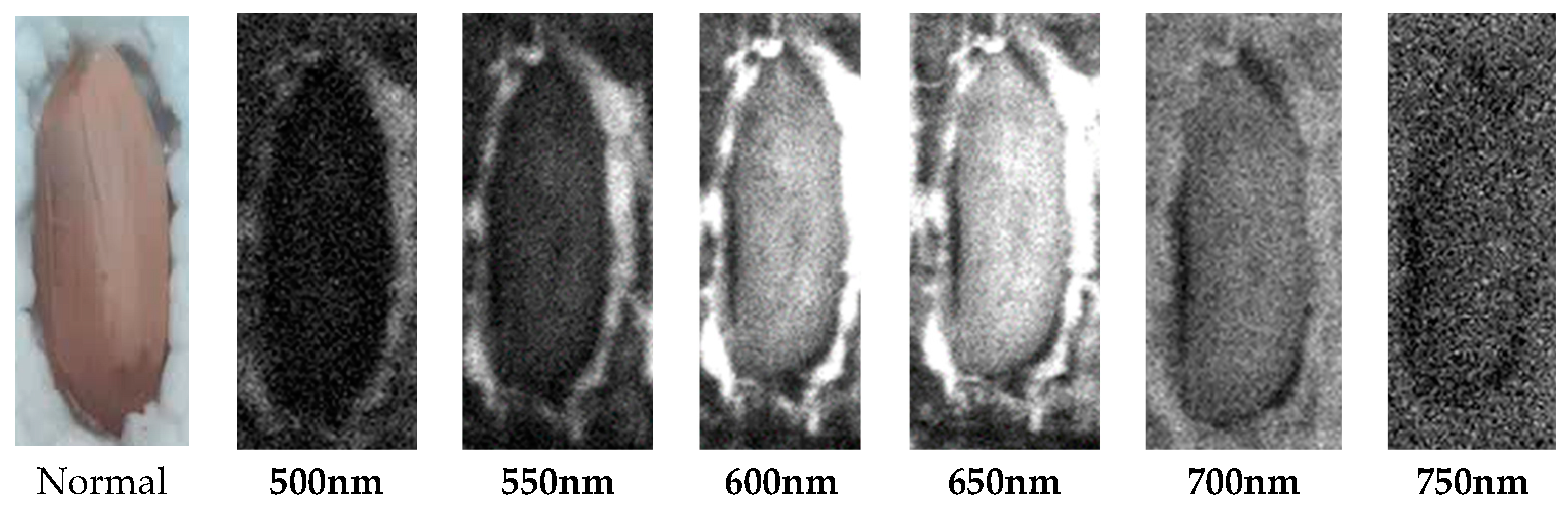
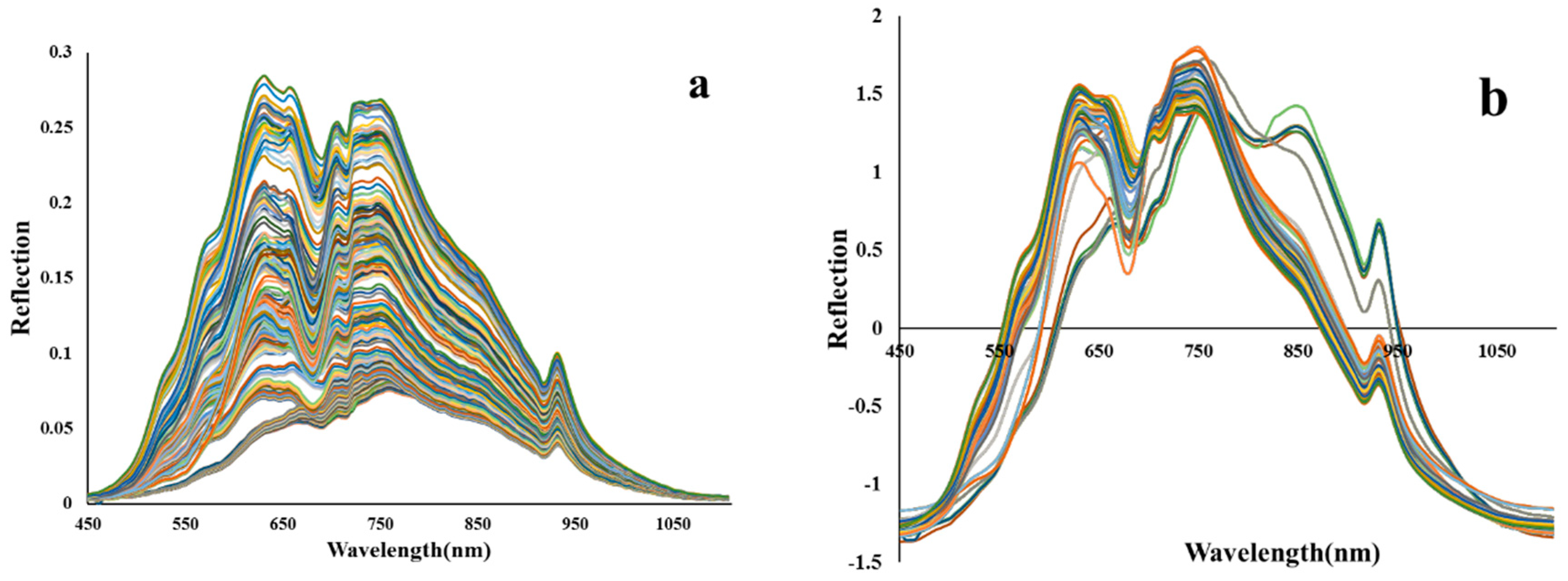
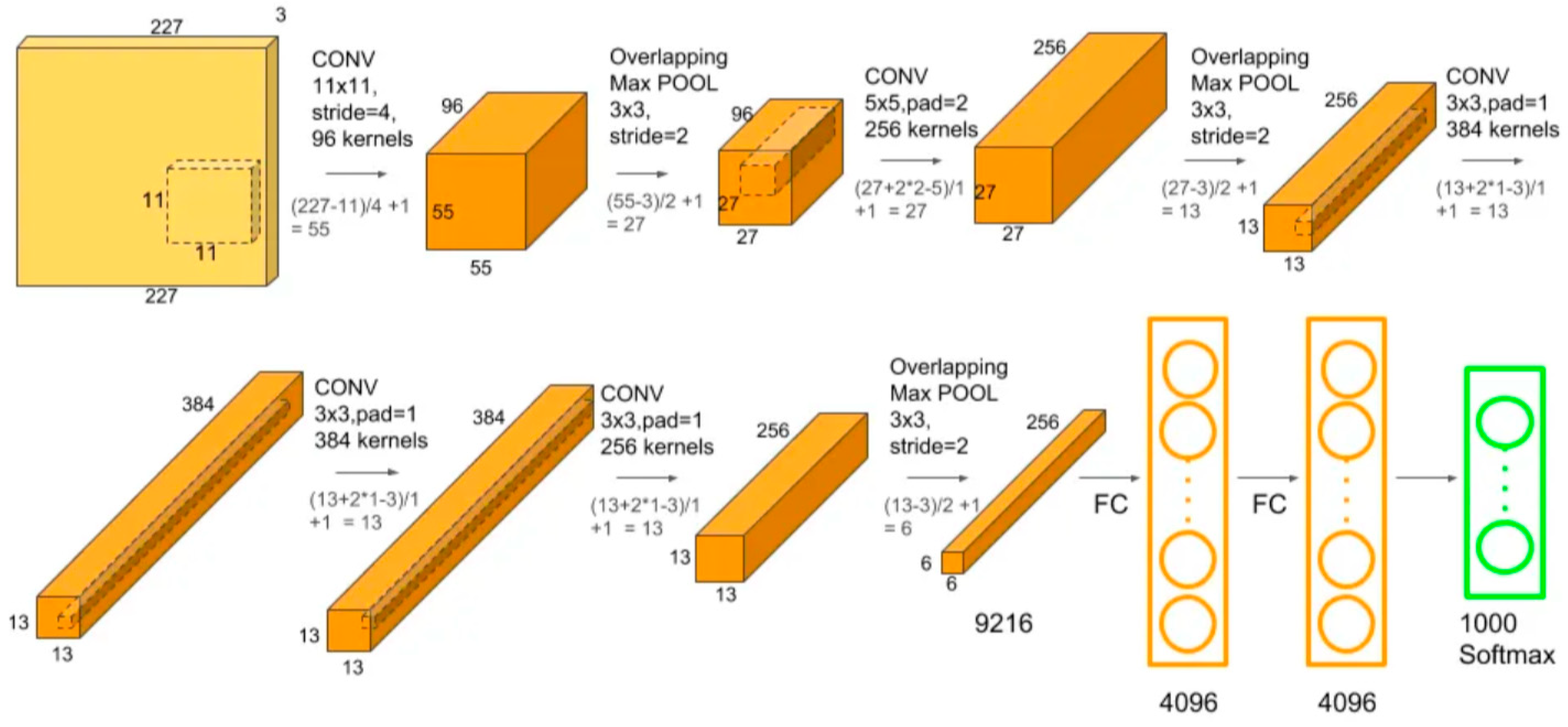
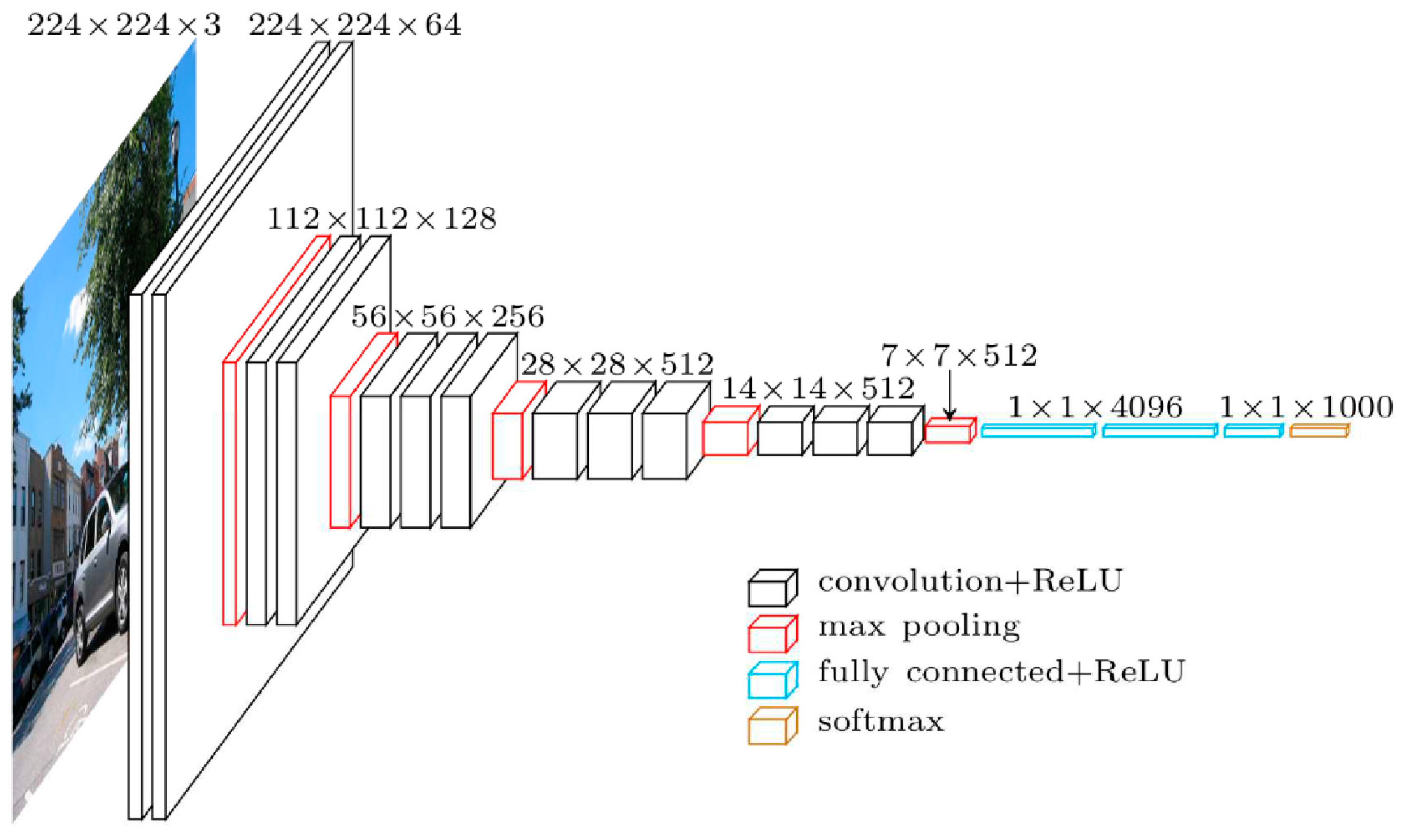

| The studied index | Equation | References |
|---|---|---|
| Germination Energy | [36] | |
| Germination Value | [35] | |
| Germination Vigour | [37] | |
| Allometric Coefficien | [38] | |
| Daily Germination Speed | [39] | |
| Mean Daily Germination | [40] |
| Variety | Period | Number | Germination percentage | Germination Energy | Mean Daily Germination | Germination Value | Daily Germination Speed | Simple vitality index |
|---|---|---|---|---|---|---|---|---|
| NC-2 | 1 | 100 | 91 | 86 | 11.3750 | 250.2500 | 0.0879 | 7.0752 |
| NC-2 | 2 | 100 | 69 | 61 | 8.6250 | 146.6250 | 0.1159 | 3.7826 |
| NC-2 | 3 | 100 | 47 | 34 | 5.8750 | 64.6250 | 0.1702 | 0.9795 |
| Florispan | 1 | 100 | 94 | 92 | 11.7500 | 329.0000 | 0.0851 | 9.5836 |
| Florispan | 2 | 100 | 72 | 63 | 9.000 | 153.0000 | 0.1111 | 4.0761 |
| Florispan | 3 | 100 | 52 | 65 | 6.5000 | 84.5000 | 0.1538 | 2.0072 |
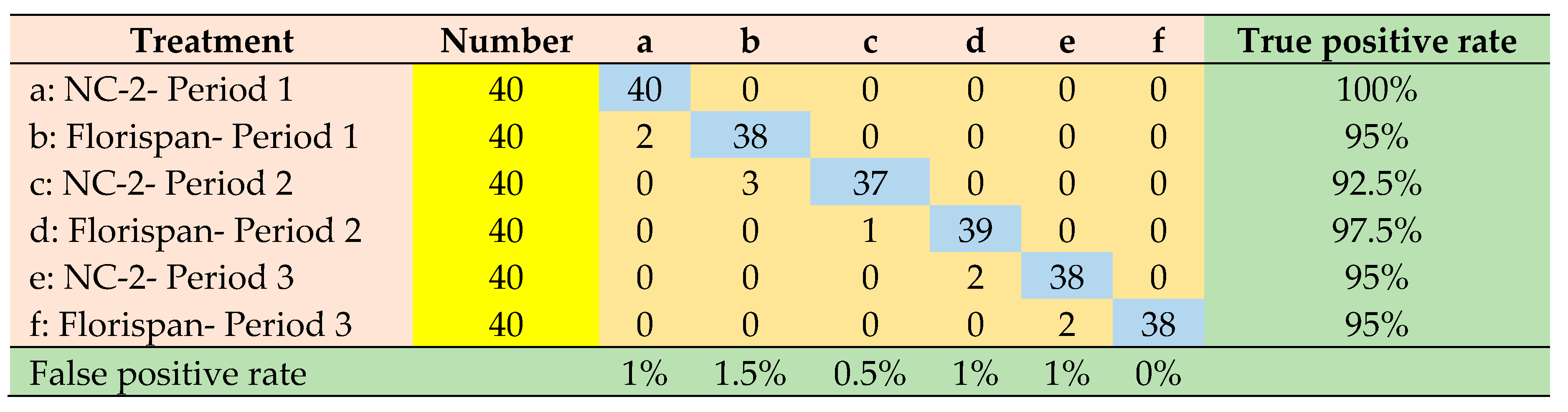
| Network architecture | Accuracy | Precision | Sensitivity | Specificity | ROC Area | Execution time |
|---|---|---|---|---|---|---|
| AlexNet | 0.985 | 0.955 | 0.954 | 0.991 | 0.992 | 2153 (s) |
| VGGNet | 0.986 | 0.959 | 0.958 | 0.992 | 0.981 | 2356 (s) |
| Statistical criteria | Pre-processing method | |||||
|---|---|---|---|---|---|---|
| Raw | MA | MA+SNV | ||||
| PC1 | PC2 | PC1 | PC2 | PC1 | PC2 | |
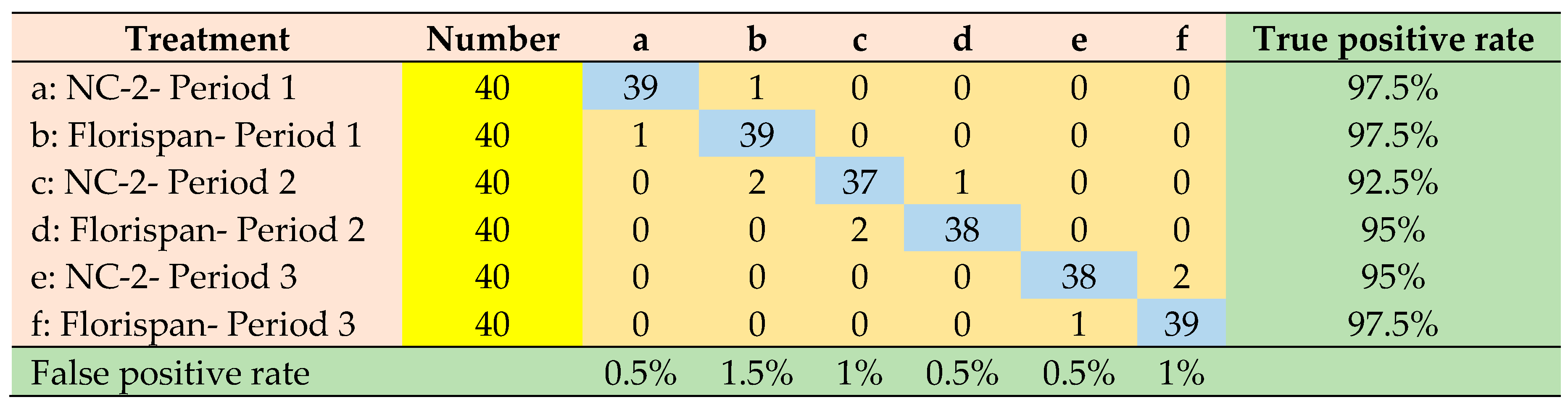
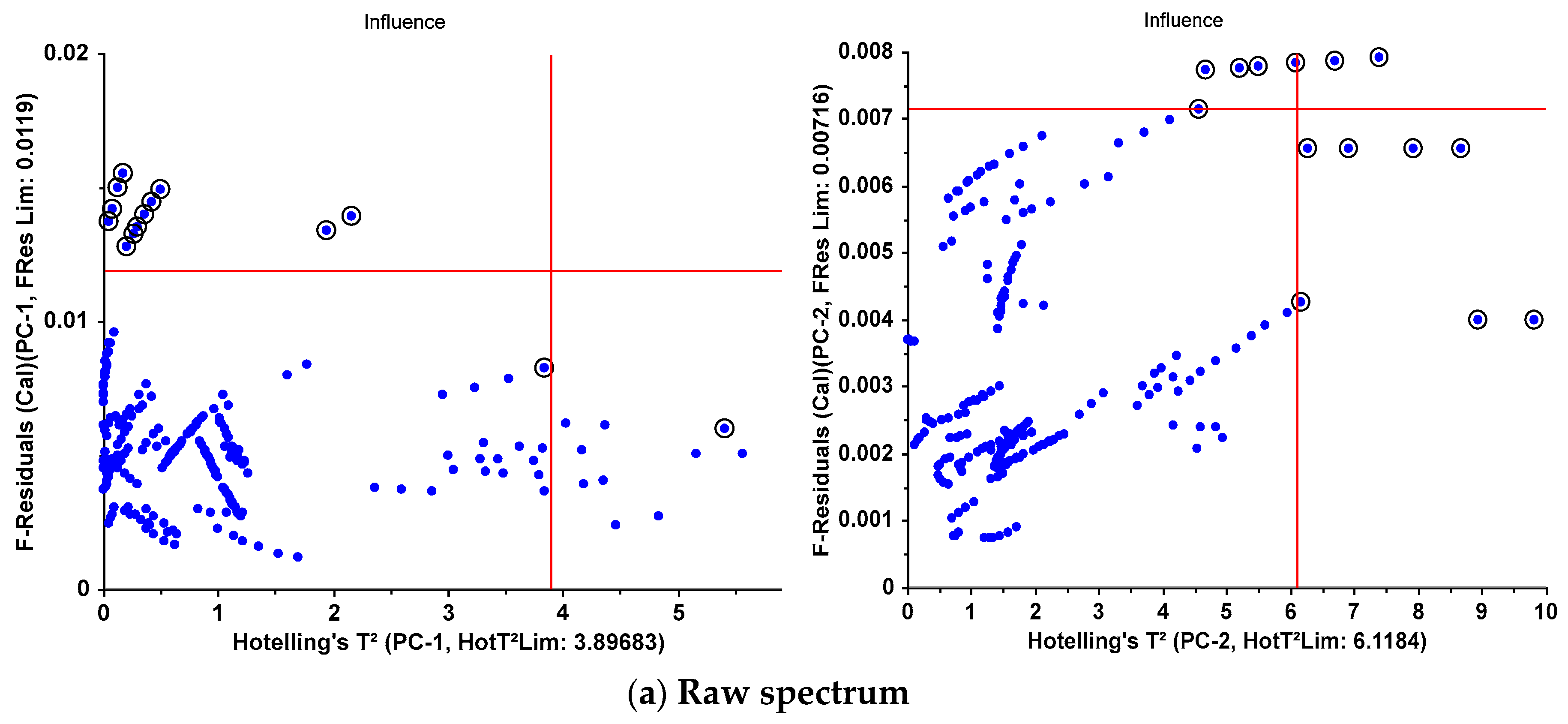
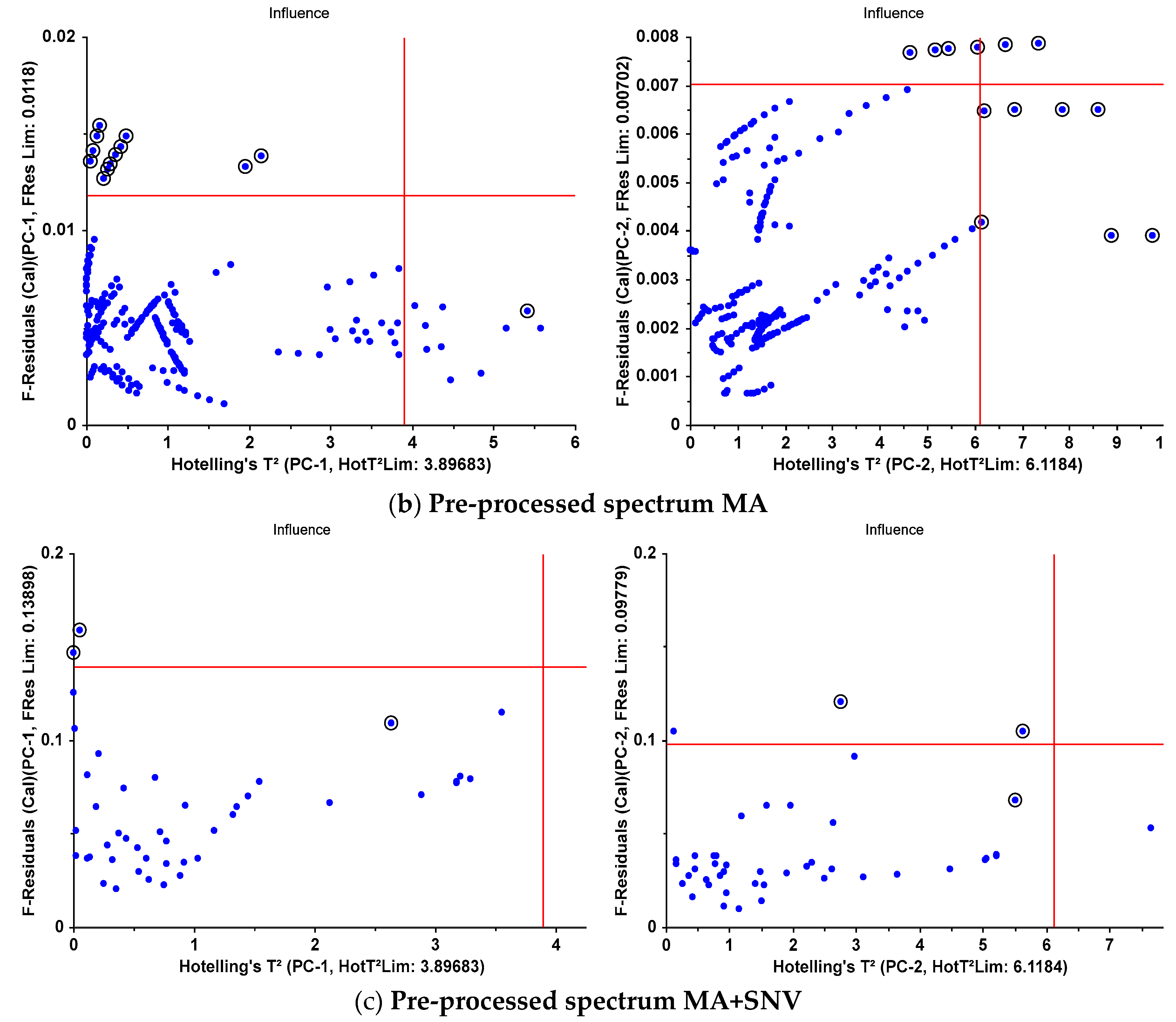
| F-Residuals | 13 | 4 | 12 | 3 | 2 | 2 |
| Hotelling’sT² | 1 | 7 | 1 | 7 | 1 | 1 |
| both | 0 | 3 | 0 | 3 | 0 | 0 |
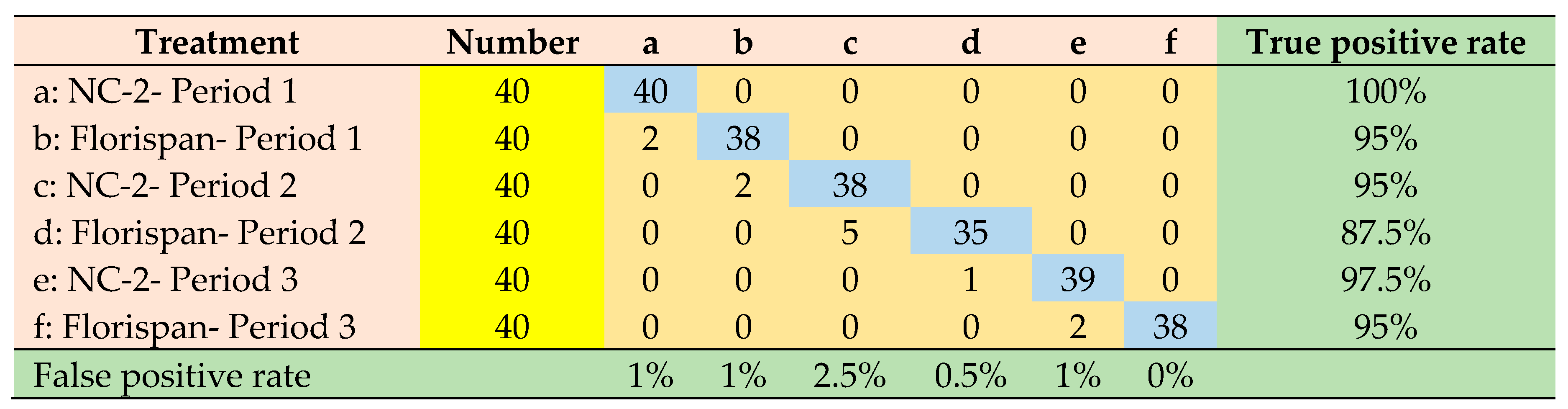
| Network architecture | Accuracy | Precision | Sensitivity | Specificity | ROC Area |
|---|---|---|---|---|---|
| SVM-PC | 0.986 | 0.959 | 0.958 | 0.991 | 0.970 |
| LDA-PC | 0.983 | 0.952 | 0.950 | 0.990 | 0.997 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





