Submitted:
19 November 2023
Posted:
21 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethics Approval and Consent to Participate
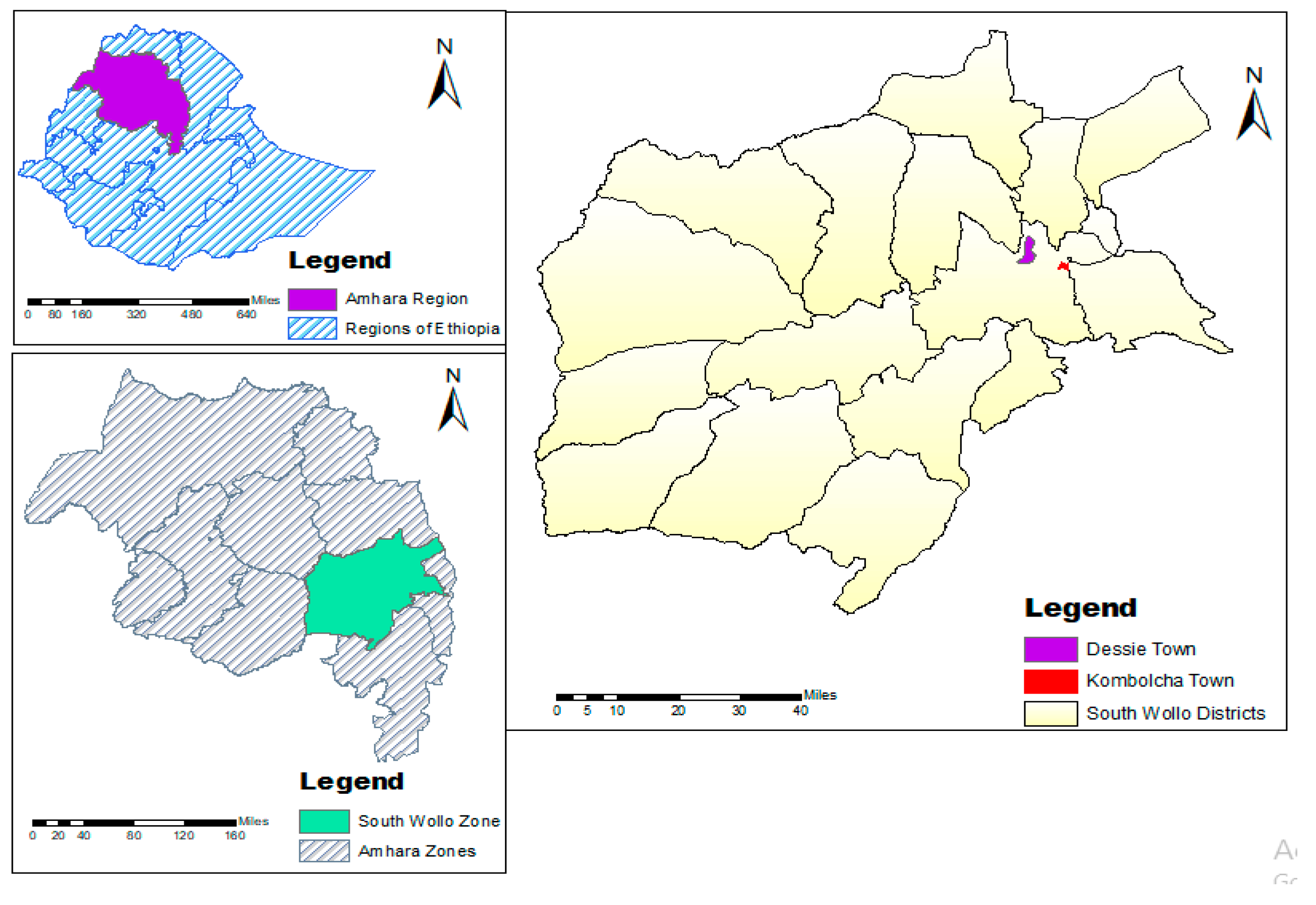
2.2. Study Area
2.3. Study Population
2.4. Study Design
2.5. Sample Size and Sample Collection
2.6. Sampling Technique and Sample Collection
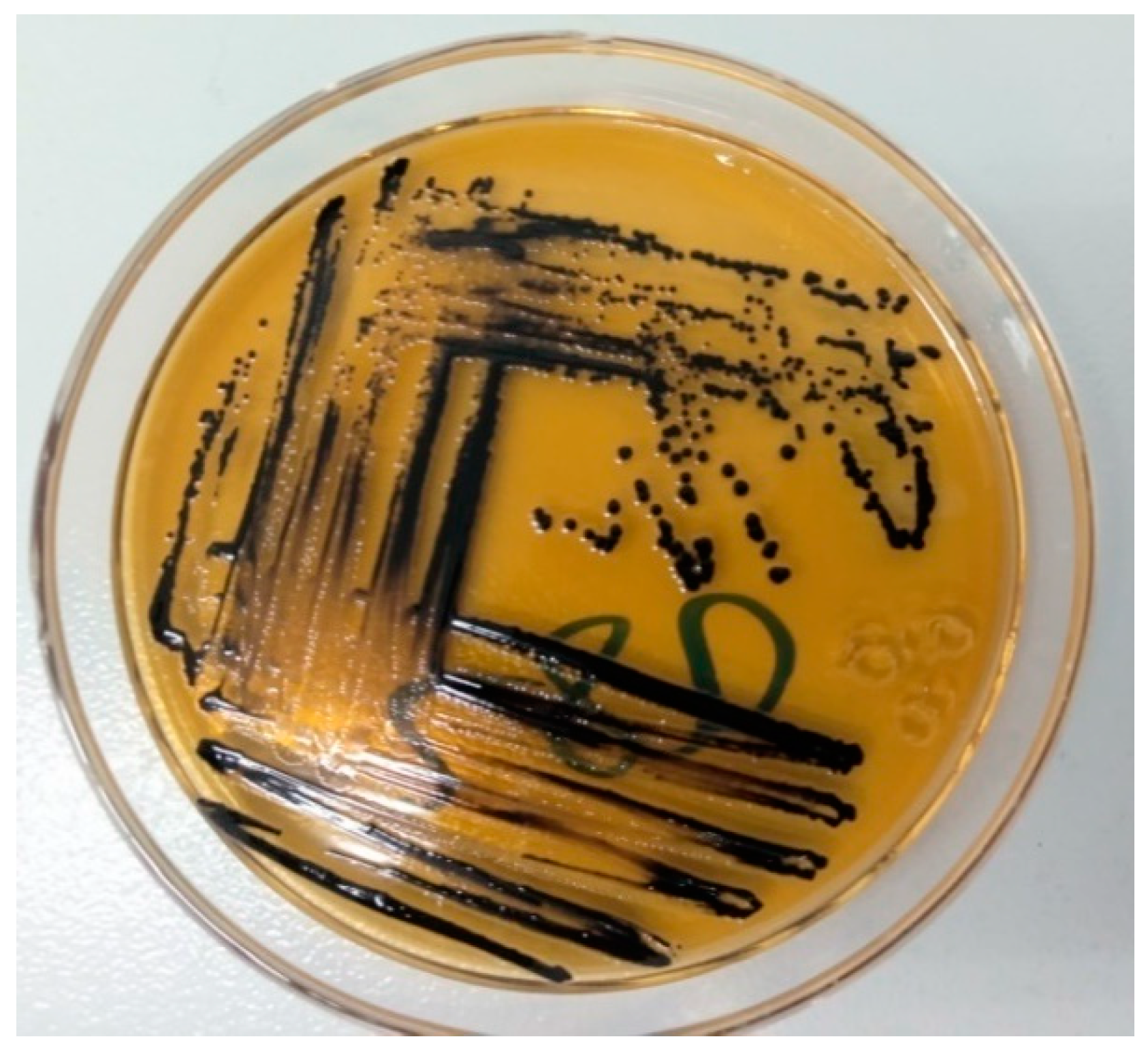
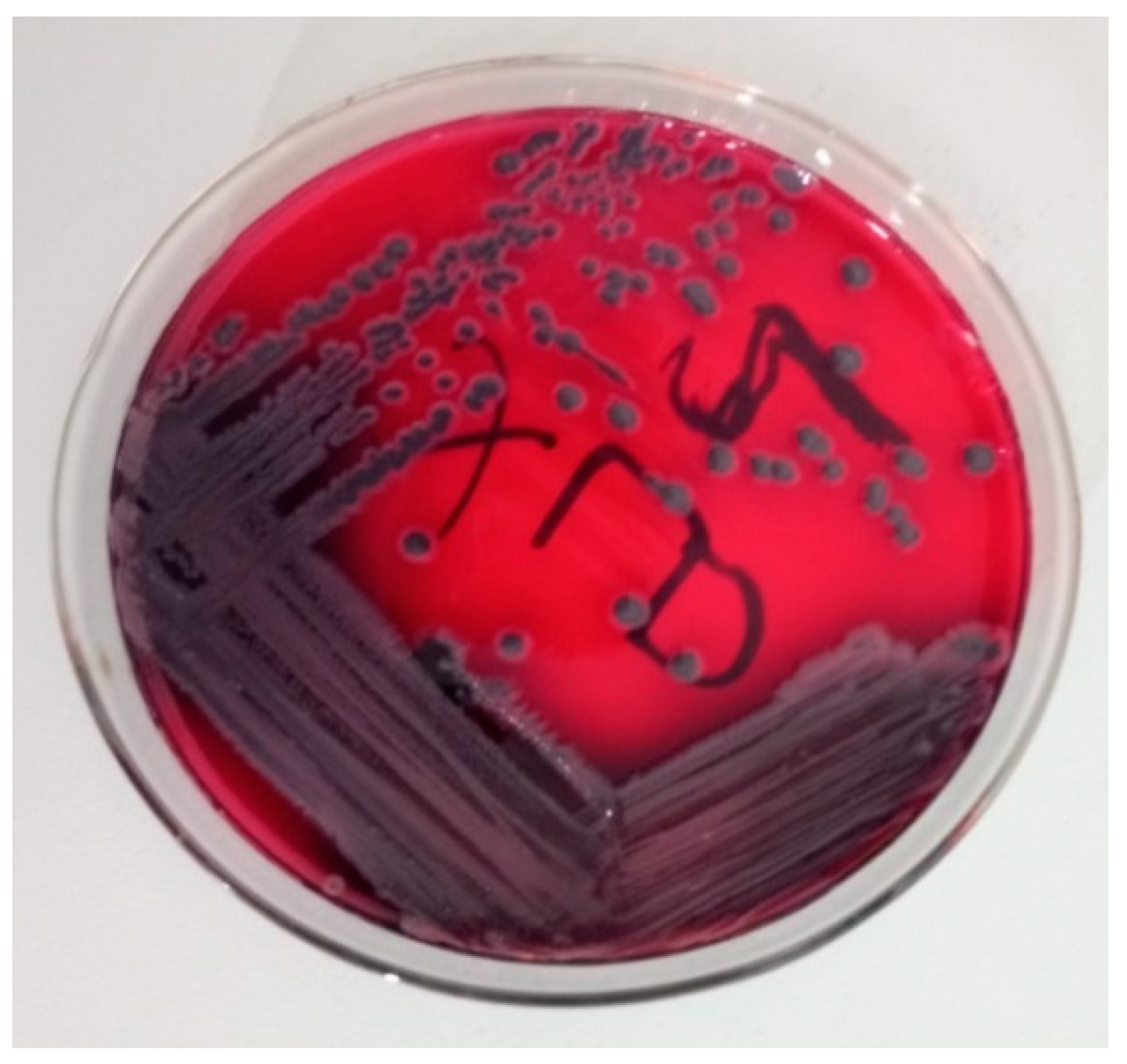
2.7. Isolation and Identification of Salmonella species
2.8. Antimicrobial Susceptibility Testing of Salmonella species
2.9. Standard Strains for Quality Control.
2.10. Data Management and Processing.
3. Results
3.1. Overall Prevalence
3.2. Prevalence of Salmonella Species among Variables of Different Sample Types
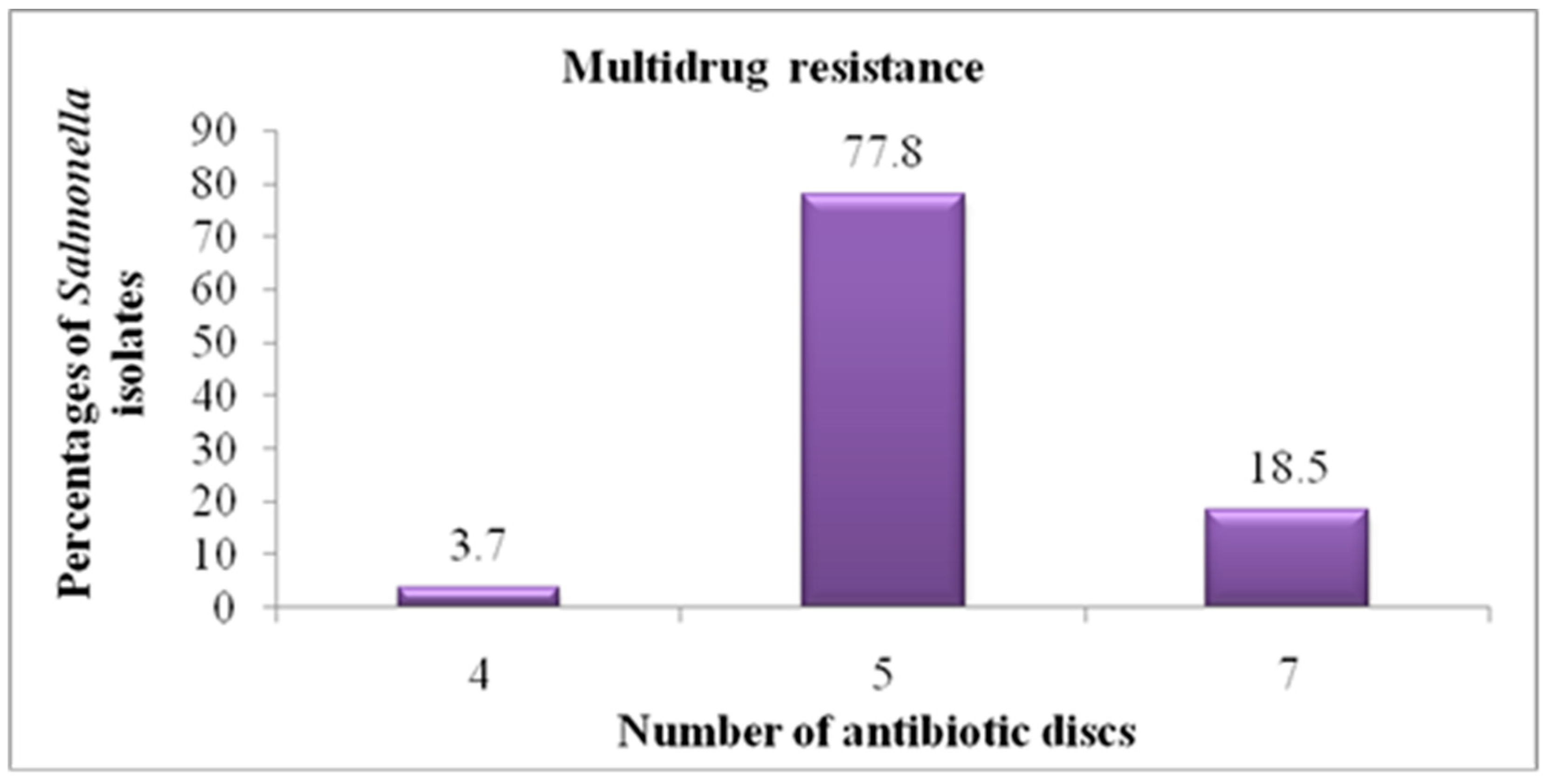
3.3. In Vitro Antimicrobial Sensitivity Pattern
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abebe, E.; Gugsa, G.; Ahmed, M. Review on Major Food-Borne Zoonotic Bacterial Pathogens. J. Trop. Med. 2020, 2020, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K. , Rajagunalan, S., Chakraborty, S., Verma, A. K., Kumar, A., Tiwari, R. and Kapoor, S. (2013): Food-borne pathogens of animal origin-diagnosis, prevention, control and their zoonotic significance: a review. Pak. J. Biol. Sci., 16(20): 1076-1085.
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Leedom, J. M. (2006): Milk of non-human origin and infectious diseases in humans. Clin. Infect. Dis., 43: 610-615.
- Oliver, S.; Jayarao, B.; Almeida, R. Foodborne Pathogens in Milk and the Dairy Farm Environment: Food Safety and Public Health Implications. Foodborne Pathog. Dis. 2005, 2, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Godefay, B. and Molla, B. (2000): Bacteriological quality of raw milk from four dairy farms and milk collection center in and around Addis Ababa. Berl. Münch. Tierärztl. Wschr., 113: 1-3.
- McAuley, C.M.; McMillan, K.; Moore, S.C.; Fegan, N.; Fox, E.M. Prevalence and characterization of foodborne pathogens from Australian dairy farm environments. J. Dairy Sci. 2014, 97, 7402–7412. [Google Scholar] [CrossRef]
- Murphy, S. C. (1996): Sources and causes of high bacteria count in raw milk: an abbreviated review. Cornell University, Ithaca, N.Y. Pp.1-4.
- Joshi, D.D.; Maharjan, M.; Johansen, M.V.; Willingham, A.L.; Sharma, M. Improving meat inspection and control in resource-poor communities: the Nepal example. Acta Trop. 2003, 87, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Madden, R.H.; Espie, W.; Moran, L.; McBride, J.; Scates, P. Occurrence of Escherichia coli O157:H7, Listeria monocytogenes, Salmonella and Campylobacter spp. on beef carcasses in Northern Ireland. Meat Sci. 2001, 58, 343–346. [Google Scholar] [CrossRef] [PubMed]
- WHO (2000): Food borne disease: A focus for health education. World Health Organization. Accessed from: https://apps.who.int/iris/handle/10665/42428.
- WHO (2013): Water- related diseases, campylobacteriosis. Reviewed by staff and experts from the Cluster on Communicable Diseases (CDS) and the Water, Sanitation and Health Unit (WSH), World Health Organization (WHO). Accessed from: https://apps.who.int/iris/bitstream/handle/10665/80751/9789241564601_eng.
- Zelalem, A.; Sisay, M.; Vipham, J.L.; Abegaz, K.; Kebede, A.; Terefe, Y. The prevalence and antimicrobial resistance profiles of bacterial isolates from meat and meat products in Ethiopia: a systematic review and meta-analysis. Int. J. Food Contam. 2019, 6, 1. [Google Scholar] [CrossRef]
- Abunna, F. , Bedashu, A., Beyene, T., Ayana, D., Wakjira, B., Feyisa, A. and Duguma, R. (2018a): Occurrence of Salmonella and its antimicrobial sensitivity test in abattoir and dairy farms in Adama town, Oromia, Ethiopia. J. Vet. Med. Res., 5(3): 1-7.
- Addis, Z. , Kebede, N., Worku, Z., Gezahegn, H., Yirsaw, A. and Kassa, T. (2011): Prevalence and antimicrobial resistance of Salmonella isolated from lactating cows and in contact humans in dairy farms of Addis Ababa: A Cross Sectional Study. BMC Infect. Dis., 11: 1-7.
- Gebremedhin, E.Z.; Soboka, G.T.; Borana, B.M.; Marami, L.M.; Sarba, E.J.; Tadese, N.D.; Ambecha, H.A. Prevalence, Risk Factors, and Antibiogram of Nontyphoidal Salmonella from Beef in Ambo and Holeta Towns, Oromia Region, Ethiopia. Int. J. Microbiol. 2021, 2021, 1–13. [Google Scholar] [CrossRef]
- Hailu, D. , Gelaw, A., Molla, W., Garedew, L., Cole, L. and Johnson, R. (2015): Prevalence and antibiotic resistance patterns of Salmonella isolates from lactating cows and in-contact humans in dairy farms, Northwest Ethiopia. J. Environ. Occup. Sci., 4(4): 171-178.
- Mossie, T. , Tadesse, G. and Dires, A. (2016): Prevalence of antimicrobial resistant Salmonellae isolated from bulk milk of dairy cows in and around Debre Zeit, Ethiopia. World Vet. J., 6(3): 110-116.
- Wolde, T. , Abate, M., Sileshi, H. and Mekonnen, Y. (2016): Prevalence and antimicrobial susceptibility profile of Salmonella species from ready-to-eat foods from catering establishments in Jigjiga City, Ethiopia. Afr. J. Microbiol. Res., 10(37): 1555-1560.
- Tadesse, G. and Tessema, S. T. (2014): A meta-analysis of the prevalence of Salmonella in food animals in Ethiopia. BMC Microbiol., 14(270): 1-9.
- Kore, K. , Asrade, B., Demissie, K. and Aragaw, K. (2017): Characterization of Salmonella isolated from apparently healthy slaughtered cattle and retail beef in Hawassa, southern Ethiopia. Prev. Vet. Med., 147: 11-16.
- Musa, A. J. , Dauda, I. K., Lawan, A. F., Diyo, D., Meshack, M. M. and Jauro, S. (2017): Prevalence and antibiotic sensitivity pattern of Salmonella isolates from milk products and water reservoirs in Maiduguri, North-Eastern Nigeria. IOSR-J.A.V.S. 10(2): 87-92.
- Wang, J.; Xue, K.; Yi, P.; Zhu, X.; Peng, Q.; Wang, Z.; Peng, Y.; Chen, Y.; Robertson, I.D.; Li, X.; et al. An Abattoir-Based Study on the Prevalence of Salmonella Fecal Carriage and ESBL Related Antimicrobial Resistance from Culled Adult Dairy Cows in Wuhan, China. Pathogens 2020, 9, 853. [Google Scholar] [CrossRef]
- Gutema, F.D.; Agga, G.E.; Abdi, R.D.; De Zutter, L.; Duchateau, L.; Gabriël, S. Prevalence and Serotype Diversity of Salmonella in Apparently Healthy Cattle: Systematic Review and Meta-Analysis of Published Studies, 2000–2017. Front. Vet. Sci. 2019, 6, 102. [Google Scholar] [CrossRef]
- EFSA (2007): Scientific opinion of the panel on biological hazards on a request from the European Commission for updating the former SCVPH opinion on Listeria monocytogenes risk related to ready-to-eat foods and scientific advice on different levels of Listeria monocytogenes in ready-to-eat foods and the related risk for human illness. E. F. S. A. J., 599: 1-42.
- White, D. G. , Zhao, S., Simjee, S., Wagner, D. D. and McDermott, P. F. (2002): Antimicrobial resistance of food-borne pathogens. Microbes Infect., 4(4): 405-412.
- Aarestrup, F.M.; Wegener, H.C.; Collignon, P. Resistance in bacteria of the food chain: epidemiology and control strategies. Expert Rev. Anti-infective Ther. 2008, 6, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Ejo, M. , Garedew, L., Alebachew, Z. and Worku, W. (2016): Prevalence and antimicrobial resistance of Salmonella isolated from animal-origin food items in Gondar, Ethiopia. Biomed. Res. Int., 2016: 1-8.
- Nair, D.V.T.; Venkitanarayanan, K.; Kollanoor Johny, A. Antibiotic-Resistant Salmonella in the Food Supply and the Potential Role of Antibiotic Alternatives for Control. Foods 2018, 7, 167. [Google Scholar] [CrossRef] [PubMed]
- Alemu, A. , Addis, M. and Degefu, H. (2020): Identification, antimicrobial resistance pattern and community knowledge, attitude and practices of Salmonella in Mizan Town, Ethiopia: cross sectional study. Health Sci. J., 17(4): 1-7.
- Molla, W.; Molla, B.; Alemayehu, D.; Muckle, A.; Cole, L.; Wilkie, E. Occurrence and antimicrobial resistance of Salmonella serovars in apparently healthy slaughtered sheep and goats of central Ethiopia. Trop. Anim. Heal. Prod. 2006, 38, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Sibhat, B. , Zewde, B. M., Zerihun, A., Muckle, A., Cole, L., Boerlin, P., Wilkie, E., Perets, A., Mistry, K. and Gebreyes, W. A. (2011): Salmonella serovars and antimicrobial resistance profiles in beef cattle, slaughterhouse personnel and slaughterhouse environment in Ethiopia. Zoonoses Public Health, 58: 102-109.
- Zarei, M.; Maktabi, S.; Ghorbanpour, M. Prevalence of Listeria monocytogenes, Vibrio parahaemolyticus, Staphylococcus aureus, and Salmonella spp. in Seafood Products Using Multiplex Polymerase Chain Reaction. Foodborne Pathog. Dis. 2012, 9, 108–112. [Google Scholar] [CrossRef] [PubMed]
- WHO (2017): An integral approach for containment of food-borne antimicrobial resistance. Accessed from: https://apps.who.int/iris/bitstream/handle/10665/255747/9789241512411-eng.pdf?sequence=1.
- Dawit, B. (2013): Economic and social vulnerability of rural - urban migrant women in Dessie Town, South Wollo Zone, Amhara Regional State, M.A Thesis.
- Muluwork, Z. (2014): An assessment of livelihood and food security of farmers displaced due to urban expansion, the case of Kombolcha town in Amhara national regional state, Ethiopia, a thesis report.
- Eskinder, Z. , Eyassu, Y. and Mitiku, H. (2010): Assessment of the impact of industrial effluents on the quality of irrigation water and changes on soil characteristics (a case of Kombolcha town), fourteenth international water technology conference, IWTC 14, 2010, Cairo, Egypt. Pp. 711-727.
- Kombolcha Town Animal Production and Health Office (2019): Annual report of dairy cattle production in Kombolcha town.
- Dessie Town Animal Production and Health Office (2019): Annual report of dairy cattle production in Dessie town.
- Thrusfield, M. (2005): Veterinary Epidemiology (3rd Ed.). UK: Black well science Ltd., A Blackwell publishing company. Pp. 230-234.
- Andrews, W. H, Jacobson, A. and Hammack, T. S. (1998): Chapter 5. Salmonella. In: Bacteriological Analytical Manual (BAM) 8th ed. Silver Spring (USA): U.S. Food and Drug Administration.
- Quinn, P. J. , Markey, B. K., Carter, M. E., Donelly, W. J. and Leonard, F. C. (2002): Veterinary microbiology and microbial disease, Blackwell Science Ltd, a Blackwell Publishing Company. Pp. 465-475.
- Merchant, I. A. and Packer, R. A. (1969): Veterinary bacteriology and virology. 7thed, the Iowa State University Press, Ames, Iowa, USA. Pp. 211-305.
- Jarvis, C. J. , Kellerman, G. E., Van-Rensburg, W. J. J. and Whitehead C. J. (1994): The bacteriology manual. 2nd edition.
- Brenner, D. J. , Krieg, N. R. and Staley, J. T. (2005): Bergey’s manual of systematic bacteriology. 2nded, Springer, New York.
- Quinn, P. J. , Carter, M. E., Markey, B. and Carter, G. R. (1999): Enterobacteriaceace. In: clinical Veterinary Microbiology. Mosby international limited. Pp. 226-460.
- Bauer, A. W. , Kirby, M., Sheris, J. D. and Turch, M. (1966): Antibiotic susceptibility testing by standard single disc method. Am. J. Clin. Pathol., 45(4): 493-496.
- CLSI (2012): Performance standards for antimicrobial susceptibility testing; M100-S22 Vol. 32 No.3, twenty-second informational supplement, Wayne PA., USA. Pp. 1-183.
- CLSI (2014): Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. m100-s24, an informational supplement for global application through the clinical and laboratory standard institute consensus process, Wayne PA, USA. Pp. 1-226.
- CLSI (2017): Performance standards for antimicrobial susceptibility Testing; M100 27th ed., An informational supplement for global application through the clinical and laboratory standard institute consensus process, Wayne PA, USA. Pp. 1-250.
- CLSI (2020): Performance standards for antimicrobial susceptibility testing; M100 30th ed., A CLSI supplement for global application. Wayne PA, USA. Pp. 1-294.
- Arabzadeh, F.; Aeini, F.; Keshavarzi, F.; Behrvash, S. Resistance to Tetracycline and Vancomycin of Staphylococcus aureus Isolates from Sanandaj Patients by Molecular Genotyping. Ann. Clin. Lab. Res. 2018, 6. [Google Scholar] [CrossRef]
- Reza, R. H. , Shipa, S. A., Naser, M. N. and Miah, F. (2020): Surveillance of Escherichia coli in a fish farm of Sylhet, Bangladesh. Bangladesh J. Zool., 48(1): 335-346.
- Tadesse, H. A. , Gidey, N. B., Workelule, K., Hailu, H., Gidey, S., Bsrat, A. and Taddele, H. (2018): Antimicrobial resistance profile of E. coli Isolated from raw cow milk and fresh fruit juice in Mekelle, Tigray, Ethiopia. Vet. Med. Int., 2018: 1-7.
- TMCC (2021): Microbiology resource center: antimicrobial susceptibility testing. Accessed from: https://www.tmcc.edu/microbiology-resource-center/lab-protocols/antimicrobial susceptibility-testing.
- Abunna, F. , Ngusie, G., Tufa, T. B., Ayana, D., Wakjira, B., Waktole, H. and Duguma, R. (2018b): Occurence and antimicrobial susceptibility profile of Salmonella from dairy farms in and around Meki Town, Oromia, Ethiopia. Biomed. J. Sci. & Tech. Res., 6(4): 5388-5395.
- Atsbha, W. T. , Weldeabezgi, T. L., Seyoum, A. K., Tafere, G. and Kassegn, H. H. (2018): Salmonella and risk factors for the contamination of cattle carcass from abattoir of Mekelle City, Ethiopia. Cogent. Food Agric., 4(1): 1-8.
- Singh, P. , Singh, R. V., Gupta, B., Tripathi, S. S., Tomar, K. S., Jain, S. and Sahni, Y. P. (2018): Prevalence study of Salmonella spp. in milk and milk products. Asian J. Dairy & Food Res., 37(1): 7-12.
- Bekele, F. and Lulu, D. (2017): Detection of Salmonella in Haramaya Univesity Slaughter House and assessment of hygienic practic among slaughter house workers, Haramaya, Ethiopia. J. Health Med. Nur., 44: 13-22.
- Abunna, F.; Ashenafi, D.; Beyene, T.; Ayana, D.; Mamo, B.; Duguma, R. Isolation, identification and antimicrobial susceptibility profiles of Salmonella isolates from dairy farms in and around Modjo town, Ethiopia. Ethiop. Vet. J. 2017, 21, 92. [Google Scholar] [CrossRef]
- Karshima, N. S. , Pam, V. A., Bata, S. I., Dung, P. A. and Paman, N. D. (2013): Isolation of Salmonella spp. from milk and locally processed milk products traded for human consumption and associated risk factors in Kanam, Plateau State, Nigeria. J. Anim. Prod. Adv., 3(3): 69-74.
- Alemu, S.; Zewde, B.M. Prevalence and antimicrobial resistance profiles of Salmonella enterica serovars isolated from slaughtered cattle in Bahir Dar, Ethiopia. Trop. Anim. Heal. Prod. 2011, 44, 595–600. [Google Scholar] [CrossRef]
- Rahman, M.A.; Rahman, A.K.M.A.; Islam, M.A.; Alam, M.M. Detection of multi-drug resistant Salmonella From milk and meat in Bangladesh. Bangl. J. Vet. Med. 2018, 16, 115–120. [Google Scholar] [CrossRef]
- Kalambhe, D.G.; Zade, N.N.; Chaudhari, S.P.; Shinde, S.V.; Khan, W.; Patil, A.R. Isolation, antibiogram and pathogenicity of Salmonella spp. recovered from slaughtered food animals in Nagpur region of Central India. Vet. World 2016, 9, 176–181. [Google Scholar] [CrossRef]
- Mulaw, G. (2017): Prevalence and antimicrobial susceptibility of Salmonella Species from lactating cows in dairy farm of Bahir Dar Town, Ethiopia. Afr. J. Microbiol. Res., 11(43): 1578-1585.
- Banti, B. H. (2018): Isolation, identification and antimicrobial susceptibility profile of Salmonella isolates from abattoir and selected dairy farms of Addis Ababa City, Ethiopia. Glob. Vet., 20(6): 285-292.
- Alemayehu, D.; Molla, B.; Muckle, A. Prevalence and antimicrobial resistance pattern of Salmonella isolates from apparently healthy slaughtered cattle in Ethiopia. Trop. Anim. Heal. Prod. 2003, 35, 309–319. [Google Scholar] [CrossRef]
- Zerabruk, K.; Retta, N.; Muleta, D.; Tefera, A.T. Assessment of Microbiological Safety and Quality of Minced Meat and Meat Contact Surfaces in Selected Butcher Shops of Addis Ababa, Ethiopia. J. Food Qual. 2019, 2019, 1–9. [Google Scholar] [CrossRef]
- Kebede, A. , Kemal, J., Alemayehu, H. and Habte-Mariam, S. (2016): Isolation, identification, and antibiotic susceptibility testing of Salmonella from slaughtered bovines and ovines in Addis Ababa Abattoir Enterprise, Ethiopia: a cross-sectional study. Int. J. Bacteriol., 2016: 1-8.
- Mhone, T. A. , Matope, G. and Saidi, P. T. (2012): Detection of Salmonella spp., Candida albicans, Aspergillus spp., and antimicrobial residues in raw and processed cow milk from selected small holder farms of Zimbabwe. Vet. Med. Int., 2012: 1-5.
- Hiko, A.; Ameni, G.; Langkabel, N.; Fries, R. Microbiological Load and Zoonotic Agents in Beef Mortadella from Addis Ababa City Supermarkets. J. Food Prot. 2015, 78, 1043–1045. [Google Scholar] [CrossRef] [PubMed]
- Shilangale, R.; Kaaya, G.; Chimwamurombe, P. Prevalence and Characterization of Salmonella Isolated from Beef in Namibia. Eur. J. Nutr. Food Saf. 2015, 5, 267–274. [Google Scholar] [CrossRef]
- Chyea, F. Y. , Abdullah, A. and Ayob, M. K. (2004): Bacteriological quality and safety of raw milkin Malaysia. Food Microbiol., 21: 535-541.
- Liyuwork, T. , Taye, B., Alemu, S., Alemayehu, H., Sisay, Z. and Negussie, H. (2013): Prevalence and antimicrobial resistance profile of Salmonella isolates from dairy products in Addis Ababa, Ethiopia. Afr. J. Microbiol. Res., 7(43): 5046-5050.
- Ketema, L. , Ketema, Z., Kiflu, B., Alemayehu, H., Terefe, Y., Ibrahim, M. and Eguale, T. (2018): Prevalence and antimicrobial susceptibility profile of Salmonella serovars isolated from slaughtered cattle in Addis Ababa, Ethiopia. Biomed Res. Int., 2018: 1-7.
- Van Kessel, J.; Karns, J.; Gorski, L.; McCluskey, B.; Perdue, M. Prevalence of Salmonellae, Listeria monocytogenes, and Fecal Coliforms in Bulk Tank Milk on US Dairies. J. Dairy Sci. 2004, 87, 2822–2830. [Google Scholar] [CrossRef]
- Mengistu, S.; Abayneh, E.; Shiferaw, D. E. coli O157:H7 and Salmonella Species: Public Health Importance and Microbial Safety in Beef at Selected Slaughter Houses and Retail Shops in Eastern Ethiopia. J. Vet. Sci. Technol. 2017, 8. [Google Scholar] [CrossRef]
- Beyene, T. , Yibeltie, H., Chebo, B., Abunna, F., Beyi, A. F., Mammo, B., Ayana, D. and Duguma, R. (2016): Identification and antimicrobial susceptibility profile of Salmonella isolated from selected dairy farms, abattoir and humans at Asella Town, Ethiopia. J. Veterinar. Sci. Techno., 7(3): 1-7.
- Reta, M. A. , Bereda, T. W. and Alemu, A. N. (2016): Bacterial contaminations of raw cow’s milk consumed at Jigjiga City of Somali Regional State, Eastern Ethiopia. Int. J. Food Contam., 3(4): 1-9.
- Azage, M. and Kibret, M. (2017): The Bacteriological quality, safety, and antibiogram of Salmonella isolates from fresh meat in retail shops of Bahir Dar City, Ethiopia. Int. J. Food Sci., 2017: 1-5.
- Tadesse, T. and Dabassa, A. (2012): Prevalence and antimicrobial resistance of Salmonella isolated from raw milk samples collected from Kersa District, Jimma Zone, Southwest Ethiopia. J. Med. Sci., 12: 224-228.
- Takele, S.; Woldemichael, K.; Gashaw, M.; Tassew, H.; Yohannes, M.; Abdissa, A. Prevalence and drug susceptibility pattern of Salmonella isolates from apparently healthy slaughter cattle and personnel working at the Jimma municipal abattoir, south-West Ethiopia. Trop. Dis. Travel Med. Vaccines 2018, 4, 13. [Google Scholar] [CrossRef]
- Ejeta, G. , Molla, B., Alemayehu, D. and Muckle, A. (2004): Salmonella serotypes isolated from minced meat beef, mutton and pork in Addis Ababa. Revue. Med. Vet., 155(11): 547-551.
- Lingathurai, S. and Vellathurai, P. (2013): Bacteriological quality and safety of raw cow milk in Madurai, South India. Webmed. Cent. Microbiol., 1:1-10.
- Hiko, A.; Irsigler, H.; Ameni, G.; Zessin, K.-H.; Fries, R. Salmonella serovars along two beef chains in Ethiopia. J. Infect. Dev. Ctries. 2016, 10, 1168–1176. [Google Scholar] [CrossRef]
- Szczawiński, J.; Szczawińska, M.E.; Łobacz, A.; Jackowska-Tracz, A. Modeling the effect of temperature on survival rate of Salmonella Enteritidis in yogurt. Pol. J. Vet. Sci. 2014, 17, 479–485. [Google Scholar] [CrossRef]
- Tanih, N. F. , Sekwadi, E., Ndip, R. N. and Bessong, P. O. (2015): Detection of pathogenic Escherichia coli and Staphylococcus aureus from cattle and pigs slaughtered in abattoirs in Vhembe District, South Africa. Sci. World J., 2015: 1-8.
- Geresu, M.A.; Regassa, S. Escherichia coli O157 : H7 from Food of Animal Origin in Arsi: Occurrence at Catering Establishments and Antimicrobial Susceptibility Profile. Sci. World J. 2021, 2021, 1–10. [Google Scholar] [CrossRef]
- Arthur, T. M. , Kalchayanand, N., Bosilevac, J. M., Brichta-Harhay, D. M., Shackelford, S. D., Bono, J. L., Wheeler, T. L. and Koohmaraie, M. (2008): Comparison of effects of antimicrobial interventions on multidrug-resistant Salmonella, susceptible Salmonella, and Escherichia coli O157:H7. J. Food Prot., 71(11): 2177-2181.
- Ekli, R.; Adzitey, F.; Huda, N. Prevalence of resistant Salmonella spp. isolated from raw meat and liver of cattle in the Wa Municipality of Ghana. IOP Conf. Series: Earth Environ. Sci. 2019, 287. [Google Scholar] [CrossRef]
- Mokgophi, T.M.; Gcebe, N.; Fasina, F.; Adesiyun, A.A. Antimicrobial Resistance Profiles of Salmonella Isolates on Chickens Processed and Retailed at Outlets of the Informal Market in Gauteng Province, South Africa. Pathogens 2021, 10, 273. [Google Scholar] [CrossRef] [PubMed]
- Qamar, A. , Ismail, T. and Akhtar, S. (2020): Prevalence and antibiotic resistance of Salmonella spp. in South Punjab-Pakistan. PLoS ONE, 15(11):1-15.
- Adzitey, F. , Asiamah, P. and Boateng, E. F. (2020): Prevalence and Antibiotic Susceptibility of Salmonella enterica isolated from Cow Milk, Milk Products and Hands of Sellers in the Tamale Metropolis of Ghana. J. Appl. Sci. Environ. Manage., 24(1): 59-64.
| Variables | No. of examined | No. of positive (%) | χ2-value | P value |
|---|---|---|---|---|
| Sample types | 6.836 | 0.233 | ||
| Udder milk | 146 | 8 (5.5) | ||
| Tank milk | 6 | 1 (16.7) | ||
| Yoghurt | 36 | 0 (0.0) | ||
| Cheese | 9 | 0 (0.0) | ||
| Beef swab | 25 | 2 (8.0) | ||
| Carcass swab | 162 | 16 (9.9) | ||
| Study sites | 0.0844 | 0.771 | ||
| Dessie | 181 | 12 (6.6) | ||
| Kombolcha | 203 | 15 (7.4) | ||
| Overall | 384 | 27 (7.0) |
| Variables | No. of examined | No. of positive (%) | χ2-value | P-value |
|---|---|---|---|---|
| Study sites | 4.477 | 0.034 | ||
| Dessie | 52 | 6 (11.5) | ||
| Kombolcha | 100 | 3 (3.0) | ||
| Sample types | 1.295 | 0.255 | ||
| Pooled Udder milk | 146 | 8 (5.5) | ||
| Bucket milk | 6 | 1 (16.7) | ||
| Farm systems | 1.534 | 0.216 | ||
| Intensive | 131 | 9 (6.9) | ||
| Semi Intensive | 21 | 0 (0.0) | ||
| Treatment history | 0.578 | 0.447 | ||
| No | 50 | 4 (8.0) | ||
| Yes | 102 | 5 (4.9) | ||
| Milking practices | 22.287 | 0.000 | ||
| Excellent | 3 | 0 (0.0) | ||
| Very good | 42 | 0 (0.0 | ||
| Good | 104 | 7 (6.7)) | ||
| Poor | 3 | 2 (66.7) | ||
| Farm hygiene | 2.759 | 0.430 | ||
| Excellent | 4 | 0 (0.0 | ||
| Very good | 52 | 1 (1.9) | ||
| Good | 85 | 7 (8.2) | ||
| Poor | 11 | 1 (9.1) | ||
| Total | 152 | 9 (5.9) |
| Variables | No. of examined | No. of positive (%) | χ2-value | P-value |
|---|---|---|---|---|
| Study sites | 3.482 | 0.062 | ||
| Dessie | 96 | 6 (6.2) | ||
| Kombolcha | 66 | 10 (15.2) | ||
| Sources | 0.571 | 0.450 | ||
| Municipal Abattoir | 105 | 9 (8.6) | ||
| ELFORA | 57 | 7 (12.3) | ||
| Hygiene of slaughtering process | 0.447 | 0.800 | ||
| Very good | 78 | 7 (9.0) | ||
| Good | 49 | 6 (12.2) | ||
| Poor | 35 | 3 (8.6) | ||
| Hygiene of butchers | 2.765 | 0.251 | ||
| Very good | 78 | 7 (9.0) | ||
| Good | 55 | 8 (14.5) | ||
| Poor | 29 | 1 (3.4) | ||
| Hygiene of slaughtering materials | 1.373 | 0.503 | ||
| Excellent | 57 | 7 (12.3) | ||
| Good | 83 | 6 (7.2) | ||
| Poor | 22 | 3 (13.6) | ||
| Total | 162 | 16 (9.9) |
| Variables | No. of examined | No. of positive (%) | χ2-value | P-value |
|---|---|---|---|---|
| Study sites | 2.355 | 0.125 | ||
| Dessie | 13 | 0 (0.0) | ||
| Kombolcha | 12 | 2 (16.7) | ||
| Where get slaughtered | 1.870 | 0.171 | ||
| Abattoir | 21 | 1 (4.8) | ||
| Field | 4 | 1 (25.0) | ||
| Hygiene of butchers | 0.845 | 0.655 | ||
| Very good | 18 | 1 (5.6) | ||
| Good | 6 | 1 (16.7) | ||
| Poor | 1 | 0 (0.0) | ||
| Hygiene of cutting utensils | 1.223 | 0.269 | ||
| Very good | 5 | 1 (20.0) | ||
| Good | 20 | 1 (5.0) | ||
| Hygiene of butcher shops | 0.570 | 0.903 | ||
| Excellent | 2 | 0 (0.0) | ||
| Very good | 13 | 1 (7.7) | ||
| Good | 8 | 1 (12.5) | ||
| Poor | 2 | 0 (0.0) | ||
| Total | 25 | 2 (8.0) |
| Antimicrobial agents | Interpretation categories | ||
|---|---|---|---|
| Sensitive | Intermediate | Resistant | |
| Erythromycin | 0 (0.0) | 0 (0.0) | 27 (100) |
| Nalidixic acid | 23 (85.2) | 4 (14.8) | 0 (0.0) |
| Kanamycin | 19 (70.4) | 8 (29.6) | 0 (0.0) |
| Gentamicin | 27(100) | 0 (0.0) | 0 (0.0) |
| Amoxicillin | 21 (77.8) | 0 (0.0) | 6 (22.2) |
| Doxycycline | 1 (3.7) | 0 (0.0) | 26 (96.3) |
| Tetracycline | 0 (0.0) | 0 (0.0) | 27(100) |
| Penicillin G | 6 (22.2) | 21 (77.8) | 0 (0.0) |
| Sulfamethoxazole-trimetoprim | 22 (81.5) | 0 (0.0) | 5 (18.5) |
| Polymyxin B | 1 (3.7) | 0 (0.0) | 26 (96.3) |
| Vancomycin | 0 (0.0) | 0 (0.0) | 27(100) |
| Ciprofloxacin | 27 (100) | 0 (0.0) | 0 (0.0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





