Submitted:
17 November 2023
Posted:
17 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Protective HLA alleles in Celiac disease
1.2. The human gastrointestinal tract is a target organ for SARS-CoV-2
1.3. COVID-19-celiac disease interplay
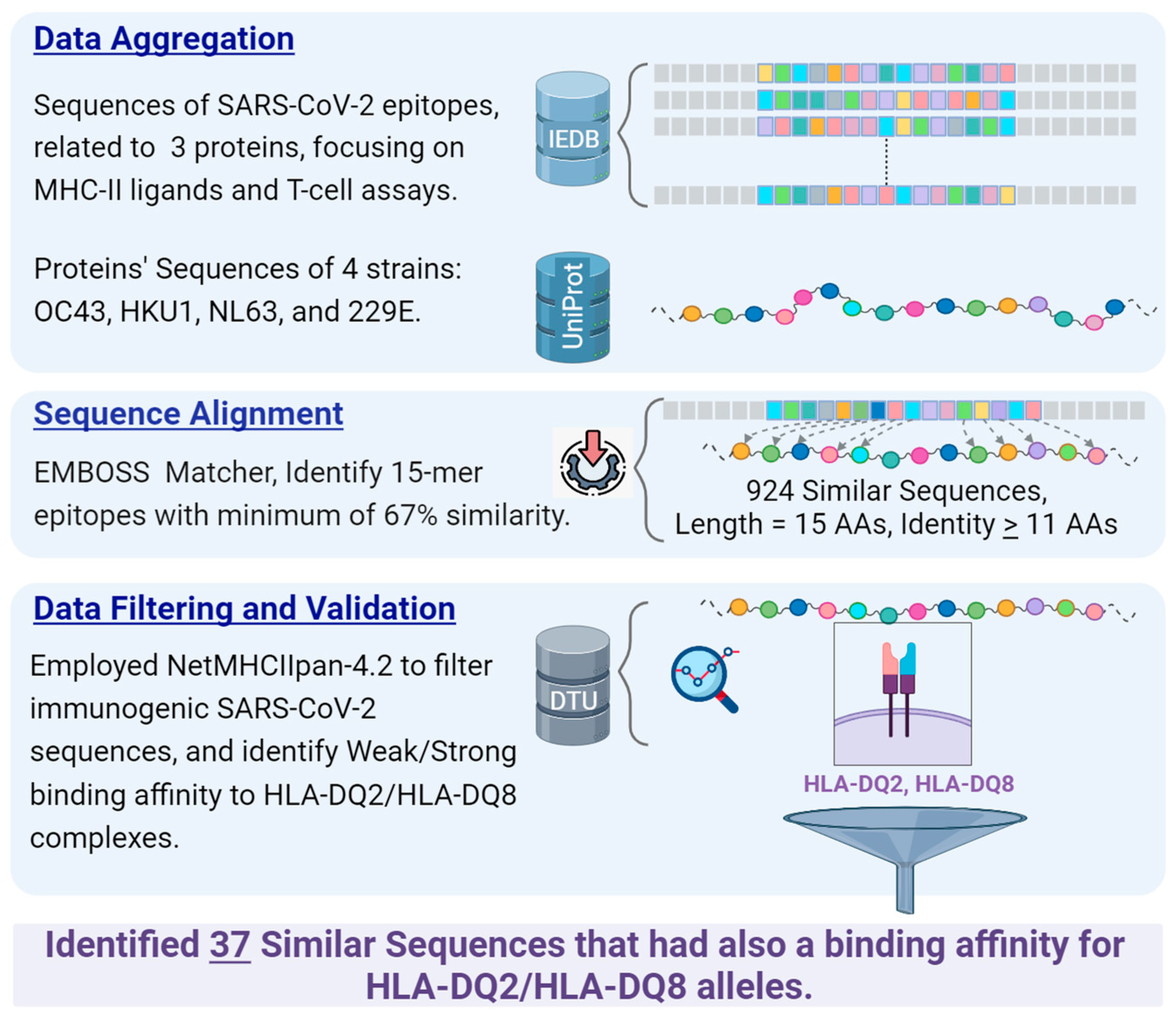
2. Materials and Methods
2.1. Data Sources
- Protein Sequences Extraction of Common Cold Coronaviruses:
- Data Retrieval from IEDB:
- Organism: SARS-CoV-2 (ID:2697049).
- Antigens: Spike glycoprotein (P0DTC2), Nucleoprotein (P0DTC9), Replicase polyprotein 1ab (P0DTD1).
- Epitope Structure: Linear Sequence.
- Assay Type: Only positive assays pertaining to T-cell epitopes and MHC ligands were considered. This refers to epitopes validated through laboratory experiments.
- MHC Restriction: Class II.
- Host: Human.
- Diseases: No specific restrictions; all diseases were considered.
2.2. Sequence Similarity Identification
2.3. Binding Affinity Prediction to HLA-DQ2, DQ8
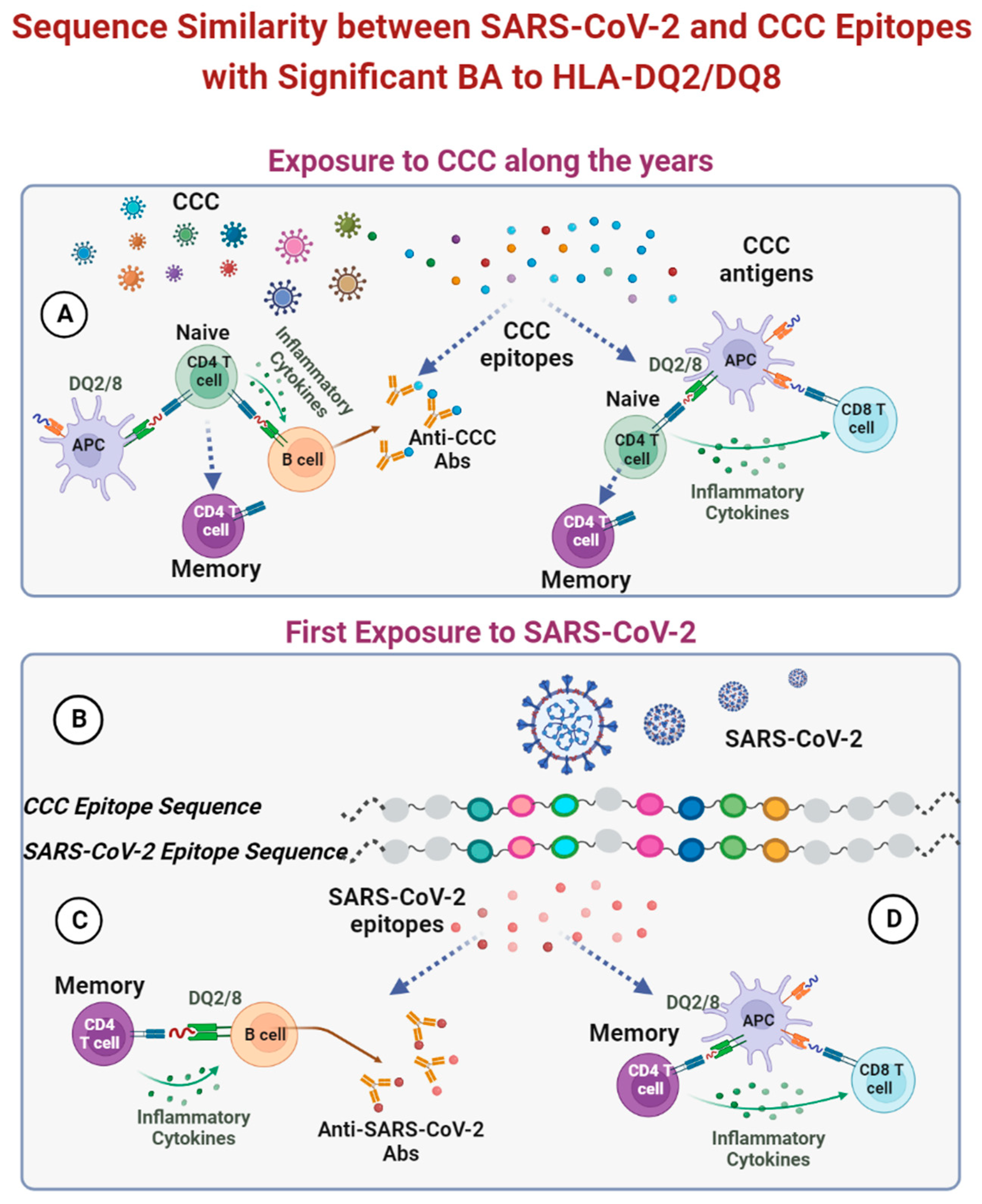
3. Results
4. Discussion
- 4.1. High affinity between SARS-CoV-2 antigens and HLA-DQ immune presentation to the T cells enhances anti-SARS-CoV-2 immunity. In fact, the HLA allele most associated with COVID-19 deterioration is HLA-A*11. However, HLA II also plays a role in the disease severity, HLA-DRB1*15:01 and HLA-DRB1*04 alleles being examples [124].. Unfortunately, the CD-associated HLA-DQs were not explored when the binding affinities of 438 HLA alleles were screened [124]. Nevertheless, asymptomatic and mildly/moderate affected patients likely develop an effective early immune response to clear the virus [125]. A reasonable explanation for the associations between CD, SARS-CoV-2 and HLA-DQ2/8 observed presently is that most of the strong HLA binders of coronavirus peptides are also, strong binders of other sequences, and hence, are likely to be a general strong binders that probably underwent selection in the past [124].
- 4.2. SARS-CoV-2 naïve people might have a certain measure of HLA-dependent immune defense, presented by antibodies cross-reactive to other CCC [126]. Most of those HLAs belong to HLA class I, hence, a minority of them are part of class II. Unfortunately, the CD-associated HLA-DQ were not explored [126]. The topic of a potential protective cross-reactivity against the COVID-19 virus in uninfected CD patients, conferred by their HLA-DQ 2/8 is a subject for further investigation.
- 4.3. Individual HLA variant has its unique repertoire of peptides with a specific sequence structure to stick in the peptide-binding groove of HLA. It appears that certain HLA haplotypes have higher preferences to present peptides with specific molecular functions [127]. This HLA preferential presentation was extrapolated to explain the protective effect of certain HLA alleles in infectious diseases, including COVID-19. Indeed, Karnaukhov V. et al reported-on HLA-A/HLA-B and HLA-A/HLA-C variants having a more distinct functional antigen's preference presentations, but the HLA-DQ2/8 ones were not explored [127]. The authors reported on HLA differential presentation of SARS-CoV-2 antigens mainly by HLA type I alleles, hence, the CD-associated HLA-DQs haplotypes might play a protective or attenuative role in COVID-19 disease. Notably, several studies reported on HLA-DQ variants associated with a dominant T cell response against the SARS-CoV-2 virus, resulting in a milder disease [122,128,129], including a higher production of antibodies post mRNA-based vaccination [130].
- 4.4. Cross reactive antibodies shared between SARS-CoV-2 and gluten? If cross-reactivity exists between the virus and gluten, those reactive antibodies might attenuate the severity of COVID-19 and protect the untreated or the non-compliant CD patients. In fact, Vojdani A, et al. reported on such cross-reactive antibodies [131]. Screening 180 different food antigens and peptides, the authors showed that SARS-CoV-2 proteins share cross-reactive epitopes with various food antigens that had not been previously explored. Wheat and alpha-gliadin were shown to cross-react with SARS-CoV-2 spike protein and nucleoprotein [131]. More so, the authors reported on sequence similarity between SARS-CoV-2 proteins and alpha-gliadin toxic peptides and glutenin, thus, reinforcing a potential effect of the COVID-19-food axis relationships. It should be stressed that the potential protective effects of the above-mentioned cross-reactive antibodies and the sequence similarity were not substantiated and should be further evaluated.
- 4.5. Increase in anti-inflammatory factors in COVID-19-infected celiac patients. Recently, Asri N et al. studied naïve CD patients for various inflammatory and anti-inflammatory markers [86]. The CD patients exposed an increase expression of anti-inflammatory molecules like CD4, CD25 (IL-2Rα) and FOXP3, compared to severe COVID-19 patients and controls. However, the HLA-DQs allelic status was not investigated. The increase anti-inflammatory profile might be beneficial to the CD patients, by lowering COVID-19 severity and attenuating the disease course. The relationship of those markers to the HLA-DQs should farther be explored.
- 4.6. HLA-DQ2/8 might be important in fighting human viruses. The mechanism of CD risk modification by HLA heterogeneity might involve differential presentation of autoantigenic sequences by HLA class II proteins. The HLA-DQ2 and DQ8 presentation of viral epitopes were reported concerning coxsackievirus specific peptides [132]. The authors speculated that the phenomenon might represent a protective adaptive mechanism to maximize anti-enterovirus responses. The same can be speculated for the COVID-19 virus and the HLA-DQ2/8 epitopic presentation in CD, alluding to the potential protective role of those HLAs in fighting SARS-CoV-2 viruses.
- 4.7. HLA class II: Evolutionary protective mechanisms for CD survival. The wide range of COVID-19 manifestations, morbidity and mortality, seen across various ethnicities and geographical distribution was suggested to be host genetic dependent [133]. This genetic adaptative diversity may apply to CD. Interestingly, selective advantage mechanisms for polymorphic genes were speculated to contribute to the evolutionally survival of the CD populations [38,39,134] (Table 1). In fact, the human HLA's genetic heterogeneity is a known major anti-infectious mechanism to fight microbes, parasites and even viruses, SARS-CoV-2 included. Although the variants of class II HLA loci were less frequently analyzed, they can impact COVID-19 outcome. Most recently, HLA class II DRB1*01:01, DRB1∗04:01 and DRB1*03:01 were reported to reduce disease duration and attenuated COVID-19 course [135,136,137]. Unfortunately, the HLA-DQ repertoire was not screened in those studies. Of note, the topic is still controversial and some studies denied the association between HLA polymorphism and COVID-19 outcome [138,139].
5. Celiac disease and long COVID-19 syndromes
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability
- UniProt Knowledgebase www.uniprot.org, reference [100].
- Pairwise Local Alignment tool, EMBOSS Matcher, at www.emboss.sourceforge.net, reference [104,105], A python script can be found at https://raw.githubusercontent.com/ebi-wp/webservice-clients/master/python/emboss_matcher.py
- Binding Affinity Prediction tool from DTU Health Tech, the NetMHCIIpan-4.2 method, at https://services.healthtech.dtu.dk/services/NetMHCIIpan-4.2/ [108]
Conflicts of Interest
Abbreviations
References
- Wang, D.; Wu, X.; Li, C.; Han, J.; Yin, J.; Gan, J. The impact of geo-environmental factors on global COVID-19 transmission: A review of evidence and methodology. 2022. [CrossRef]
- Begou, P.; Kassomenos, P. The ecosyndemic framework of the global environmental change and the COVID-19 pandemic. Sci. Total Environ. 2023, 857. [Google Scholar] [CrossRef] [PubMed]
- Zguro, K.; Fallerini, C.; Fava, F.; Furini, S.; Renieri, A. Host genetic basis of COVID-19: from methodologies to genes. Eur. J. Hum. Genet. 2022, 30, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.S.; Chen, B.; Ze, B.; Zhou, W.H. Human genetic basis of severe or critical illness in COVID-19. Front. Cell. Infect. Microbiol. 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. jin; Dong, X.; Liu, G. hui; Gao, Y. dong Risk and Protective Factors for COVID-19 Morbidity, Severity, and Mortality. Clin. Rev. Allergy Immunol. 2023, 64, 90–107. [Google Scholar] [CrossRef] [PubMed]
- Dobrijević, Z.; Robajac, D.; Gligorijević, N.; Šunderić, M.; Penezić, A.; Miljuš, G.; Nedić, O. The association of ACE1, ACE2, TMPRSS2, IFITM3 and VDR polymorphisms with COVID-19 severity: A systematic review and meta-analysis. EXCLI J. 2022, 21, 818–839. [Google Scholar] [CrossRef] [PubMed]
- Migliorini, F.; Torsiello, E.; Spiezia, F.; Oliva, F.; Tingart, M.; Maffulli, N. Association between HLA genotypes and COVID-19 susceptibility, severity and progression: a comprehensive review of the literature. Eur. J. Med. Res. 2021, 26. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Hollenbach, J.A. The immunogenetics of COVID-19. Immunogenetics 2023, 75, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Ghazy, A.A.; Alrasheedi, A.N.; Elashri, M.; Moussa, H.H.; Rashwan, E.K.; Amer, I.; El Sharawy, S.; Elgamal, S.; Tawfik, S.; Abdelnasser, M.; et al. Relevance of HLA-DP/DQ and INF-λ4 Polymorphisms to COVID-19 Outcomes. Br. J. Biomed. Sci. 2023, 80. [Google Scholar] [CrossRef] [PubMed]
- Pishesha, N.; Harmand, T.J.; Ploegh, H.L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 2022, 22, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Arab, F.; Mollazadeh, S.; Ghayourbabaei, F.; Moghbeli, M.; Saburi, E. The role of HLA genotypes in understanding the pathogenesis of severe COVID-19. Egypt. J. Med. Hum. Genet. 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Lin, X.; Fu, B.; Xiong, Y.; Zaky, M.Y.; Wu, H. Functional studies of HLA and its role in SARS-CoV-2: Stimulating T cell response and vaccine development. Life Sci. 2023, 315. [Google Scholar] [CrossRef] [PubMed]
- Dotan, A.; Muller, S.; Kanduc, D.; David, P.; Halpert, G.; Shoenfeld, Y. The SARS-CoV-2 as an instrumental trigger of autoimmunity. Autoimmun. Rev. 2021, 20. [Google Scholar] [CrossRef] [PubMed]
- Dotan, A.; Mahroum, N.; Bogdanos, D.P.; Shoenfeld, Y. COVID-19 as an infectome paradigm of autoimmunity. J. Allergy Clin. Immunol. 2022, 149, 63–64. [Google Scholar] [CrossRef] [PubMed]
- Baiocchi, G.C.; Vojdani, A.; Rosenberg, A.Z.; Vojdani, E.; Halpert, G.; Ostrinski, Y.; Zyskind, I.; Filgueiras, I.S.; Schimke, L.F.; Marques, A.H.C.; et al. Cross-sectional analysis reveals autoantibody signatures associated with COVID-19 severity. J. Med. Virol. 2023, 95. [Google Scholar] [CrossRef] [PubMed]
- Lavi, Y.; Vojdani, A.; Halpert, G.; Sharif, K.; Ostrinski, Y.; Zyskind, I.; Lattin, M.T.; Zimmerman, J.; Silverberg, J.I.; Rosenberg, A.Z.; et al. Dysregulated Levels of Circulating Autoantibodies against Neuronal and Nervous System Autoantigens in COVID-19 Patients. Diagnostics 2023, Vol. 13, Page 687 2023, 13, 687. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.; Yen-Ting Chen, T.; Wang, S.I.; Hung, Y.M.; Chen, H.Y.; Wei, C.C.J. Risk of autoimmune diseases in patients with COVID-19: A retrospective cohort study. EClinicalMedicine 2023, 56. [Google Scholar] [CrossRef] [PubMed]
- Miyadera, H.; Tokunaga, K. Associations of human leukocyte antigens with autoimmune diseases: challenges in identifying the mechanism. J. Hum. Genet. 2015, 60, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Jara, L.J.; Vera-Lastra, O.; Mahroum, N.; Pineda, C.; Shoenfeld, Y. Autoimmune post-COVID vaccine syndromes: does the spectrum of autoimmune/inflammatory syndrome expand? Clin. Rheumatol. 2022, 41, 1603–1609. [Google Scholar] [CrossRef] [PubMed]
- Dotan, A.; David, P.; Arnheim, D.; Shoenfeld, Y. The autonomic aspects of the post-COVID19 syndrome. Autoimmun. Rev. 2022, 21. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.K.; Guandalini, S.; Semrad, C.; Kupfer, S.S. A Clinician’s Guide to Celiac Disease HLA Genetics. Am. J. Gastroenterol. 2019, 114, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Del Pozzo, G.; Farina, F.; Picascia, S.; Laezza, M.; Vitale, S.; Gianfrani, C. HLA class II genes in precision-based care of childhood diseases: what we can learn from celiac disease. Pediatr. Res. 2021, 89, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Greco, N.; Meacci, A.; Mora, B.; Vestri, A.; Picarelli, A. Coeliac disease in the COVID-19 pandemic: does HLA have a protective effect? Ann. Med. 2022, 54, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, H.; Oka, S.; Tsuchiya, N.; Shimada, K.; Hashimoto, A.; Tohma, S.; Kawasaki, A. The role of common protective alleles HLA-DRB1*13 among systemic autoimmune diseases. Genes Immun. 2017, 18, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt, A.; Carvalho, C.; Leal, B.; Brás, S.; Lopes, D.; Martins Da Silva, A.; Santos, E.; Torres, T.; Almeida, I.; Farinha, F.; et al. The Protective Role of HLA-DRB1(∗)13 in Autoimmune Diseases. J. Immunol. Res. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Rabaan, A.A.; Mutair, A. Al; Aljeldah, M.; Shammari, B.R.A.; Sulaiman, T.; Alshukairi, A.N.; Alfaresi, M.; Al-Jishi, J.M.; Al Bati, N.A.; Al-Mozaini, M.A.; et al. Genetic Variants and Protective Immunity against SARS-CoV-2. Genes (Basel). 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Lani, R.; Senin, N.A.; AbuBakar, S.; Hassandarvish, P. Knowledge of SARS-CoV-2 Epitopes and Population HLA Types Is Important in the Design of COVID-19 Vaccines. Vaccines 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Troshina, E.; Yukina, M.; Nuralieva, N.; Vasilyev, E.; Rebrova, O.; Akhmatova, R.; Ikonnikova, A.; Savvateeva, E.; Gryadunov, D.; Melnichenko, G.; et al. Association of Alleles of Human Leukocyte Antigen Class II Genes and Severity of COVID-19 in Patients of the “Red Zone” of the Endocrinology Research Center, Moscow, Russia. Dis. (Basel, Switzerland) 2022, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Doño, S.; Sánchez-González, R.A.; Trujillo-Vizuet, M.G.; Zamudio-Castellanos, F.Y.; García-Silva, R.; Bulos-Rodríguez, P.; Vazquez-Guzmán, C.A.; Cárdenas-Ramos, X.; de León Rodríguez, D.; Elías, F.; et al. Protective HLA alleles against severe COVID-19: HLA-A*68 as an ancestral protection allele in Tapachula-Chiapas, Mexico. Clin. Immunol. 2022, 238. [Google Scholar] [CrossRef] [PubMed]
- Mocci, S.; Littera, R.; Tranquilli, S.; Provenzano, A.; Mascia, A.; Cannas, F.; Lai, S.; Giuressi, E.; Chessa, L.; Angioni, G.; et al. A Protective HLA Extended Haplotype Outweighs the Major COVID-19 Risk Factor Inherited From Neanderthals in the Sardinian Population. Front. Immunol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Suslova, T.A.; Vavilov, M.N.; Belyaeva, S. V.; Evdokimov, A. V.; Stashkevich, D.S.; Galkin, A.; Kofiadi, I.A. Distribution of HLA-A, -B, -C, -DRB1, -DQB1, -DPB1 allele frequencies in patients with COVID-19 bilateral pneumonia in Russians, living in the Chelyabinsk region (Russia). Hum. Immunol. 2022, 83, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Deb, P.; Zannat, K. e.; Talukder, S.; Bhuiyan, A.H.; Jilani, M.S.A.; Saif-Ur-Rahman, K.M. Association of HLA gene polymorphism with susceptibility, severity, and mortality of COVID-19: A systematic review. HLA 2022, 99, 281–312. [Google Scholar] [CrossRef] [PubMed]
- Bubnova, L.; Pavlova, I.; Terentieva, M.; Glazanova, T.; Belyaeva, E.; Sidorkevich, S.; Bashketova, N.; Chkhingeria, I.; Kozhemyakina, M.; Azarov, D.; et al. HLA Genotypes in Patients with Infection Caused by Different Strains of SARS-CoV-2. Int. J. Environ. Res. Public Health 2022, 19. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.H.L.; Lau, K.M.; Li, L.; Cheng, S.H.; Chan, W.Y.; Hui, P.K.; Zee, B.; Leung, C.B.; Sung, J.J.Y. Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J. Infect. Dis. 2004, 190, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; David, J.K.; Maden, S.K.; Wood, M.A.; Weeder, B.R.; Nellore, A.; Thompson, R.F. Human leukocyte antigen susceptibility map for SARS-CoV-2. J. Virol. 2020, 94, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tavasolian, F.; Rashidi, M.; Hatam, G.R.; Jeddi, M.; Hosseini, A.Z.; Mosawi, S.H.; Abdollahi, E.; Inman, R.D. HLA, Immune Response, and Susceptibility to COVID-19. Front. Immunol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Ben Shachar, S.; Barda, N.; Manor, S.; Israeli, S.; Dagan, N.; Carmi, S.; Balicer, R.; Zisser, B.; Louzoun, Y. MHC Haplotyping of SARS-CoV-2 Patients: HLA Subtypes Are Not Associated with the Presence and Severity of COVID-19 in the Israeli Population. J. Clin. Immunol. 2021, 41, 1154–1161. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A. The last two millennias echo-catastrophes are the driving forces for the potential genetic advantage mechanisms in celiac disease. Med. Hypotheses 2011, 77, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A. Balanced polymorphism: a survival advantage in celiac disease. Med. Hypotheses 2011, 77, 1–2. [Google Scholar] [CrossRef] [PubMed]
- King, J.A.; Jeong, J.; Underwood, F.E.; Quan, J.; Panaccione, N.; Windsor, J.W.; Coward, S.; Debruyn, J.; Ronksley, P.E.; Shaheen, A.A.; et al. Incidence of Celiac Disease Is Increasing Over Time: A Systematic Review and Meta-analysis. Am. J. Gastroenterol. 2020, 115, 507–525. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.; Jeremias, P.; Matthias, T. the World Incidence of Celiac Disease Is Increasing: a Review. Int. J. Recent Sci. Res. 2015, 6, 5491–5496. [Google Scholar]
- Hadley, D.; Hagopian, W.; Liu, E.; She, J.X.; Simell, O.; Akolkar, B.; Ziegler, A.G.; Rewers, M.; Krischer, J.P.; Chen, W.M.; et al. HLA-DPB1*04:01 Protects Genetically Susceptible Children from Celiac Disease Autoimmunity in the TEDDY Study. Am. J. Gastroenterol. 2015, 110, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Balamtekin, N.; Baysoy, G.; Tan, Ç.; Kızılkan, N.U.; Demir, H.; Temizel, İ.N.S.; Özen, H.; Yüce, A.; Tezcan, İ.; Gürakan, F. The HLA groups and their relationship with clinical features in Turkish children and adolescents with celiac disease. Turk. J. Pediatr. 2021, 63, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Del Prado, M.Y.; Olivares López, J.L.; Lázaro Almarza, A.; Lasierra Díaz, M.P. HLA system. Phenotypic and gene frequencies in celiac and healthy subjects from the same geographical area. Rev. Esp. Enfermedades Dig. 2001, 93, 110–113. [Google Scholar]
- Silva, E.M.B.T.; Fernandes, M.I.M.; Galvão, L.C.; Sawamura, R.; Donadi, E.A. Human Leukocyte Antigen Class II Alleles in White Brazilian Patients With Celiac Disease. J. Pediatr. Gastroenterol. Nutr. 2000, 31, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Laadhar, L.; Toumi, A.; Kallel-Sellami, M.; Zitouni, M.; Bouraoui, S.; Maherzi, A.; Makni, S.; Ben Hariz, M. HLA class II polymorphism in children with coeliac disease in Tunisia: is there any influence on clinical manifestation? Eur. J. Gastroenterol. Hepatol. 2009, 21, 1286–1290. [Google Scholar] [CrossRef] [PubMed]
- Boy, M.F.; Balestrieri, A.; Cherchi, M. V.; Usai, P.; La Nasa, G. Distribution of HLA-DPBL, -DQB1 -DQA1 Alleles Among Sardinian Celiac Patients. Dis. Markers 1994, 12, 199–204. [Google Scholar] [CrossRef]
- Lopez-Vazquez, A. MHC class I chain related gene A (MICA) modulates the development of coeliac disease in patients with the high risk heterodimer DQA1*0501/DQB1*0201. Gut 2002, 50, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Bilbao, J.R.; Martín-Pagola, A.; Vitoria, J.C.; Zubillaga, P.; Ortiz, L.; Castaño, L. HLA-DRB1 and MHC class 1 chain-related A haplotypes in Basque families with celiac disease. Tissue Antigens 2002, 60, 71–76. [Google Scholar] [CrossRef] [PubMed]
- BILBAO, J.R.; MARTÍN-PAGOLA, A.; PÉREZ DE NANCLARES, G.; CALVO, B.; VITORIA, J.C.; VÁZQUEZ, F.; CASTAÑO, L. HLA-DRB1 and MICA in Autoimmunity. Ann. N. Y. Acad. Sci. 2003, 1005, 314–318. [Google Scholar] [CrossRef]
- Bravo, F.P.-; Araya, M.; Mondragón, A.; Rı́os, G.; Alarcón, T.; Roessler, J..; Santos, J.. Genetic differences in HLA-DQA1∗ and DQB1∗ allelic distributions between celiac and control children in Santiago, Chile. Hum. Immunol. 1999, 60, 262–267. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T.; Wusterhausen, P. Autoimmunity in celiac disease: Extra-intestinal manifestations. Autoimmun. Rev. 2019, 18, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.; Matthias, T. Extra intestinal manifestations of CD: Common pathways in the gut- remote organs’ axes. Int. J. Celiac Dis. 2017, 5, 24–27. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T. GUT-the Trojan Horse in remote organs’ Autoimmunity. J. Clin. Cell. Immunol. 2016, 7, 1–10. [Google Scholar] [CrossRef]
- Catassi, C.; Verdu, E.F.; Bai, J.C.; Lionetti, E. Coeliac disease. Lancet (London, England) 2022, 399, 2413–2426. [Google Scholar] [CrossRef]
- Lerner, A. New therapeutic strategies for celiac disease. Autoimmun. Rev. 2010, 9, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Ghazanfar, H.; Kandhi, S.; Shin, D.; Muthumanickam, A.; Gurjar, H.; Qureshi, Z.A.; Shaban, M.; Farag, M.; Haider, A.; Budhathoki, P.; et al. Impact of COVID-19 on the Gastrointestinal Tract: A Clinical Review. Cureus 2022. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A. Are my patients with celiac disease at higher risk of COVID-19 virus. Int. J. Celiac Dis. 2020, 8, 35–38. [Google Scholar] [CrossRef]
- Lerner, A. Covid-19 and the human gut: A new runner on the tract. Int. J. Celiac Dis. 2020, 8, 64–67. [Google Scholar] [CrossRef]
- Lerner, A. The COVID-19 vaccination debate: CoV-2 in celiac disease: A Pathogen or just along for the ride? Int. J. Celiac Dis. 2021, 9, 6–9. [Google Scholar] [CrossRef]
- Samasca, G.; Lerner, A. Celiac disease in the COVID-19 pandemic. J. Transl. Autoimmun. 2021, 4. [Google Scholar] [CrossRef]
- Clerbaux, L.-A.; Mayasich, S.A.; Muñoz, A.; Soares, H.; Petrillo, M.; Albertini, M.C.; Lanthier, N.; Grenga, L.; Amorim, M.-J. Gut as an Alternative Entry Route for SARS-CoV-2: Current Evidence and Uncertainties of Productive Enteric Infection in COVID-19. J. Clin. Med. 2022, 11, 5691. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.; McCarty, M.F. The aging bowel dysfunction and elderly vulnerability towards COVID-19 infection. Life (Basel, Switzerland) 2021, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- McCarty, M.F.; Lerner, A. Perspective: Prospects for Nutraceutical Support of Intestinal Barrier Function. Adv. Nutr. 2021, 12, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Lian, J.S.; Hu, J.H.; Gao, J.; Zheng, L.; Zhang, Y.M.; Hao, S.R.; Jia, H.Y.; Cai, H.; Zhang, X.L.; et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 2020, 69, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.W.; Wu, X.X.; Jiang, X.G.; Xu, K.J.; Ying, L.J.; Ma, C.L.; Li, S.B.; Wang, H.Y.; Zhang, S.; Gao, H.N.; et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: Retrospective case series. BMJ 2020, 368. [Google Scholar] [CrossRef] [PubMed]
- FANG, D.; MA, J.; GUAN, J.; WANG, M.; SONG, Y.; TIAN, D.; LI, P. Manifestations of Digestive system in hospitalized patients with novel coronavirus pneumonia in Wuhan, China: a single-center, descriptive study. Chinese J. Dig. 2020, E005–E005. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, Z.; Gong, H.; Xu, D.; Wang, J.; Li, Z.; Cui, X.; Xiao, J.; Meng, T.; Zhou, W.; et al. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxiv 2020, 2020.01.30.927806. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H.B.; Lyu, J.R.; Lei, X.M.; Li, W.; Wu, G.; Lyu, J.; Dai, Z.M. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int. J. Infect. Dis. 2020, 96, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Rong, L.; Nian, W.; He, Y. Review article: gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment. Pharmacol. Ther. 2020, 51, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, R.S.; Qu, G.Q. Gross Examination Report of a COVID-19 Death Autopsy. J. Forensic Med. 2020, 36, 21–23. [Google Scholar] [CrossRef]
- YANG, Z.; LI, G.; DAI, X.; LIU, G.; LI, G.; JIE, Y. Three cases of novel coronavirus pneumonia with viral nucleic acids still positive in stool after throat swab detection turned negative. Chinese J. Dig. 2020, E002–E002. [Google Scholar] [CrossRef]
- Zhang, J.C.; Wang, S. Bin; Xue, Y.D. Fecal specimen diagnosis 2019 novel coronavirus–infected pneumonia. J. Med. Virol. 2020, 92, 680–682. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Taki, K.; Gahlot, R.; Sharma, A.; Dhangar, K. A chronicle of SARS-CoV-2: Part-I-Epidemiology, diagnosis, prognosis, transmission and treatment. 2020. [CrossRef]
- Natarajan, A.; Zlitni, S.; Brooks, E.F.; Vance, S.E.; Dahlen, A.; Hedlin, H.; Park, R.M.; Han, A.; Schmidtke, D.T.; Verma, R.; et al. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med (New York, N.Y.) 2022, 3, 371–387.e9. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.L.; Baluja, M.Q.; Graham, D.W.; Corbishley, A.; McDonald, J.E.; Malham, S.K.; Hillary, L.S.; Connor, T.R.; Gaze, W.H.; Moura, I.B.; et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020, 749. [Google Scholar] [CrossRef] [PubMed]
- Lavania, M.; Joshi, M.S.; Ranshing, S.S.; Potdar, V.A.; Shinde, M.; Chavan, N.; Jadhav, S.M.; Sarkale, P.; Mohandas, S.; Sawant, P.M.; et al. Prolonged shedding of SARS-CoV-2 in feces of COVID-19 positive patients: Trends in genomic variation in first and second wave. Front. Med. 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- Lavania, M.; Joshi, M.S.; Ranshing, S.S.; Potdar, V.A.; Shinde, M.; Chavan, N.; Jadhav, S.M.; Sarkale, P.; Mohandas, S.; Sawant, P.M.; et al. Prolonged Shedding of SARS-CoV-2 in Feces of COVID-19 Positive Patients: Trends in Genomic Variation in First and Second Wave. Front. Med. 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- Gandhi P, A.; Singh, T. Feco-Oral Transmission of SARS-CoV-2. Asia-Pacific J. public Heal. 2020, 32, 370. [Google Scholar] [CrossRef] [PubMed]
- Targoński, R.; Gąsecka, A.; Prowancki, A.; Targoński, R. An alternative to airborne droplet transmission route of SARS-CoV-2, the feco-oral route, as a factor shaping COVID-19 pandemic. Med. Hypotheses 2022, 166. [Google Scholar] [CrossRef] [PubMed]
- Foladori, P.; Cutrupi, F.; Segata, N.; Manara, S.; Pinto, F.; Malpei, F.; Bruni, L.; La Rosa, G. SARS-CoV-2 from faeces to wastewater treatment: What do we know? A review. Sci. Total Environ. 2020, 743. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, S.; Levy, J.I.; De Hoff, P.; Humphrey, G.; Birmingham, A.; Jepsen, K.; Farmer, S.; Tubb, H.M.; Valles, T.; Tribelhorn, C.E.; et al. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature 2022, 609, 101–108. [Google Scholar] [CrossRef]
- Schiepatti, A.; Alimenti, E.; Maimaris, S.; Nicolardi, M.L.; La Barbera, F.M.; Baiardi, P.; Biagi, F. Prevalence, incidence and clinical features of SARS-CoV-2 infection in adult coeliac patients. Eur. J. Gastroenterol. Hepatol. 2021, 33, 1361–1366. [Google Scholar] [CrossRef]
- Gokden, Y.; Hot, S.; Adas, M.; Ogutmen Koc, D.; Atak, S.; Hot, A.B. Celiac disease and COVID-19 pandemic: Should we worry? Acta Gastroenterol. Belg. 2020, 83, 517–525. [Google Scholar] [PubMed]
- Gholam-Mostafaei, F.S.; Asri, N.; Parvani, N.; khamene, E.A.; Barzegar, F.; Rostami-Nejad, M.; Rezaei-Tavirani, M.; Shahbazkhani, B.; Jahani-Sherafat, S.; Rostami, K.; et al. Prevalence and outcome of COVID-19 among Iranian celiac patients. Gastroenterol. Hepatol. from Bed to Bench 2022, 15, 153–157. [Google Scholar] [CrossRef]
- Asri, N.; Nazemalhosseini Mojarad, E.; Mirjalali, H.; Mohebbi, S.R.; Baghaei, K.; Rostami-Nejad, M.; Yadegar, A.; Rezaei-Tavirani, M.; Asadzadeh Aghdaei, H.; Rostami, K.; et al. Toward finding the difference between untreated celiac disease and COVID-19 infected patients in terms of CD4, CD25 (IL-2 Rα), FOXP3 and IL-6 expressions as genes affecting immune homeostasis. BMC Gastroenterol. 2021, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tian, A.; Yang, D.; Zhang, M.; Chen, L.; Wen, J.; Chen, P. Celiac Disease and the Susceptibility of COVID-19 and the Risk of Severe COVID-19: A Mendelian Randomization Study. Clin. Transl. Gastroenterol. 2022, 13, e00480. [Google Scholar] [CrossRef] [PubMed]
- Zhen, J.; Stefanolo, J.P.; Temprano, M. de la P.; Tedesco, S.; Seiler, C.; Caminero, A.F.; de-Madaria, E.; Huguet, M.M.; Vivas, S.; Niveloni, S.I.; et al. The Risk of Contracting COVID-19 Is Not Increased in Patients With Celiac Disease. Clin. Gastroenterol. Hepatol. 2021, 19, 391–393. [Google Scholar] [CrossRef] [PubMed]
- Hadi, Y.B.; Sohail, A.H.; Lakhani, D.A.; Naqvi, S.F.; Kupec, J.T.; Pervez, A. Outcomes of SARS-CoV-2 infection in patients with celiac disease: a multicenter research network study. Ann. Gastroenterol. 2022, 35, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Rathore, S.S.; Velasquez-Botero, F.; Nieto-Salazar, M.A.; Flowers, T.C.; Hasan, J.; Parashar, A.K.; Tanveer, K.; Aneis, H.; Buremoh, A.I.; Yusuf, K.; et al. Prevalence and clinical outcomes of COVID-19 in patients with pre-existing celiac disease: A systematic review and meta-analysis. Rev. Med. Virol. 2023, 33. [Google Scholar] [CrossRef] [PubMed]
- Crocco, M.; Calvi, A.; Canzoneri, F.; Malerba, F.; Zampatti, N.; Chiaro, A.; Arrigo, S.; Gandullia, P.; Proietti, S.; Bonassi, S. The Influence of SARS-CoV-2 Pandemic on the Diagnosis of Celiac Disease and Clinical Practice in Pediatric Gastroenterology. Nutrients 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Elli, L.; Barisani, D.; Vaira, V.; Bardella, M.T.; Topa, M.; Vecchi, M.; Doneda, L.; Scricciolo, A.; Lombardo, V.; Roncoroni, L. How to manage celiac disease and gluten-free diet during the COVID-19 era: proposals from a tertiary referral center in a high-incidence scenario. BMC Gastroenterol. 2020, 20, 387. [Google Scholar] [CrossRef]
- Monzani, A.; Lionetti, E.; Felici, E.; Fransos, L.; Azzolina, D.; Rabbone, I.; Catassi, C. Adherence to the Gluten-Free Diet during the Lockdown for COVID-19 Pandemic: A Web-Based Survey of Italian Subjects with Celiac Disease. Nutrients 2020, 12, 3467. [Google Scholar] [CrossRef]
- Catassi, C.; Verdu, E.F.; Bai, J.C.; Lionetti, E. Coeliac disease. Lancet 2022, 399, 2413–2426. [Google Scholar] [CrossRef] [PubMed]
- Catassi, G.N.; Vallorani, M.; Cerioni, F.; Lionetti, E.; Catassi, C. A negative fallout of COVID-19 lockdown in Italy: Life-threatening delay in the diagnosis of celiac disease. Dig. Liver Dis. 2020, 52, 1092–1093. [Google Scholar] [CrossRef] [PubMed]
- Cakir, M.; Guven, B.; Issi, F.; Ozkaya, E. New-onset celiac disease in children during COVID-19 pandemic. Acta Paediatr. 2022, 111, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, Y.; Prévost, J.; Ullah, I.; Lu, M.; Gong, S.Y.; Tauzin, A.; Gasser, R.; Vézina, D.; Anand, S.P.; et al. Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern. Cell Rep. 2022, 38. [Google Scholar] [CrossRef] [PubMed]
- Trovato, C.M.; Montuori, M.; Pietropaoli, N.; Oliva, S. COVID-19 and celiac disease: A pathogenetic hypothesis for a celiac outbreak. Int. J. Clin. Pract. 2021, 75. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Faiq, M.A.; Pareek, V.; Raza, K.; Narayan, R.K.; Prasoon, P.; Kumar, P.; Kulandhasamy, M.; Kumari, C.; Kant, K.; et al. Relevance of SARS-CoV-2 related factors ACE2 and TMPRSS2 expressions in gastrointestinal tissue with pathogenesis of digestive symptoms, diabetes-associated mortality, and disease recurrence in COVID-19 patients. Med. Hypotheses 2020, 144. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Fleri, W.; Paul, S.; Dhanda, S.K.; Mahajan, S.; Xu, X.; Peters, B.; Sette, A. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Tarke, A.; Sidney, J.; Kidd, C.K.; Dan, J.M.; Ramirez, S.I.; Yu, E.D.; Mateus, J.; da Silva Antunes, R.; Moore, E.; Rubiro, P.; et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Reports Med. 2021, 2. [Google Scholar] [CrossRef]
- Verhagen, J.; van der Meijden, E.D.; Lang, V.; Kremer, A.E.; Völkl, S.; Mackensen, A.; Aigner, M.; Kremer, A.N. Human CD4+ T cells specific for dominant epitopes of SARS-CoV-2 Spike and Nucleocapsid proteins with therapeutic potential. Clin. Exp. Immunol. 2021, 205, 363–378. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50. [Google Scholar] [CrossRef]
- Rice, P.; Longden, L.; Bleasby, A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Mateus, J.; Grifoni, A.; Tarke, A.; Sidney, J.; Ramirez, S.I.; Dan, J.M.; Burger, Z.C.; Rawlings, S.A.; Smith, D.M.; Phillips, E.; et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science 2020, 370. [Google Scholar] [CrossRef] [PubMed]
- Aboulaghras, S.; Piancatelli, D.; Taghzouti, K.; Balahbib, A.; Alshahrani, M.M.; Al Awadh, A.A.; Goh, K.W.; Ming, L.C.; Bouyahya, A.; Oumhani, K. Meta-Analysis and Systematic Review of HLA DQ2/DQ8 in Adults with Celiac Disease. Int. J. Mol. Sci. 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.B.; Kaabinejadian, S.; Yari, H.; Peters, B.; Barra, C.; Gragert, L.; Hildebrand, W.; Nielsen, M. Machine learning reveals limited contribution of trans-only encoded variants to the HLA-DQ immunopeptidome. Commun. Biol. 2023, 6. [Google Scholar] [CrossRef] [PubMed]
- Obermair, F.J.; Renoux, F.; Heer, S.; Lee, C.H.; Cereghetti, N.; Loi, M.; Maestri, G.; Haldner, Y.; Wuigk, R.; Iosefson, O.; et al. High-resolution profiling of MHC II peptide presentation capacity reveals SARS-CoV-2 CD4 T cell targets and mechanisms of immune escape. Sci. Adv. 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Le Bert, N.; Tan, A.T.; Kunasegaran, K.; Tham, C.Y.L.; Hafezi, M.; Chia, A.; Chng, M.H.Y.; Lin, M.; Tan, N.; Linster, M.; et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 2020, 584, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Sciurti, M.; Fornaroli, F.; Gaiani, F.; Bonaguri, C.; Leandro, G.; Di Mario, F.; De’angelis, G.L. Genetic susceptibilty and celiac disease: What role do HLA haplotypes play? Acta Biomed. 2018, 89, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Sahin, Y.; Mermer, S. Frequency of celiac disease and distribution of HLA-DQ2/DQ8 haplotypes among siblings of children with celiac disease. World J. Clin. Pediatr. 2022, 11, 351–359. [Google Scholar] [CrossRef]
- Megiorni, F.; Pizzuti, A. HLA-DQA1 and HLA-DQB1 in Celiac disease predisposition: practical implications of the HLA molecular typing. J. Biomed. Sci. 2012, 19. [Google Scholar] [CrossRef]
- Oliveira-Cortez, A.; Melo, A.C.; Chaves, V.E.; Condino-Neto, A.; Camargos, P. Do HLA class II genes protect against pulmonary tuberculosis? A systematic review and meta-analysis. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1567–1580. [Google Scholar] [CrossRef] [PubMed]
- Walker-Sperlin, V.; Digitale, J.C.; Viard, M.; Martin, M.P.; Bashirova, A.; Yuki, Y.; Ramsuran, V.; Kulkarni, S.; Naranbhai, V.; Li, H.; et al. Genetic variation that determines TAPBP expression levels associates with the course of malaria in an HLA allotype-dependent manner. Proc. Natl. Acad. Sci. U. S. A. 2022, 119. [Google Scholar] [CrossRef]
- Zhang, Y.; Chikata, T.; Kuse, N.; Murakoshi, H.; Gatanaga, H.; Oka, S.; Takiguchi, M. Immunological Control of HIV-1 Disease Progression by Rare Protective HLA Allele. J. Virol. 2022, 96. [Google Scholar] [CrossRef] [PubMed]
- Crux, N.B.; Elahi, S. Human Leukocyte Antigen (HLA) and Immune Regulation: How Do Classical and Non-Classical HLA Alleles Modulate Immune Response to Human Immunodeficiency Virus and Hepatitis C Virus Infections? Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Ou, G.; Liu, X.; Xu, H.; Ji, X.; Liu, X.; Wang, J. Variation and expression of HLA-DPB1 gene in HBV infection. Immunogenetics 2021, 73, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Castelli, E.C.; de Castro, M. V.; Naslavsky, M.S.; Scliar, M.O.; Silva, N.S.B.; Pereira, R.N.; Ciriaco, V.A.O.; Castro, C.F.B.; Mendes-Junior, C.T.; Silveira, E. de S.; et al. MUC22, HLA-A, and HLA-DOB variants and COVID-19 in resilient super-agers from Brazil. Front. Immunol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- SHAKOOR, S.; ISMAIL, A.; SABRAN, M.R.; MOHTARRUDIN, N.; KAKA, U.; NADEEM, M. In-vivo study of synthetic and natural food colors effect on biochemical and immunity parameters. Food Sci. Technol. 2021. [Google Scholar] [CrossRef]
- Langton, D.J.; Bourke, S.C.; Lie, B.A.; Reiff, G.; Natu, S.; Darlay, R.; Burn, J.; Echevarria, C. The influence of HLA genotype on the severity of COVID-19 infection. HLA 2021, 98, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Augusto, D.G.; Hollenbach, J.A. HLA variation and antigen presentation in COVID-19 and SARS-CoV-2 infection. 2022, 6, 1–8.
- Augusto, D.G.; Murdolo, L.D.; Chatzileontiadou, D.S.M.; Sabatino, J.J.; Yusufali, T.; Peyser, N.D.; Butcher, X.; Kizer, K.; Guthrie, K.; Murray, V.W.; et al. A common allele of HLA is associated with asymptomatic SARS-CoV-2 infection. Nature 2023, 620, 128–136. [Google Scholar] [CrossRef]
- Barquera, R.; Collen, E.; Di, D.; Buhler, S.; Teixeira, J.; Llamas, B.; Nunes, J.M.; Sanchez-Mazas, A. Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA 2020, 96, 277–298. [Google Scholar] [CrossRef]
- Purbey, P.K.; Roy, K.; Gupta, S.; Paul, M.K. Mechanistic insight into the protective and pathogenic immune-responses against SARS-CoV-2. Mol. Immunol. 2023, 156, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Buckley, P.R.; Lee, C.H.; Pereira Pinho, M.; Ottakandathil Babu, R.; Woo, J.; Antanaviciute, A.; Simmons, A.; Ogg, G.; Koohy, H. HLA-dependent variation in SARS-CoV-2 CD8 + T cell cross-reactivity with human coronaviruses. Immunology 2022, 166, 78–103. [Google Scholar] [CrossRef] [PubMed]
- Karnaukhov, V.; Paes, W.; Woodhouse, I.B.; Partridge, T.; Nicastri, A.; Brackenridge, S.; Shcherbinin, D.; Chudakov, D.M.; Zvyagin, I. V.; Ternette, N.; et al. HLA variants have different preferences to present proteins with specific molecular functions which are complemented in frequent haplotypes. Front. Immunol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.S.; Lee, Y.H.; Jo, H.A.; Baek, I.C.; Kim, S.M.; Sohn, H.J.; Kim, T.G. Comprehensive Analysis of CD4+ T Cell Response Cross-Reactive to SARS-CoV-2 Antigens at the Single Allele Level of HLA Class II. Front. Immunol. 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Naemi, F.M.A.; Al-adwani, S.; Al-khatabi, H.; Al-nazawi, A. Association between the HLA genotype and the severity of COVID-19 infection among South Asians. J. Med. Virol. 2021, 93, 4430–4437. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Bautista, J.F.; Sampedro, A.; Gómez-Vicente, E.; Rodríguez-Granger, J.; Reguera, J.A.; Cobo, F.; Ruiz-Cabello, F.; López-Nevot, M.Á. HLA Class II Polymorphism and Humoral Immunity Induced by the SARS-CoV-2 mRNA-1273 Vaccine. Vaccines 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Vojdani, A.; Vojdani, E.; Melgar, A.L.; Redd, J. Reaction of SARS-CoV-2 antibodies with other pathogens, vaccines, and food antigens. Front. Immunol. 2022, 13, 5254. [Google Scholar] [CrossRef]
- Ellis, R.J.; Varela-Calvino, R.; Tree, T.I.M.; Peakman, M. HLA Class II molecules on haplotypes associated with type 1 diabetes exhibit similar patterns of binding affinities for coxsackievirus P2C peptides. Immunology 2005, 116, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Adli, A.; Rahimi, M.; Khodaie, R.; Hashemzaei, N.; Hosseini, S.M. Role of genetic variants and host polymorphisms on COVID-19: From viral entrance mechanisms to immunological reactions. J. Med. Virol. 2022, 94, 1846–1865. [Google Scholar] [CrossRef]
- David, J.; Mody, B. Potential selective advantage mechanism for polymorphic genetics in celiac disease. Med. Hypotheses 2011, 77, 3–4. [Google Scholar] [CrossRef]
- Fischer, J.C.; Schmidt, A.G.; Bölke, E.; Uhrberg, M.; Keitel, V.; Feldt, T.; Jensen, B.; Häussinger, D.; Adams, O.; Schneider, E.M.; et al. Association of HLA genotypes, AB0 blood type and chemokine receptor 5 mutant CD195 with the clinical course of COVID-19. Eur. J. Med. Res. 2021, 26. [Google Scholar] [CrossRef] [PubMed]
- Littera, R.; Campagna, M.; Deidda, S.; Angioni, G.; Cipri, S.; Melis, M.; Firinu, D.; Santus, S.; Lai, A.; Porcella, R.; et al. Human Leukocyte Antigen Complex and Other Immunogenetic and Clinical Factors Influence Susceptibility or Protection to SARS-CoV-2 Infection and Severity of the Disease Course. The Sardinian Experience. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Parker, R.; Partridge, T.; Wormald, C.; Kawahara, R.; Stalls, V.; Aggelakopoulou, M.; Parker, J.; Powell Doherty, R.; Ariosa Morejon, Y.; Lee, E.; et al. Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells. Cell Rep. 2021, 35. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Bautista, J.F.; Rodriguez-Nicolas, A.; Rosales-Castillo, A.; López-Ruz, M.Á.; Martín-Casares, A.M.; Fernández-Rubiales, A.; Anderson, P.; Garrido, F.; Ruiz-Cabello, F.; López-Nevot, M.Á. Study of HLA-A, -B, -C, -DRB1 and -DQB1 polymorphisms in COVID-19 patients. J. Microbiol. Immunol. Infect. 2022, 55, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Copley, H.C.; Gragert, L.; Leach, A.R.; Kosmoliaptsis, V. Influence of HLA Class II Polymorphism on Predicted Cellular Immunity Against SARS-CoV-2 at the Population and Individual Level. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Greenhalgh, T.; Sivan, M.; Delaney, B.; Evans, R.; Milne, R. Long covid-an update for primary care. BMJ 2022, 378. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Piazza, M.; Di Cicco, M.; Pecoraro, L.; Ghezzi, M.; Peroni, D.; Comberiati, P. Long COVID-19 in Children: From the Pathogenesis to the Biologically Plausible Roots of the Syndrome. Biomolecules 2022, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lerner, A.; O’Bryan, T.; Matthias, T. Navigating the Gluten-Free Boom: The Dark Side of Gluten Free Diet. Front. Pediatr. 2019, 7. [Google Scholar] [CrossRef]
- Lerner, A.; Matthias, T. Gluten-free diet tough alley in torrid time. Int. J. Celiac Dis. 2017, 5, 50–55. [Google Scholar] [CrossRef]
- Vojdani, A.; Vojdani, E.; Saidara, E.; Maes, M. Persistent SARS-CoV-2 Infection, EBV, HHV-6 and Other Factors May Contribute to Inflammation and Autoimmunity in Long COVID. Viruses 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Shariati, A.; Aslani, H.R.; Shayesteh, M.R.H.; Taghipour, A.; Nasser, A.; Safari, H.; Alizade-Sani, M.; Dehghan, A.; Azimi, T. Are Viruses and Parasites Linked to Celiac Disease? A Question that Still has no Definite Answer. Curr. Pharm. Biotechnol. 2019, 20, 1181–1193. [Google Scholar] [CrossRef] [PubMed]
- Calabretto, M.; Di Carlo, D.; Falasca, F.; Mazzuti, L.; Meacci, A.; Donato, G.; Greco, N.; Mezzatesta, L.; Morrone, A.; Turriziani, O.; et al. Analysis of viral nucleic acids in duodenal biopsies from adult patients with celiac disease. Eur. J. Gastroenterol. Hepatol. 2022, 34, 1107–1110. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef] [PubMed]
| Protective HLA alleles | Country | References |
|---|---|---|
| HLA-DPB1*04:01 | Germany, Finland, Sweden, and the United States | [42] |
| HLA C14, DR11, DR15, DQ3 | Turkey | [43] |
| Cw4 and DQ1 | Spain | [44] |
| HLADQB1*06 | Brazil | [45] |
| DRB1*13, DQA1*0102, DQB1*06 | Tunisia | [46] |
| DQB1*0502/DQA1*0102 | Sardinia | [47] |
| MICA-A5.1 | Spain | [48] |
| MICA-A9 | Basque country, Spain | [49,50] |
| DQA1*0101, DQA1*0201, DQB1*0301 | Chile | [51] |
| Gastrointestinal aspects | Enteric features | References |
|---|---|---|
| Symptoms | Diarrhea, nausea, vomiting | [65,66,67] |
| ACE2 expression | All along the GIT | [68,69] |
| Inflammation and damage | Lymphocytic infiltration, edema, necrosis, degeneration, cellular shedding | [70,71] |
| Viral particles, nucleocapsid proteins | In the stomach, duodenum and colonic cells | [70,71,72,73] |
| Stool shading | SARS-CoV-2 RNA in stool | [74,75,76,77] |
| Gut replication | SARS-CoV-2 multiplication in the gut | [76] |
| Feco-Oral Transmission | Infectious virus is recovered from stool and urine samples. | [76,78,79,80] |
| Sewage, Wastewater transmission | SARS-CoV-2 particles, RNA and infectivity | [81,82] |
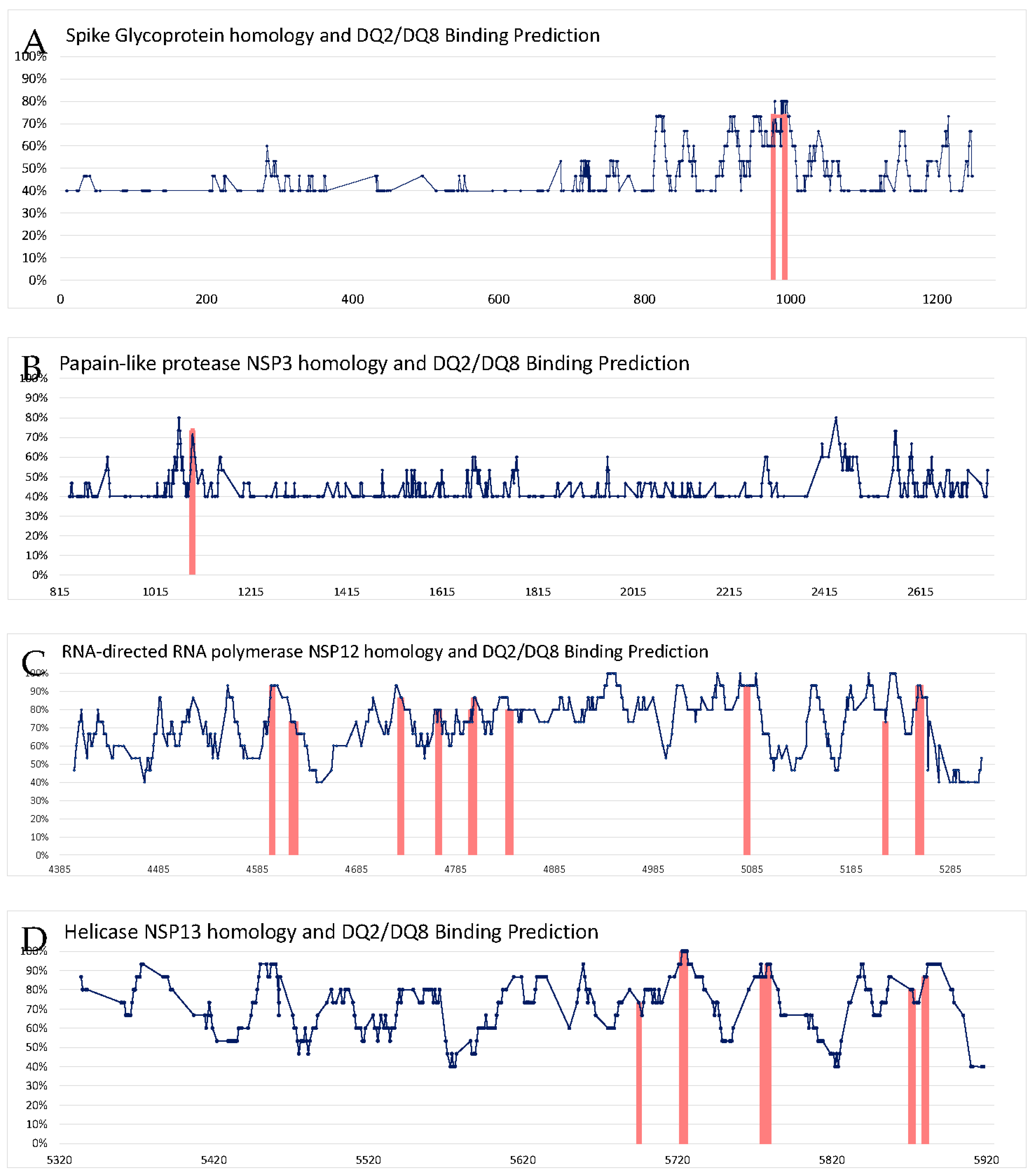
| SARS-CoV-2 Parent Protein | CCC protein ID | Epitope Sequence CCC vs SARS-CoV-2 |
Identity | % Identity | start | end | HLA-DQA10301-DQB10302 *rank | HLA-DQA10501-DQB10201 *rank |
|---|---|---|---|---|---|---|---|---|
| NSP3 Replicase polyprotein 1ab P0DTD1 | P0C6X5 |
SKDYISSNGPLKVG SDDYIATNGPLKVG |
11/15 | 73.4% | 1086 | 1100 | 4.28 | |
| NSP12 Replicase polyprotein 1ab P0DTD1 | P0C6X2 P0C6X6 P0C6X1 P0C6X5 |
VVGVLTLDNQDLNGN IVGVLTLDNQDLNGN |
14/15 | 93.3% | 4593 | 4607 | 4.33 | |
| P0C6X2 |
DFIQTAPGFGVAVAD DFIQTTPGSGVPVVD |
11/15 | 73.3% | 4613 | 4627 | 3.38 | ||
|
FIQTAPGFGVAVADS FIQTTPGSGVPVVDS |
11/15 | 73.3% | 4614 | 4628 | 1.15 | 4.18 | ||
|
IQTAPGFGVAVADSY IQTTPGSGVPVVDSY |
11/15 | 73.3% | 4615 | 4629 | 0.76 | 2.75 | ||
|
QTAPGFGVAVADSYY QTTPGSGVPVVDSYY |
11/15 | 73.3% | 4616 | 4630 | 0.63 | 2.24 | ||
| P0C6X2 P0C6X6 |
RQIFVDGVPFVVSIG RKIFVDGVPFVVSTG |
13/15 | 86.7% | 4723 | 4737 | 4.34 | ||
|
KDLLLYAADPAMHVA KELLVYAADPAMHAA |
12/15 | 80.0% | 4761 | 4775 | 4.3 | |||
| P0C6X6 |
TSGVKFQTVKPGNFN TNNVAFQTVKPGNFN |
12/15 | 80.0% | 4794 | 4808 | 3.03 | ||
|
SGVKFQTVKPGNFNQ NNVAFQTVKPGNFNK |
11/15 | 73.3% | 4795 | 4809 | 1.6 | |||
| P0C6X2 P0C6X6 |
VKFQTVKPGNFNQD VAFQTVKPGNFNKD |
12/15 | 80.0% | 4796 | 4810 | 0.7 | ||
|
VKFQTVKPGNFNQDF VAFQTVKPGNFNKDF |
13/15 | 86.7% | 4797 | 4811 | 2.98 | |||
|
FFFTQDGNAAITDYN FFFAQDGNAAISDYD |
12/15 | 80.0% | 4832 | 4846 | 2.9 | |||
|
FFTQDGNAAITDYNY FFAQDGNAAISDYDY |
12/15 | 80.0% | 4833 | 4847 | 1.98 | |||
|
FTQDGNAAITDYNYY FAQDGNAAISDYDYY |
12/15 | 80.0% | 4834 | 4848 | 2.12 | |||
| P0C6X5 P0C6X1 P0C6X2 P0C6X6 |
SSGDATTAFANSVFN SSGDATTAYANSVFN |
14/15 | 93.3% | 5073 | 5087 | 4.91 | 2.73 | |
| P0C6X2 |
DYVYLPYPDPS DYVYLPYPDPS |
11/15 | 73.3% | 5213 | 5227 | 4.02 | ||
| P0C6X5 P0C6X1 P0C6X2 P0C6X6 |
LLIERFVSLAIDAYP LMIERFVSLAIDAYP |
14/15 | 93.3% | 5246 | 5260 | 3.36 | 1.42 | |
|
LIERFVSLAIDAYPL MIERFVSLAIDAYPL |
14/15 | 93.3% | 5247 | 5261 | 4.39 | 1.75 | ||
|
IERFVSLAIDAYPL IERFVSLAIDAYPL |
14/15 | 93.3% | 5248 | 5262 | 1.53 | |||
|
ERYVSLAIDAYPLSK ERFVSLAIDAYPLTK |
13/15 | 86.7% | 5249 | 5263 | 3.4 | |||
| NSP13 Replicase polyprotein 1ab P0DTD1 | P0C6X5 P0C6X1 |
PEVNADIVVVDEVSM PETTADIVVFDEISM |
11/15 | 73.3% | 5688 | 5702 | 1.08 | |
| P0C6X2 P0C6X6 |
RAKHYVYIGDPAQLP RAKHYVYIGDPAQLP |
15/15 | 100.0% | 5716 | 5730 | 3.24 | ||
| P0C6X5 P0C6X1 P0C6X2 P0C6X6 |
AKHYVYIGDPAQLPA AKHYVYIGDPAQLPA |
15/15 | 100.0% | 5717 | 5731 | 2.45 | ||
|
KHYVYIGDPAQLPAP KHYVYIGDPAQLPAP |
15/15 | 100.0% | 5718 | 5732 | 3.52 | |||
|
CPKEIVDTVSALVYE CPAEIVDTVSALVYD |
13/15 | 86.7% | 5768 | 5782 | 3.27 | |||
|
PKEIVDTVSALVYEN PAEIVDTVSALVYDN |
13/15 | 86.7% | 5769 | 5783 | 2.67 | 1.3 | ||
|
EIVDTVSALVYENK EIVDTVSALVYDNK |
13/15 | 86.7% | 5770 | 5784 | 1.09 | 0.65 | ||
|
EIVDTVSALVYENKL EIVDTVSALVYDNKL |
14/15 | 93.3% | 5771 | 5785 | 1.61 | 2.77 | ||
| P0C6X6 P0C6X2 |
IVETVSALVYDNKLK IVDTVSALVYDNKLK |
14/15 | 93.3% | 5772 | 5786 | 4.2 | ||
| P0C6X5 P0C6X1 P0C6X2 P0C6X6 |
EYDYVIYSQTAETAH EYDYVIFTQTTETAH |
12/15 | 80.0% | 5864 | 5878 | 3.76 | ||
| P0C6X1 P0C6X2 P0C6X6 |
YDYVIYSQTAETAHS YDYVIFTQTTETAHS |
12/15 | 80.0% | 5865 | 5879 | 3.07 | ||
| P0C6X1 P0C6X5 P0C6X2 P0C6X6 |
TAETAHSVNVNRFNV TTETAHSCNVNRFNV |
13/15 | 86.7% | 5873 | 5887 | 3.43 | ||
|
ETAHSVNVNRFNVA ETAHSCNVNRFNVA |
13/15 | 86.7% | 5874 | 5888 | 2.98 | |||
| Spike glycoprotein P0DTC2 | Q5MQD0 |
FGAISSSLQEILSR FGAISSVLNDILSR |
11/15 | 73.4% | 969 | 983 | 4.86 | |
| P36334 Q5MQD0 |
DALEAQVQIDRLING DKVEAEVQIDRLITG |
11/15 | 73.3% | 985 | 999 | 4.45 | 4.48 | |
|
DALEAQVQIDRLING DPPEAEVQIDRLITG |
11/15 | 73.3% | 985 | 999 | 2.93 | 3.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





