Submitted:
10 November 2023
Posted:
14 November 2023
You are already at the latest version
Abstract
Keywords:
1. OPSs Can Serve as Physical Barriers to Phage Adsorption
1.1. Exopolysaccharides and surface polysaccharides of enterobacteria
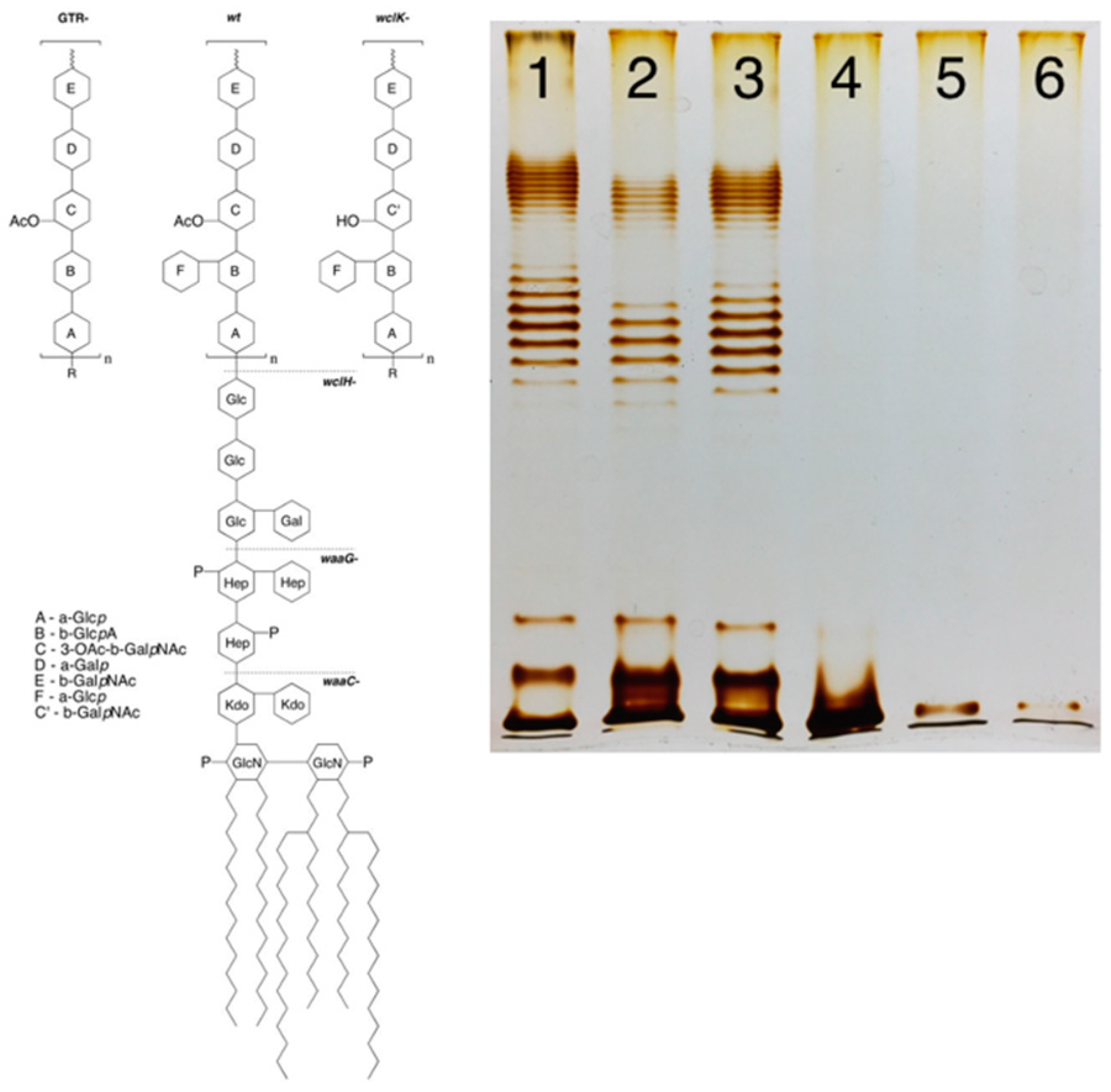
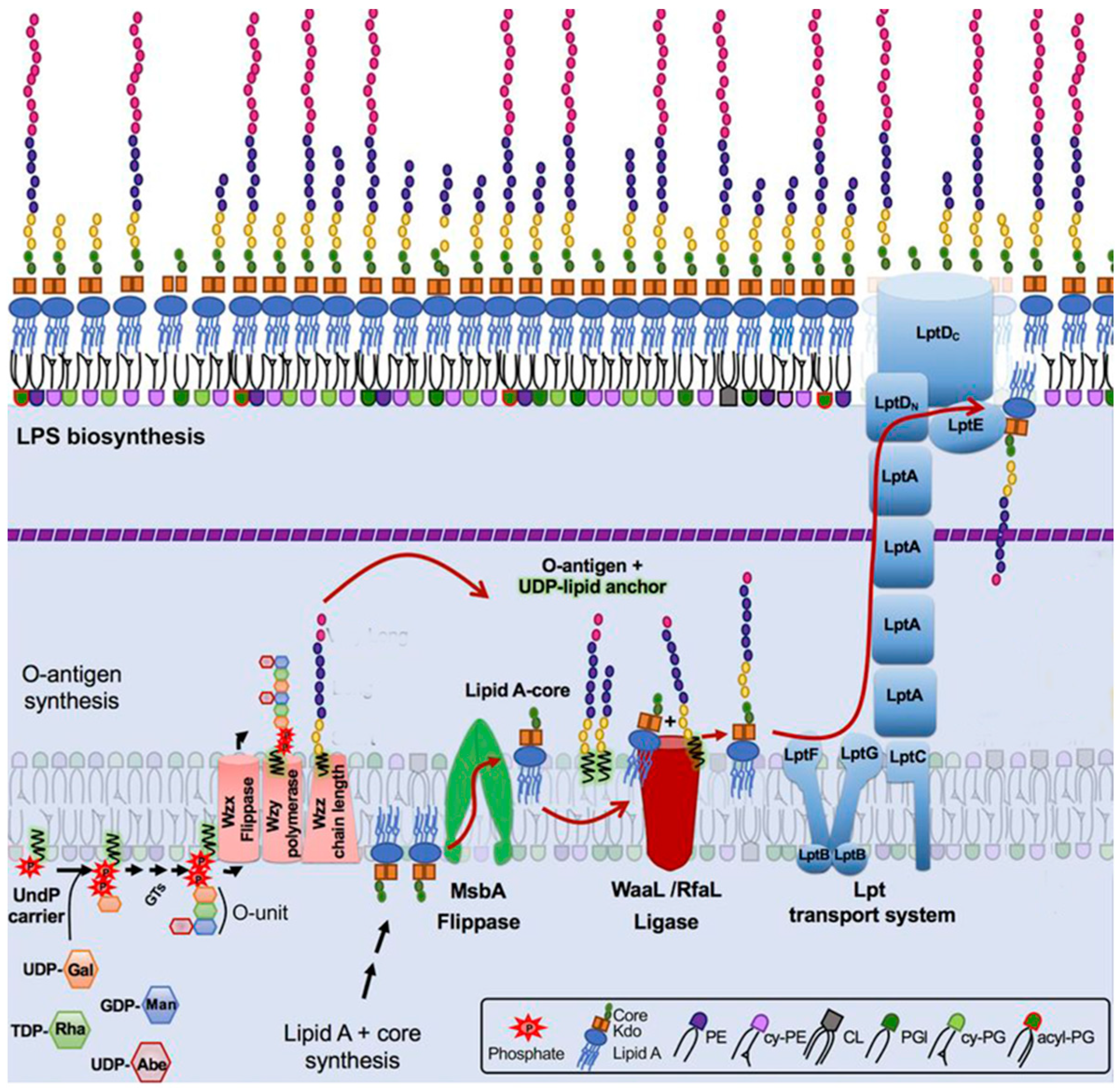
1.2. OPS synthesis
1.3. Other surface polysaccharides of E. coli
2. OPS-mediated OM protection
2.1. OPS shields outer membrane surface
3. Phenomenology of modulation of phage-host interactions by O antigen
3.1. Strategies of the host cell recognition by bacteriophages and properties of phage resistant mutants.
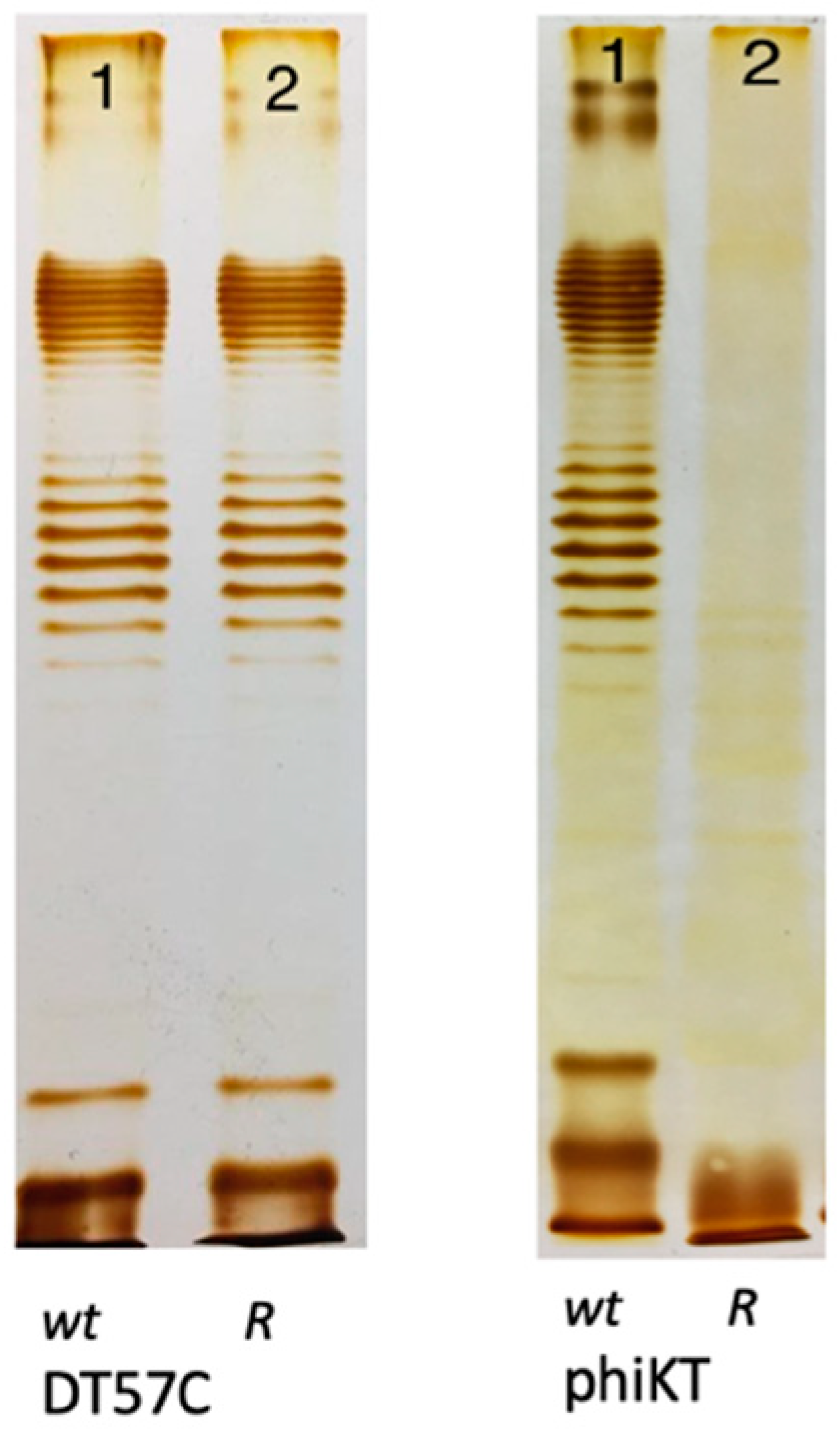
4. How strong is the O antigen barrier in Enterobacteria?
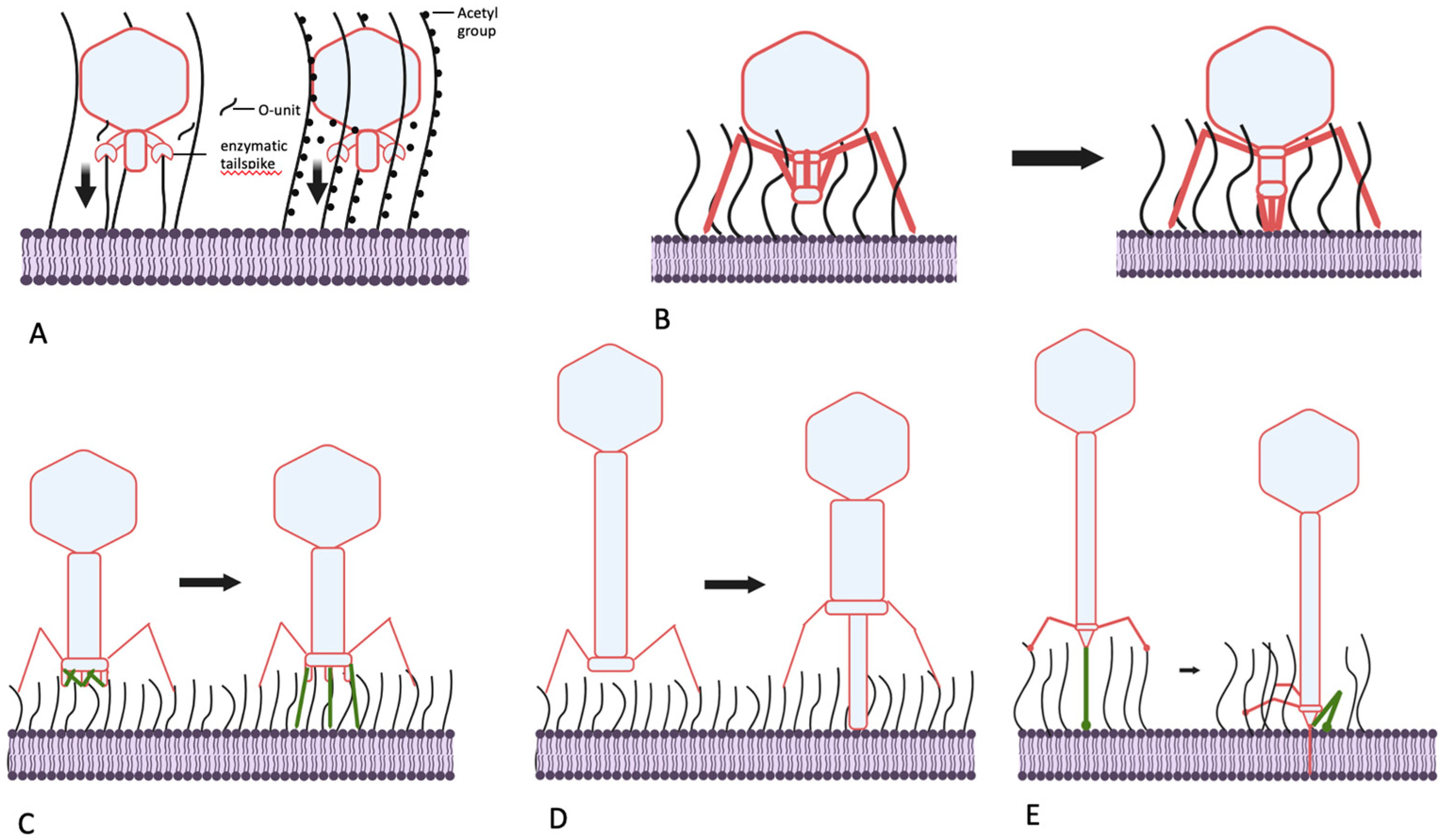
5. Mechanisms used by bacteriophages to penetrate O antigen barrier.
5.1. Podoviruses: cut or pull?
5.2. Can podoviruses push?
5.3. Myoviruses – clutch and push
5.4. Siphoviruses – grab and drag.
6. Conclusions and perspectives
Acknowledgments
Reference
- Kaur, N.; Dey, P. Bacterial exopolysaccharides as emerging bioactive macromolecules: from fundamentals to applications. Res Microbiol 2023, 174, 104024. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, M.; Walker, S. Envelope Structures of Gram-Positive Bacteria. Curr Top Microbiol Immunol 2017, 404, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol Spectr 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Aftalion, M.; Tidhar, A.; Vagima, Y.; Gur, D.; Zauberman, A.; Holtzman, T.; Makovitzki, A.; Chitlaru, T.; Mamroud, E.; Levy, Y. Rapid Induction of Protective Immunity against Pneumonic Plague by Yersinia pestis Polymeric F1 and LcrV Antigens. Vaccines (Basel) 2023, 11. [Google Scholar] [CrossRef] [PubMed]
- Secor, P.R.; Burgener, E.B.; Kinnersley, M.; Jennings, L.K.; Roman-Cruz, V.; Popescu, M.; Van Belleghem, J.D.; Haddock, N.; Copeland, C.; Michaels, L.A.; et al. Pf Bacteriophage and Their Impact on Pseudomonas Virulence, Mammalian Immunity, and Chronic Infections. Front Immunol 2020, 11, 244. [Google Scholar] [CrossRef] [PubMed]
- Ray, R.R. Role of Virus on Oral Biofilm: Inducer or Eradicator? Appl Biochem Biotechnol 2023. [Google Scholar] [CrossRef] [PubMed]
- Meneses, L.; Brandao, A.C.; Coenye, T.; Braga, A.C.; Pires, D.P.; Azeredo, J. A systematic review of the use of bacteriophages for in vitro biofilm control. Eur J Clin Microbiol Infect Dis 2023, 42, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Bacteriophage Adsorption: Likelihood of Virion Encounter with Bacteria and Other Factors Affecting Rates. Antibiotics (Basel) 2023, 12. [Google Scholar] [CrossRef] [PubMed]
- Caffalette, C.A.; Kuklewicz, J.; Spellmon, N.; Zimmer, J. Biosynthesis and Export of Bacterial Glycolipids. Annu Rev Biochem 2020, 89, 741–768. [Google Scholar] [CrossRef]
- Liu, B.; Furevi, A.; Perepelov, A.V.; Guo, X.; Cao, H.; Wang, Q.; Reeves, P.R.; Knirel, Y.A.; Wang, L.; Widmalm, G. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol Rev 2020, 44, 655–683. [Google Scholar] [CrossRef]
- Amor, K.; Heinrichs, D.E.; Frirdich, E.; Ziebell, K.; Johnson, R.P.; Whitfield, C. Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect Immun 2000, 68, 1116–1124. [Google Scholar] [CrossRef]
- Lundstedt, E.; Kahne, D.; Ruiz, N. Assembly and Maintenance of Lipids at the Bacterial Outer Membrane. Chem Rev 2021, 121, 5098–5123. [Google Scholar] [CrossRef] [PubMed]
- Konovalova, A.; Kahne, D.E.; Silhavy, T.J. Outer Membrane Biogenesis. Annu Rev Microbiol 2017, 71, 539–556. [Google Scholar] [CrossRef] [PubMed]
- Kulikov, E.E.; Golomidova, A.K.; Prokhorov, N.S.; Ivanov, P.A.; Letarov, A.V. High-throughput LPS profiling as a tool for revealing of bacteriophage infection strategies. Scientific reports 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cian, M.B.; Giordano, N.P.; Masilamani, R.; Minor, K.E.; Dalebroux, Z.D. Salmonella enterica Serovar Typhimurium Uses PbgA/YejM To Regulate Lipopolysaccharide Assembly during Bacteremia. Infect Immun 2019, 88. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.K.; Mitchell, A.M. Enterobacterial Common Antigen: Synthesis and Function of an Enigmatic Molecule. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C.; Williams, D.M.; Kelly, S.D. Lipopolysaccharide O-antigens-bacterial glycans made to measure. J Biol Chem 2020, 295, 10593–10609. [Google Scholar] [CrossRef]
- Weckener, M.; Woodward, L.S.; Clarke, B.R.; Liu, H.; Ward, P.N.; Le Bas, A.; Bhella, D.; Whitfield, C.; Naismith, J.H. The lipid linked oligosaccharide polymerase Wzy and its regulating co-polymerase, Wzz, from enterobacterial common antigen biosynthesis form a complex. Open Biol 2023, 13, 220373. [Google Scholar] [CrossRef] [PubMed]
- Kiino, D.R.; Rothman-Denes, L.B. Genetic analysis of bacteriophage N4 adsorption. Journal of bacteriology 1989, 171, 4595–4602. [Google Scholar] [CrossRef]
- Thongsomboon, W.; Serra, D.O.; Possling, A.; Hadjineophytou, C.; Hengge, R.; Cegelski, L. Phosphoethanolamine cellulose: A naturally produced chemically modified cellulose. Science 2018, 359, 334–338. [Google Scholar] [CrossRef]
- Ross, P.; Mayer, R.; Benziman, M. Cellulose biosynthesis and function in bacteria. Microbiol Rev 1991, 55, 35–58. [Google Scholar] [CrossRef] [PubMed]
- Boehm, A.; Steiner, S.; Zaehringer, F.; Casanova, A.; Hamburger, F.; Ritz, D.; Keck, W.; Ackermann, M.; Schirmer, T.; Jenal, U. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol Microbiol 2009, 72, 1500–1516. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Q.; Reeves, P.R. The variation of O antigens in gram-negative bacteria. Subcell Biochem 2010, 53, 123–152. [Google Scholar] [CrossRef]
- Rainard, P.; Reperant-Ferter, M.; Gitton, C.; Germon, P. Shielding Effect of Escherichia coli O-Antigen Polysaccharide on J5-Induced Cross-Reactive Antibodies. mSphere 2021, 6. [Google Scholar] [CrossRef]
- Dominguez-Medina, C.C.; Perez-Toledo, M.; Schager, A.E.; Marshall, J.L.; Cook, C.N.; Bobat, S.; Hwang, H.; Chun, B.J.; Logan, E.; Bryant, J.A.; et al. Outer membrane protein size and LPS O-antigen define protective antibody targeting to the Salmonella surface. Nature communications 2020, 11, 851. [Google Scholar] [CrossRef] [PubMed]
- Bentley, A.T.; Klebba, P.E. Effect of lipopolysaccharide structure on reactivity of antiporin monoclonal antibodies with the bacterial cell surface. J Bacteriol 1988, 170, 1063–1068. [Google Scholar] [CrossRef]
- Pluschke, G.; Mayden, J.; Achtman, M.; Levine, R.P. Role of the capsule and the O antigen in resistance of O18:K1 Escherichia coli to complement-mediated killing. Infect Immun 1983, 42, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Golomidova, A.K.; Efimov, A.D.; Kulikov, E.E.; Kuznetsov, A.S.; Belalov, I.S.; Letarov, A.V. O antigen restricts lysogenization of non-O157 Escherichia coli strains by Stx-converting bacteriophage phi24B. Sci Rep 2021, 11, 3035. [Google Scholar] [CrossRef] [PubMed]
- Krzyzewska-Dudek, E.; Kotimaa, J.; Kapczynska, K.; Rybka, J.; Meri, S. Lipopolysaccharides and outer membrane proteins as main structures involved in complement evasion strategies of non-typhoidal Salmonella strains. Mol Immunol 2022, 150, 67–77. [Google Scholar] [CrossRef]
- Heiman, C.M.; Maurhofer, M.; Calderon, S.; Dupasquier, M.; Marquis, J.; Keel, C.; Vacheron, J. Pivotal role of O-antigenic polysaccharide display in the sensitivity against phage tail-like particles in environmental Pseudomonas kin competition. ISME J 2022, 16, 1683–1693. [Google Scholar] [CrossRef]
- Vanacore, A.; Vitiello, G.; Wanke, A.; Cavasso, D.; Clifton, L.A.; Mahdi, L.; Campanero-Rhodes, M.A.; Solis, D.; Wuhrer, M.; Nicolardi, S.; et al. Lipopolysaccharide O-antigen molecular and supramolecular modifications of plant root microbiota are pivotal for host recognition. Carbohydr Polym 2022, 277, 118839. [Google Scholar] [CrossRef] [PubMed]
- Kyликoв, E.E.; Maeвcкa, Д.; Пpoxopoв, H.C.; Гoлoмидoвa, A.K.; Taтapcкий, E.B.; Лeтapoв, A.B. Bлияниe O-aцeтилиpoвaния O-aнтигeнa липoпoлиcaxapидa Escherichia coli нa нecпeциφичecкyю бapьepнyю φyнкцию внeшнeй мeмбpaны. Mикpoбиoлoгия 2017, 86, 284–291. [Google Scholar]
- Bao, Y.; Zhang, H.; Huang, X.; Ma, J.; Logue, C.M.; Nolan, L.K.; Li, G. O-specific polysaccharide confers lysozyme resistance to extraintestinal pathogenic Escherichia coli. Virulence 2018, 9, 666–680. [Google Scholar] [CrossRef] [PubMed]
- Vila, J.; Saez-Lopez, E.; Johnson, J.R.; Romling, U.; Dobrindt, U.; Canton, R.; Giske, C.G.; Naas, T.; Carattoli, A.; Martinez-Medina, M.; et al. Escherichia coli: an old friend with new tidings. FEMS Microbiol Rev 2016, 40, 437–463. [Google Scholar] [CrossRef] [PubMed]
- Luthje, P.; Brauner, A. Virulence factors of uropathogenic E. coli and their interaction with the host. Adv Microb Physiol 2014, 65, 337–372. [Google Scholar] [CrossRef] [PubMed]
- Wildschutte, H.; Lawrence, J.G. Differential Salmonella survival against communities of intestinal amoebae. Microbiology (Reading) 2007, 153, 1781–1789. [Google Scholar] [CrossRef] [PubMed]
- van der Ley, P.; de Graaff, P.; Tommassen, J. Shielding of Escherichia coli outer membrane proteins as receptors for bacteriophages and colicins by O-antigenic chains of lipopolysaccharide. Journal of bacteriology 1986, 168, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Letarov, A.; Kulikov, E. Adsorption of bacteriophages on bacterial cells. Biochemistry (Moscow) 2017, 82, 1632–1658. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Oliveira, H.; Melo, L.D.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol 2016, 100, 2141–2151. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol Lett 2016, 363. [Google Scholar] [CrossRef]
- Golomidova, A.K.; Kulikov, E.E.; Prokhorov, N.S.; Guerrero-Ferreira Rcapital Es, C.; Knirel, Y.A.; Kostryukova, E.S.; Tarasyan, K.K.; Letarov, A.V. Branched Lateral Tail Fiber Organization in T5-Like Bacteriophages DT57C and DT571/2 is Revealed by Genetic and Functional Analysis. Viruses 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Knirel, Y.A.; Ivanov, P.A.; Senchenkova, S.N.; Naumenko, O.I.; Ovchinnikova, O.O.; Shashkov, A.S.; Golomidova, A.K.; Babenko, V.V.; Kulikov, E.E.; Letarov, A.V. Structure and gene cluster of the O antigen of Escherichia coli F17, a candidate for a new O-serogroup. Int J Biol Macromol 2019, 124, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Kulikov, E.E.; Golomidova, A.K.; Prokhorov, N.S.; Ivanov, P.A.; Letarov, A.V. High-throughput LPS profiling as a tool for revealing of bacteriophage infection strategies. Sci Rep 2019, 9, 2958. [Google Scholar] [CrossRef] [PubMed]
- Knirel, Y.A.; Prokhorov, N.S.; Shashkov, A.S.; Ovchinnikova, O.G.; Zdorovenko, E.L.; Liu, B.; Kostryukova, E.S.; Larin, A.K.; Golomidova, A.K.; Letarov, A.V. Variations in O-antigen biosynthesis and O-acetylation associated with altered phage sensitivity in Escherichia coli 4s. J Bacteriol 2015, 197, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Prokhorov, N.S.; Riccio, C.; Zdorovenko, E.L.; Shneider, M.M.; Browning, C.; Knirel, Y.A.; Leiman, P.G.; Letarov, A.V. Function of bacteriophage G7C esterase tailspike in host cell adsorption. Molecular microbiology 2017, 105, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Lukianova, A.A.; Evseev, P.V.; Shneider, M.M.; Dvoryakova, E.A.; Tokmakova, A.D.; Shpirt, A.M.; Kabilov, M.R.; Obraztsova, E.A.; Shashkov, A.S.; Ignatov, A.N.; et al. Pectobacterium versatile Bacteriophage Possum: A Complex Polysaccharide-Deacetylating Tail Fiber as a Tool for Host Recognition in Pectobacterial Schitoviridae. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Lukianova, A.A.; Shneider, M.M.; Evseev, P.V.; Shpirt, A.M.; Bugaeva, E.N.; Kabanova, A.P.; Obraztsova, E.A.; Miroshnikov, K.K.; Senchenkova, S.N.; Shashkov, A.S.; et al. Morphologically Different Pectobacterium brasiliense Bacteriophages PP99 and PP101: Deacetylation of O-Polysaccharide by the Tail Spike Protein of Phage PP99 Accompanies the Infection. Frontiers in microbiology 2019, 10, 3147. [Google Scholar] [CrossRef] [PubMed]
- Efimov, A.D.; Golomidova, A.K.; Kulikov, E.E.; Belalov, I.S.; Ivanov, P.A.; Letarov, A.V. RB49-like Bacteriophages Recognize O Antigens as One of the Alternative Primary Receptors. International Journal of Molecular Sciences 2022, 23, 11329. [Google Scholar] [CrossRef]
- Sellner, B.; Prakapaite, R.; van Berkum, M.; Heinemann, M.; Harms, A.; Jenal, U. A New Sugar for an Old Phage: a c-di-GMP-Dependent Polysaccharide Pathway Sensitizes Escherichia coli for Bacteriophage Infection. mBio 2021, 12, e0324621. [Google Scholar] [CrossRef]
- Junkermeier, E.H.; Hengge, R. A Novel Locally c-di-GMP-Controlled Exopolysaccharide Synthase Required for Bacteriophage N4 Infection of Escherichia coli. mBio 2021, 12, e0324921. [Google Scholar] [CrossRef]
- Heller, K.; Braun, V. Accelerated adsorption of bacteriophage T5 to Escherichia coli F, resulting from reversible tail fiber-lipopolysaccharide binding. Journal of bacteriology 1979, 139, 32–38. [Google Scholar] [CrossRef]
- Heller, K.; Braun, V. Polymannose O-antigens of Escherichia coli, the binding sites for the reversible adsorption of bacteriophage T5+ via the L-shaped tail fibers. J Virol 1982, 41, 222–227. [Google Scholar] [CrossRef]
- Heller, K.J.; Bryniok, D. O antigen-dependent mutant of bacteriophage T5. J Virol 1984, 49, 20–25. [Google Scholar] [CrossRef]
- Golomidova, A.K.; Naumenko, O.I.; Senchenkova, S.N.; Knirel, Y.A.; Letarov, A.V. The O-polysaccharide of Escherichia coli F5, which is structurally related to that of E. coli O28ab, provides cells only weak protection against bacteriophage attack. Arch Virol 2019, 164, 2783–2787. [Google Scholar] [CrossRef] [PubMed]
- Golomidova, A.K.; Kulikov, E.E.; Prokhorov, N.S.; Guerrero-Ferreira, R.C.; Ksenzenko, V.N.; Tarasyan, K.K.; Letarov, A.V. Complete genome sequences of T5-related Escherichia coli bacteriophages DT57C and DT571/2 isolated from horse feces. Arch Virol 2015, 160, 3133–3137. [Google Scholar] [CrossRef]
- Golomidova, A.K.; Kulikov, E.E.; Babenko, V.V.; Ivanov, P.A.; Prokhorov, N.S.; Letarov, A.V. Escherichia coli bacteriophage Gostya9, representing a new species within the genus T5virus. Arch Virol 2019, 164, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Kulikov, E.E.; Golomidova, A.K.; Efimov, A.D.; Belalov, I.S.; Letarova, M.A.; Zdorovenko, E.L.; Knirel, Y.A.; Dmitrenok, A.S.; Letarov, A.V. Equine Intestinal O-Seroconverting Temperate Coliphage Hf4s: Genomic and Biological Characterization. Appl Environ Microbiol 2021, 87, e0112421. [Google Scholar] [CrossRef] [PubMed]
- Maffei, E.; Shaidullina, A.; Burkolter, M.; Heyer, Y.; Estermann, F.; Druelle, V.; Sauer, P.; Willi, L.; Michaelis, S.; Hilbi, H.; et al. Systematic exploration of Escherichia coli phage-host interactions with the BASEL phage collection. PLoS Biol 2021, 19, e3001424. [Google Scholar] [CrossRef]
- James, C.E.; Stanley, K.N.; Allison, H.E.; Flint, H.J.; Stewart, C.S.; Sharp, R.J.; Saunders, J.R.; McCarthy, A.J. Lytic and lysogenic infection of diverse Escherichia coli and Shigella strains with a verocytotoxigenic bacteriophage. Applied and environmental microbiology 2001, 67, 4335–4337. [Google Scholar] [CrossRef]
- Yerigeri, K.; Kadatane, S.; Mongan, K.; Boyer, O.; Burke, L.L.G.; Sethi, S.K.; Licht, C.; Raina, R. Atypical Hemolytic-Uremic Syndrome: Genetic Basis, Clinical Manifestations, and a Multidisciplinary Approach to Management. J Multidiscip Healthc 2023, 16, 2233–2249. [Google Scholar] [CrossRef]
- Rodriguez-Rubio, L.; Haarmann, N.; Schwidder, M.; Muniesa, M.; Schmidt, H. Bacteriophages of Shiga Toxin-Producing Escherichia coli and Their Contribution to Pathogenicity. Pathogens 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Latka, A.; Leiman, P.G.; Drulis-Kawa, Z.; Briers, Y. Modeling the Architecture of Depolymerase-Containing Receptor Binding Proteins in Klebsiella Phages. Front Microbiol 2019, 10, 2649. [Google Scholar] [CrossRef] [PubMed]
- Topka-Bielecka, G.; Dydecka, A.; Necel, A.; Bloch, S.; Nejman-Falenczyk, B.; Wegrzyn, G.; Wegrzyn, A. Bacteriophage-Derived Depolymerases against Bacterial Biofilm. Antibiotics (Basel) 2021, 10. [Google Scholar] [CrossRef]
- Reuter, M.; Kruger, D.H. Approaches to optimize therapeutic bacteriophage and bacteriophage-derived products to combat bacterial infections. Virus Genes 2020, 56, 136–149. [Google Scholar] [CrossRef]
- Knecht, L.E.; Veljkovic, M.; Fieseler, L. Diversity and Function of Phage Encoded Depolymerases. Frontiers in microbiology 2019, 10, 2949. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.; Sao-Jose, C. Enzymes and Mechanisms Employed by Tailed Bacteriophages to Breach the Bacterial Cell Barriers. Viruses 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl Microbiol Biotechnol 2017, 101, 3103–3119. [Google Scholar] [CrossRef]
- Baxa, U.; Weintraub, A.; Seckler, R. Self-Competitive Inhibition of the Bacteriophage P22 Tailspike Endorhamnosidase by O-Antigen Oligosaccharides. Biochemistry 2020, 59, 4845–4855. [Google Scholar] [CrossRef]
- Ouyang, R.; Costa, A.R.; Cassidy, C.K.; Otwinowska, A.; Williams, V.C.J.; Latka, A.; Stansfeld, P.J.; Drulis-Kawa, Z.; Briers, Y.; Pelt, D.M.; et al. High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers. Nat Commun 2022, 13, 7241. [Google Scholar] [CrossRef]
- Kutter, E.; Raya, R.; Carlson, C. Molecular mechanisms of phage infection. In Bacteriophages: biology and applications; Kutter, E., Sulakvelidze, A., Eds.; CRC Press: Boca Raton, London, New York, Washington D. C., 2004; pp. 165–222. [Google Scholar]
- Volozhantsev, N.V.; Borzilov, A.I.; Shpirt, A.M.; Krasilnikova, V.M.; Verevkin, V.V.; Denisenko, E.A.; Kombarova, T.I.; Shashkov, A.S.; Knirel, Y.A.; Dyatlov, I.A. Comparison of the therapeutic potential of bacteriophage KpV74 and phage-derived depolymerase (beta-glucosidase) against Klebsiella pneumoniae capsular type K2. Virus Res 2022, 322, 198951. [Google Scholar] [CrossRef]
- Drobiazko, A.Y.; Kasimova, A.A.; Evseev, P.V.; Shneider, M.M.; Klimuk, E.I.; Shashkov, A.S.; Dmitrenok, A.S.; Chizhov, A.O.; Slukin, P.V.; Skryabin, Y.P.; et al. Capsule-Targeting Depolymerases Derived from Acinetobacter baumannii Prophage Regions. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef]
- Parent, K.N.; Erb, M.L.; Cardone, G.; Nguyen, K.; Gilcrease, E.B.; Porcek, N.B.; Pogliano, J.; Baker, T.S.; Casjens, S.R. OmpA and OmpC are critical host factors for bacteriophage Sf6 entry in Shigella. Molecular microbiology 2014, 92, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Dover, J.A.; Parent, K.N.; Doore, S.M. Host Range Expansion of Shigella Phage Sf6 Evolves through Point Mutations in the Tailspike. J Virol 2022, 96, e0092922. [Google Scholar] [CrossRef] [PubMed]
- Andres, D.; Hanke, C.; Baxa, U.; Seul, A.; Barbirz, S.; Seckler, R. Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J Biol Chem 2010, 285, 36768–36775. [Google Scholar] [CrossRef]
- Wang, C.; Tu, J.; Liu, J.; Molineux, I.J. Structural dynamics of bacteriophage P22 infection initiation revealed by cryo-electron tomography. Nat Microbiol 2019, 4, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Babenko, V.V.; Golomidova, A.K.; Ivanov, P.A.; Letarova, M.A.; Kulikov, E.E.; Manolov, A.I.; Prokhorov, N.S.; Kostrukova, E.S.; Matyushkina, D.M.; Prilipov, A.G.; et al. Phages associated with horses provide new insights into the dominance of lateral gene transfer in virulent bacteriophages evolution in natural systems. bioRxiv 2019, 542787. [Google Scholar] [CrossRef]
- Kiino, D.R.; Singer, M.S.; Rothman-Denes, L.B. Two overlapping genes encoding membrane proteins required for bacteriophage N4 adsorption. J Bacteriol 1993, 175, 7081–7085. [Google Scholar] [CrossRef]
- McPartland, J.; Rothman-Denes, L.B. The tail sheath of bacteriophage N4 interacts with the Escherichia coli receptor. J Bacteriol 2009, 191, 525–532. [Google Scholar] [CrossRef]
- Dolgalev, G.V.; Safonov, T.A.; Arzumanian, V.A.; Kiseleva, O.I.; Poverennaya, E.V. Estimating Total Quantitative Protein Content in Escherichia coli, Saccharomyces cerevisiae, and HeLa Cells. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Golomidova, A.K.; Kulikov, E.E.; Kudryavtseva, A.V.; Letarov, A.V. Complete genome sequence of Escherichia coli bacteriophage PGT2. Genome announcements 2018, 6. [Google Scholar] [CrossRef]
- Hu, B.; Margolin, W.; Molineux, I.J.; Liu, J. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science 2013, 339, 576–579. [Google Scholar] [CrossRef] [PubMed]
- Swanson, N.A.; Lokareddy, R.K.; Li, F.; Hou, C.D.; Leptihn, S.; Pavlenok, M.; Niederweis, M.; Pumroy, R.A.; Moiseenkova-Bell, V.Y.; Cingolani, G. Cryo-EM structure of the periplasmic tunnel of T7 DNA-ejectosome at 2.7 A resolution. Mol Cell 2021, 81, 3145–3159. [Google Scholar] [CrossRef] [PubMed]
- Swanson, N.A.; Hou, C.D.; Cingolani, G. Viral Ejection Proteins: Mosaically Conserved, Conformational Gymnasts. Microorganisms 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Siborova, M.; Fuzik, T.; Prochazkova, M.; Novacek, J.; Benesik, M.; Nilsson, A.S.; Plevka, P. Tail proteins of phage SU10 reorganize into the nozzle for genome delivery. Nat Commun 2022, 13, 5622. [Google Scholar] [CrossRef]
- Fournel, P.; Page, Y.; Vergnon, J.M.; Guichenez, P.; Emonot, A. [Farmer's lung disease of the lesional pulmonary edema type]. Presse Med 1986, 15, 847. [Google Scholar] [PubMed]
- Taslem Mourosi, J.; Awe, A.; Guo, W.; Batra, H.; Ganesh, H.; Wu, X.; Zhu, J. Understanding Bacteriophage Tail Fiber Interaction with Host Surface Receptor: The Key "Blueprint" for Reprogramming Phage Host Range. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Trojet, S.N.; Caumont-Sarcos, A.; Perrody, E.; Comeau, A.M.; Krisch, H.M. The gp38 adhesins of the T4 superfamily: a complex modular determinant of the phage's host specificity. Genome Biol Evol 2011, 3, 674–686. [Google Scholar] [CrossRef]
- Dunne, M.; Denyes, J.M.; Arndt, H.; Loessner, M.J.; Leiman, P.G.; Klumpp, J. Salmonella Phage S16 Tail Fiber Adhesin Features a Rare Polyglycine Rich Domain for Host Recognition. Structure 2018, 26, 1573–1582. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.M.; Prokhorov, N.S.; Guerrero-Ferreira, R.C.; Shneider, M.M.; Browning, C.; Goldie, K.N.; Stahlberg, H.; Leiman, P.G. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 2016, 533, 346–352. [Google Scholar] [CrossRef]
- Hu, B.; Margolin, W.; Molineux, I.J.; Liu, J. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proceedings of the National Academy of Sciences of the United States of America 2015, 112, E4919–4928. [Google Scholar] [CrossRef]
- Efimov, A.; Kulikov, E.; Golomidova, A.; Belalov, I.; Babenko, V.; Letarov, A. Isolation and sequencing of three RB49-like bacteriophages infecting O antigen-producing E. coli strains [version 1; peer review: 1 approved, 1 approved with reservations]. F1000Research 2021, 1113. [Google Scholar] [CrossRef]
- Efimov, A.D.; Golomidova, A.K.; Kulikov, E.E.; Belalov, I.S.; Ivanov, P.A.; Letarov, A.V. RB49-like Bacteriophages Recognize O Antigens as One of the Alternative Primary Receptors. Int J Mol Sci 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, L.; Abdelgader, S.A.; Yu, L.; Xu, J.; Yao, H.; Lu, C.; Zhang, W. Alterations in gp37 Expand the Host Range of a T4-Like Phage. Appl Environ Microbiol 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Shi, D.; Li, Y.; Xiao, Y.; Chen, M.; Chen, L.; Du, H.; Zhang, W. Recombination of T4-like Phages and Its Activity against Pathogenic Escherichia coli in Planktonic and Biofilm Forms. Virol Sin 2020, 35, 651–661. [Google Scholar] [CrossRef]
- Hogins, J.; Sudarshan, S.; Zimmern, P.; Reitzer, L. Facile transduction with P1 phage in Escherichia coli associated with urinary tract infections. J Microbiol Methods 2023, 208, 106722. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.W.; Fa-Arun, J.; Wang, B. The Role of O-antigen in P1 Transduction of Shigella flexneri and Escherichia coli with its Alternative S' Tail Fibre. J Mol Biol 2022, 434, 167829. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.D.; Waldor, M.K. Enterohemorrhagic Escherichia coli O157:H7 gal mutants are sensitive to bacteriophage P1 and defective in intestinal colonization. Infect Immun 2007, 75, 1661–1666. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Wang, L.; Zhou, J.; Xiao, H.; Liu, H. In Situ Structures of the Ultra-Long Extended and Contracted Tail of Myoviridae Phage P1. Viruses 2023, 15. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, C.Y.; Shiomi, D.; Niki, H.; Margolin, W. Visualization of bacteriophage P1 infection by cryo-electron tomography of tiny Escherichia coli. Virology 2011, 417, 304–311. [Google Scholar] [CrossRef]
- Linares, R.; Arnaud, C.A.; Degroux, S.; Schoehn, G.; Breyton, C. Structure, function and assembly of the long, flexible tail of siphophages. Curr Opin Virol 2020, 45, 34–42. [Google Scholar] [CrossRef]
- Davidson, A.R.; Cardarelli, L.; Pell, L.G.; Radford, D.R.; Maxwell, K.L. Long noncontractile tail machines of bacteriophages. Advances in experimental medicine and biology 2012, 726, 115–142. [Google Scholar] [CrossRef] [PubMed]
- Rabsch, W.; Ma, L.; Wiley, G.; Najar, F.Z.; Kaserer, W.; Schuerch, D.W.; Klebba, J.E.; Roe, B.A.; Laverde Gomez, J.A.; Schallmey, M.; et al. FepA- and TonB-dependent bacteriophage H8: receptor binding and genomic sequence. Journal of bacteriology 2007, 189, 5658–5674. [Google Scholar] [CrossRef] [PubMed]
- Linares, R.; Arnaud, C.A.; Effantin, G.; Darnault, C.; Epalle, N.H.; Boeri Erba, E.; Schoehn, G.; Breyton, C. Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation. Sci Adv 2023, 9, eade9674. [Google Scholar] [CrossRef] [PubMed]
- Degroux, S.; Effantin, G.; Linares, R.; Schoehn, G.; Breyton, C. Deciphering Bacteriophage T5 Host Recognition Mechanism and Infection Trigger. J Virol 2023, 97, e0158422. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, B.; Silale, A.; Basle, A.; Brandner, A.F.; Mader, S.L.; Khalid, S. Structural basis for host recognition and superinfection exclusion by bacteriophage T5. Proc Natl Acad Sci U S A 2022, 119, e2211672119. [Google Scholar] [CrossRef] [PubMed]
- Isaeva, A.S.; Kulikov, E.E.; Tarasyan, K.K.; Letarov, A.V. A novel high-resolving method for genomic PCR-fingerprinting of Enterobacteria. Acta naturae 2010, 2, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Golomidova, A.; Kulikov, E.; Isaeva, A.; Manykin, A.; Letarov, A. The diversity of coliphages and coliforms in horse feces reveals a complex pattern of ecological interactions. Applied and environmental microbiology 2007, 73, 5975–5981. [Google Scholar] [CrossRef] [PubMed]
- Romling, U. The power of unbiased phenotypic screens - cellulose as a first receptor for the Schitoviridae phage S6 of Erwinia amylovora. Environ Microbiol 2022, 24, 3316–3321. [Google Scholar] [CrossRef] [PubMed]
- Knecht, L.E.; Heinrich, N.; Born, Y.; Felder, K.; Pelludat, C.; Loessner, M.J.; Fieseler, L. Bacteriophage S6 requires bacterial cellulose for Erwinia amylovora infection. Environ Microbiol 2022, 24, 3436–3450. [Google Scholar] [CrossRef]
- Letarov, A.V.; Letarova, M.A. The Burden of Survivors: How Can Phage Infection Impact Non-Infected Bacteria? Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Luthe, T.; Kever, L.; Thormann, K.; Frunzke, J. Bacterial multicellular behavior in antiviral defense. Curr Opin Microbiol 2023, 74, 102314. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



