Submitted:
10 November 2023
Posted:
13 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Seed selection and aging treatment
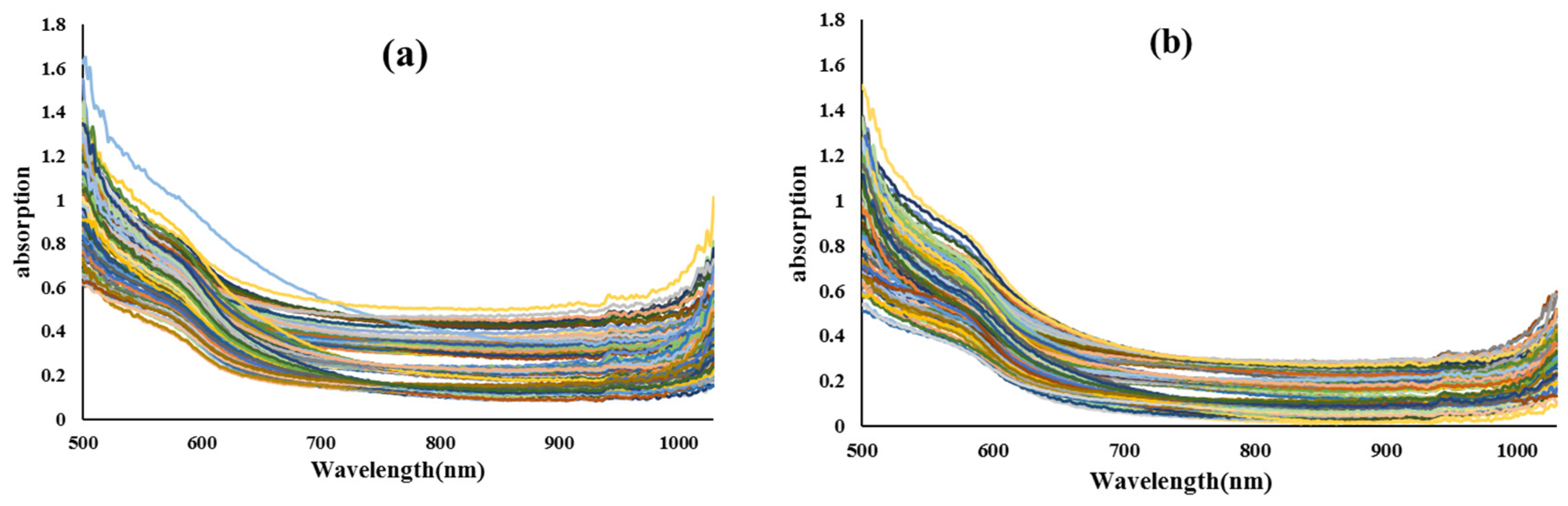
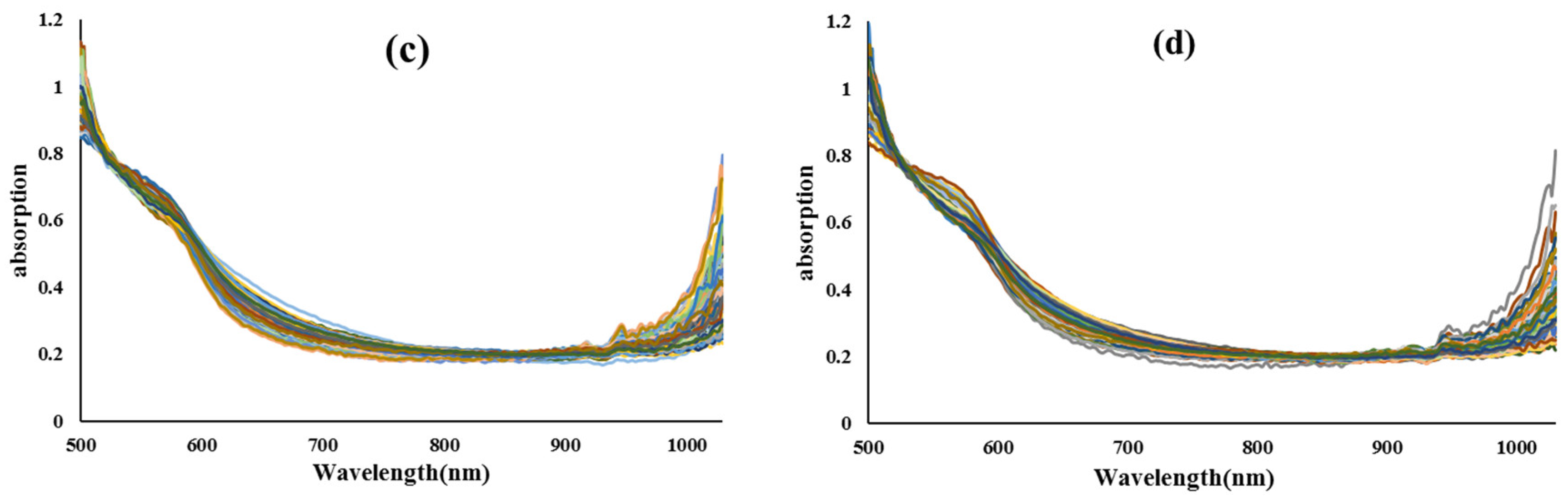
2.2. Preparation of Vis/NIR Spectra from Samples
2.3. Standard germination test
2.4. Preprocessing of Vis/NIR spectra
2.5. Methods of choosing the optimal wavelength
2.6. Modeling methods to predict seed viability
2.7. Evaluation criteria of optimal wavelength selection algorithms and machine learning models
3. Results and Discussion
3.1. Examination of seed viability indices
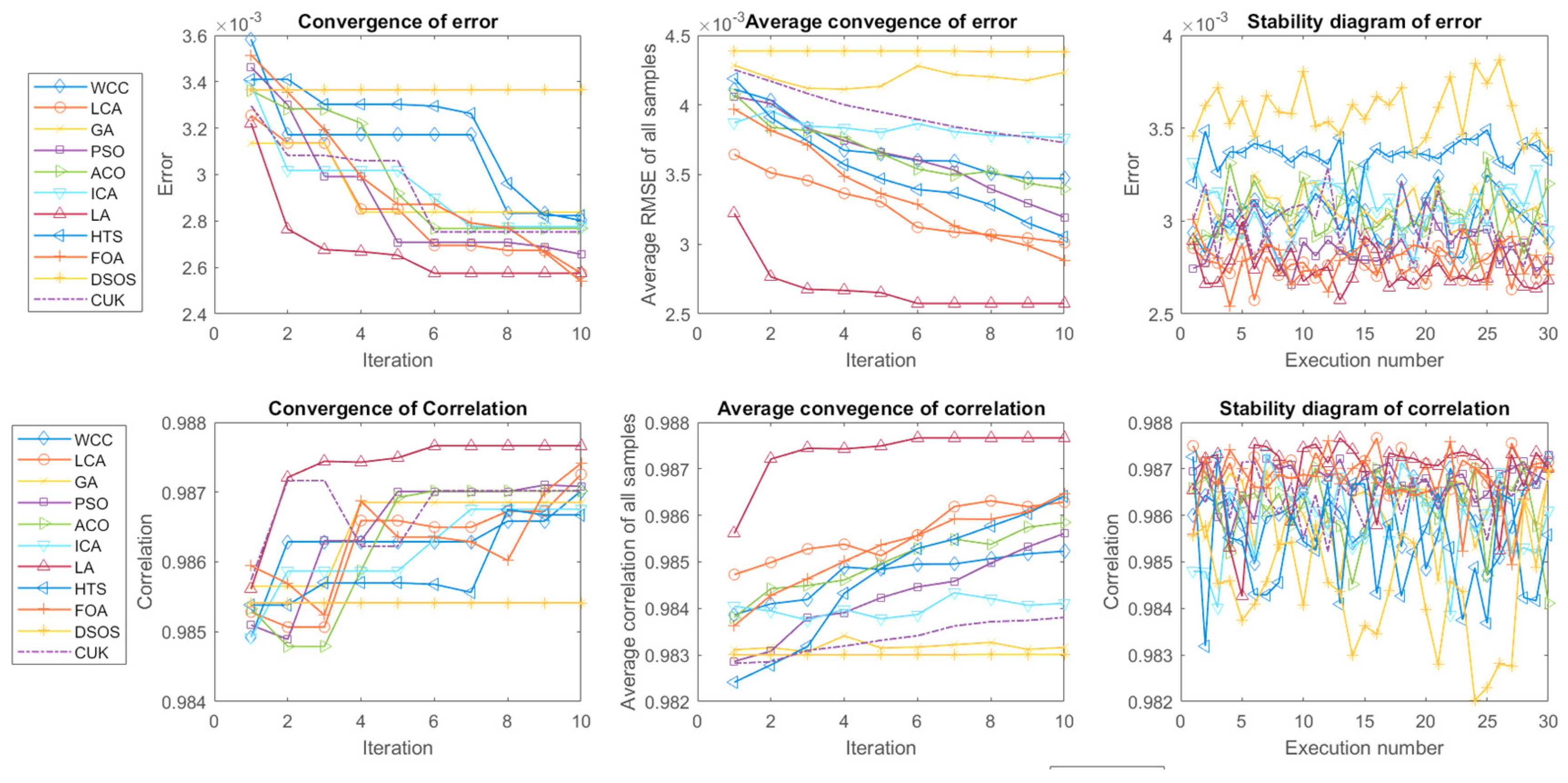
3.2. Comparison of the efficiency of optimal wavelength selection methods
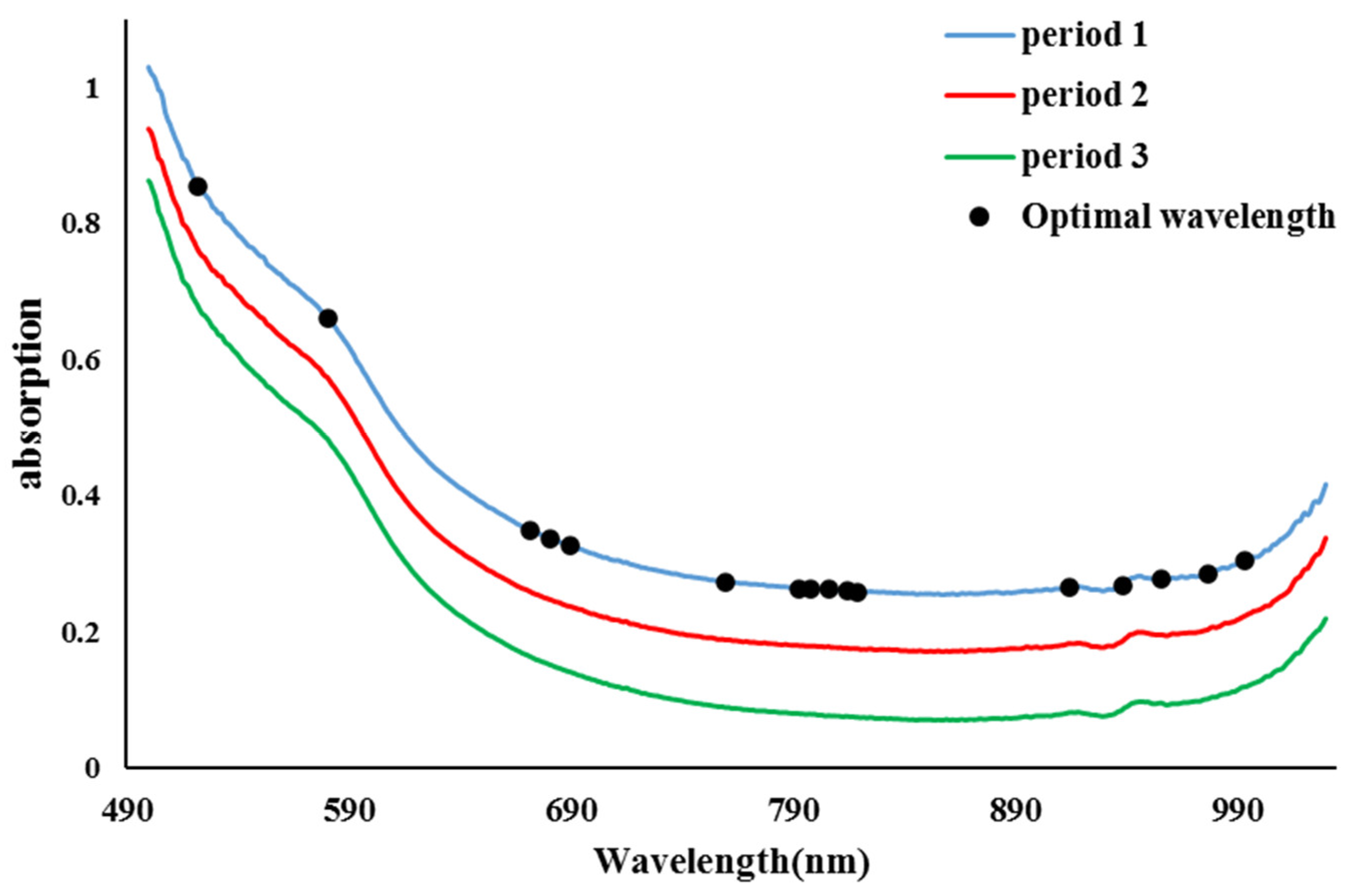
3.3. Examination of averages of Vis/NIR absorption spectra and evaluation of the location of the selected wavelength
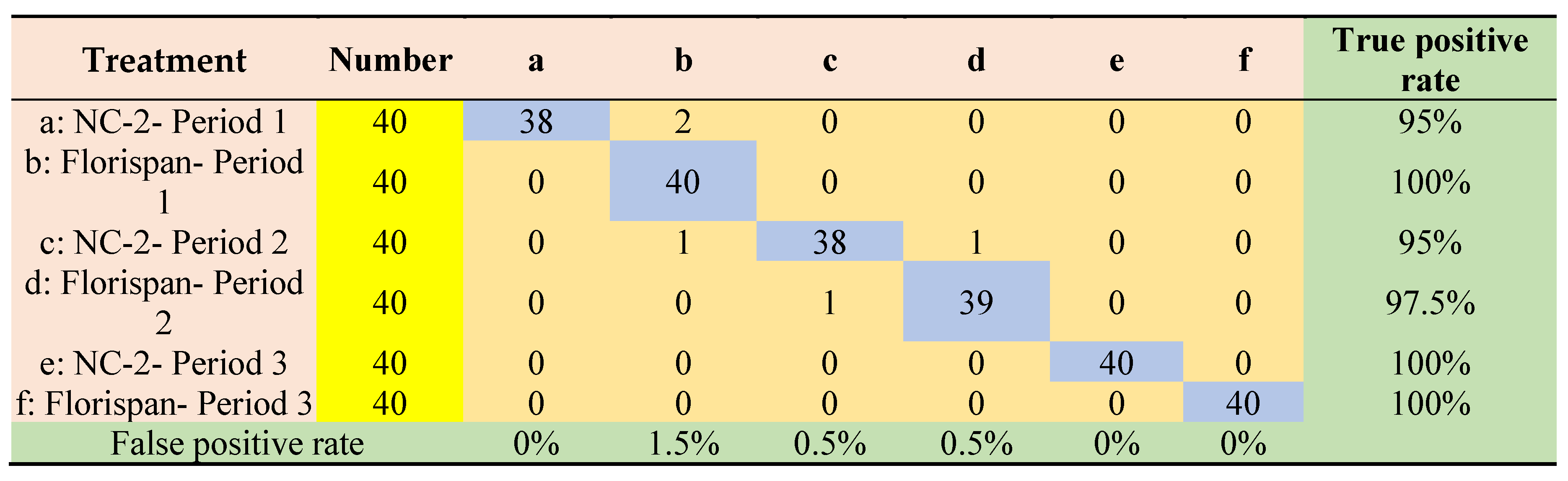
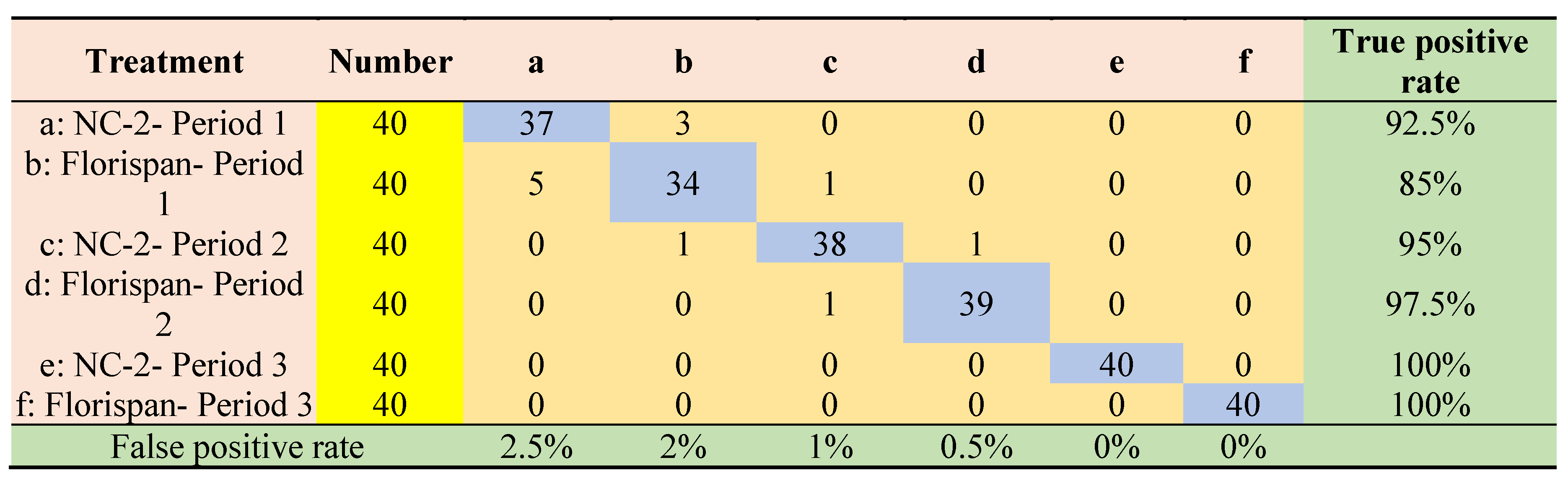
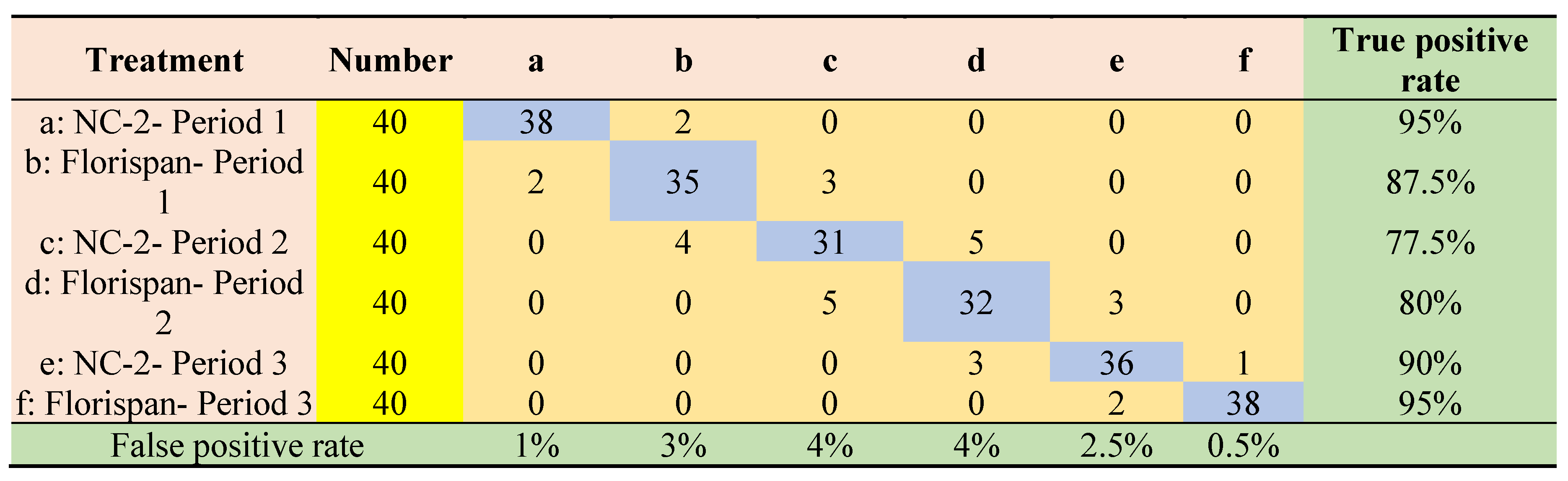
3.4. Results of seed viability classification modeling based on selected wavelengths by machine learning methods
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zou, S.; Tseng, Y.C.; Zare, A.; Rowland, D.L.; Tillman, B.L.; Yoon, S.C. Peanut maturity classification using hyperspectral imagery. biosystems engineering 2019, 188, 165–177. [Google Scholar] [CrossRef]
- Deepa, R.; Anandhi, A.; Bailey, N.O.; Grace, J.M., III; Betiku, O.C.; Muchovej, J.J. Potential Environmental Impacts of Peanut Using Water Footprint Assessment: A Case Study in Georgia. Agronomy 2022, 12, 930. [Google Scholar] [CrossRef]
- Wang, X.; Chen, C.Y.; Dang, P.; Carter, J.; Zhao, S.; Lamb, M.C.; Feng, Y. Variabilities in symbiotic nitrogen fixation and carbon isotope discrimination among peanut (Arachis hypogaea L.) genotypes under drought stress. Journal of Agronomy and Crop Science 2023, 209, 228–241. [Google Scholar] [CrossRef]
- Tan, X.L.; Azam-Ali, S.; Goh, E.V.; Mustafa, M.; Chai, H.H.; Ho, W.K.; Massawe, F. Bambara groundnut: An underutilized leguminous crop for global food security and nutrition. Frontiers in Nutrition 2020, 7, 601496. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Cho, B.K. Assessment of seed quality using non-destructive measurement techniques: a review. Seed Science Research 2016, 26, 285–305. [Google Scholar] [CrossRef]
- Kaur, N.; Erickson, T.E.; Ball, A.S.; Ryan, M.H. A review of germination and early growth as a proxy for plant fitness under petrogenic contamination—knowledge gaps and recommendations. Science of The Total Environment 2017, 603, 728–744. [Google Scholar] [CrossRef] [PubMed]
- Bastos LL, D.S.; Calvi, G.P.; Lima Júnior MD, J.V.; Ferraz ID, K. Degree of seed desiccation sensitivity of the Amazonian palm Oenocarpus bacaba depends on the criterion for germination. Acta Amazonica 2021, 51, 85–90. [Google Scholar] [CrossRef]
- Carrera-Castaño, G.; Calleja-Cabrera, J.; Pernas, M.; Gómez, L.; Oñate-Sánchez, L. An updated overview on the regulation of seed germination. Plants 2020, 9, 703. [Google Scholar] [CrossRef] [PubMed]
- Marcos Filho, J. Seed vigor testing: an overview of the past, present and future perspective. Scientia agricola 2015, 72, 363–374. [Google Scholar] [CrossRef]
- Moghaddam, S.S.; Rahimi, A.; Noorhosseini, S.A.; Heydarzadeh, S.; Mirzapour, M. Effect of seed priming with salicylic acid on germinability and seedling vigor fenugreek (Trigonella Foenum-Graecum). Yuzuncu Yıl University Journal of Agricultural Sciences 2018, 28, 192–199. [Google Scholar]
- Ebrahimi, M.; Miri, E. Effect of humic acid on seed germination and seedling growth of Borago officinalis and Cichorium intybus. Ecopersia 2016, 4, 1239–1249. [Google Scholar] [CrossRef]
- Mohajeri, F.; Taghvaei, M.; Ramrudi, M.; Galavi, M. Effect of priming duration and concentration on germination behaviors of (Phaseolus vulgaris L.) seeds. Int. J. Ecol. Environ. Conserv 2016, 22, 603–609. [Google Scholar]
- Xia, Y.; Xu, Y.; Li, J.; Zhang, C.; Fan, S. Recent advances in emerging techniques for non-destructive detection of seed viability: A review. Artificial Intelligence in Agriculture 2019, 1, 35–47. [Google Scholar] [CrossRef]
- Li, A.; Yao, C.; Xia, J.; Wang, H.; Cheng, Q.; Penty, R.; Pan, S. Advances in cost-effective integrated spectrometers. Light: Science & Applications 2022, 11, 174. [Google Scholar]
- Abasi, S.; Minaei, S.; Jamshidi, B.; Fathi, D. Dedicated non-destructive devices for food quality measurement: A review. Trends in Food Science & Technology 2018, 78, 197–205. [Google Scholar]
- El-Mesery, H.S.; Mao, H.; Abomohra AE, F. Applications of non-destructive technologies for agricultural and food products quality inspection. Sensors 2019, 19, 846. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Hashim, N. Non-destructive methods for detection of food quality. In Future Foods (pp. 645-667). Academic Press, 2022.
- Wei, X.; Xu, N.; Wu, D.; He, Y. Determination of branched-amino acid content in fermented Cordyceps sinensis mycelium by using FT-NIR spectroscopy technique. Food and Bioprocess Technology 2014, 7, 184–190. [Google Scholar] [CrossRef]
- Xia, Y.; Huang, W.; Fan, S.; Li, J.; Chen, L. Effect of spectral measurement orientation on online prediction of soluble solids content of apple using Vis/NIR diffuse reflectance. Infrared Physics & Technology 2019, 97, 467–477. [Google Scholar]
- Kusumaningrum, D.; Lee, H.; Lohumi, S.; Mo, C.; Kim, M.S.; Cho, B.K. Non-destructive technique for determining the viability of soybean (Glycine max) seeds using FT-NIR spectroscopy. Journal of the Science of Food and Agriculture 2018, 98, 1734–1742. [Google Scholar] [CrossRef]
- Ambrose, A.; Kandpal, L.M.; Kim, M.S.; Lee, W.H.; Cho, B.K. High speed measurement of corn seed viability using hyperspectral imaging. Infrared Physics & Technology 2016, 75, 173–179. [Google Scholar]
- Shrestha, S.; Deleuran, L.C.; Gislum, R. Separation of viable and non-viable tomato (Solanum lycopersicum L.) seeds using single seed near-infrared spectroscopy. Computers and Electronics in Agriculture 2017, 142, 348–355. [Google Scholar] [CrossRef]
- Lakshmanan, M.K.; Boelt, B.; Gislum, R. A chemometric method for the viability analysis of spinach seeds by near infrared spectroscopy with variable selection using successive projections algorithm. Journal of Near Infrared Spectroscopy 2023, 31, 24–32. [Google Scholar] [CrossRef]
- Li, L.; Chen, S.; Deng, M.; Gao, Z. Optical techniques in non-destructive detection of wheat quality: A review. Grain & Oil Science and Technology 2022, 5, 44–57. [Google Scholar]
- Rostami, M.; Berahmand, K.; Forouzandeh, S. A novel method of constrained feature selection by the measurement of pairwise constraints uncertainty. Journal of Big Data 2020, 7, 1–21. [Google Scholar] [CrossRef]
- Rostami, M.; Berahmand, K.; Nasiri, E.; Forouzandeh, S. Review of swarm intelligence-based feature selection methods. Engineering Applications of Artificial Intelligence 2021, 100, 104210. [Google Scholar] [CrossRef]
- Gokalp, O.; Tasci, E.; Ugur, A. A novel wrapper feature selection algorithm based on iterated greedy metaheuristic for sentiment classification. Expert Systems with Applications 2020, 146, 113176. [Google Scholar] [CrossRef]
- Tang, X.; Dai, Y.; Xiang, Y. Feature selection based on feature interactions with application to text categorization. Expert Systems with Applications 2019, 120, 207–216. [Google Scholar] [CrossRef]
- Liu, Y.; Nie, F.; Gao, Q.; Gao, X.; Han, J.; Shao, L. Flexible unsupervised feature extraction for image classification. Neural Networks 2019, 115, 65–71. [Google Scholar] [CrossRef]
- Chen, R.C.; Dewi, C.; Huang, S.W.; Caraka, R.E. Selecting critical features for data classification based on machine learning methods. Journal of Big Data 2020, 7, 52. [Google Scholar] [CrossRef]
- Zou, Z.; Chen, J.; Zhou, M.; Zhao, Y.; Long, T.; Wu, Q.; Xu, L. Prediction of peanut seed vigor based on hyperspectral images. Food Science and Technology 2022, 42. [Google Scholar] [CrossRef]
- Hampton, J.G.; Tekrony, D.M. Handbook of vigour test methods; The International Seed Testing Association: Zurich (Switzerland), 1995. [Google Scholar]
- ISTA. International rules for seed testing. Annexes. Seed Science and Technology Journal 2009, 49, 86-41. [Google Scholar]
- Panwar, P.; Bhardwaj, S.D. Handbook of practical forestry. Agrobios (India), 2005.
- ISTA. The germination test. InternationalSeed Testing Association. Seed Science and Technology 1979, 4, 23–28. [Google Scholar]
- Maguire, J.D. Speed of germination-aid in selection and evaluation for seedling emergence and vigor. Crop Sci. 1962, 2, 176–177. [Google Scholar] [CrossRef]
- Hunter, E.A.; Glasbey, C.A.; Naylor RE, L. The analysis of data from germination tests. The Journal of Agricultural Science 1984, 102, 207–213. [Google Scholar] [CrossRef]
- Xu, Y.; Zhong, P.; Jiang, A.; Shen, X.; Li, X.; Xu, Z.; Lei, H. Raman spectroscopy coupled with chemometrics for food authentication: A review. TrAC Trends in Analytical Chemistry 2020, 131, 116017. [Google Scholar] [CrossRef]
- Tie-cheng, B.; Tao, W.; You-qi, C.; Mercatoris, B. Comparison of near-infrared spectrum pretreatment methods for Jujube leaf moisture content detection in the sand and dust area of Southern Xinjiang. Spectroscopy and Spectral Analysis 2019, 39, 1323–1328. [Google Scholar]
- Pang, L.; Wang, J.; Men, S.; Yan, L.; Xiao, J. Hyperspectral imaging coupled with multivariate methods for seed vitality estimation and forecast for Quercus variabilis. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 2021, 245, 118888. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.W.; Tang, S.S.; Ma, S.X.; Ma, Y.Y.; Hao, Z.Q.; Guo, Y.M.; Zeng, X.Y. Accuracy and stability improvement for meat species identification using multiplicative scatter correction and laser-induced breakdown spectroscopy. Optics Express 2018, 26, 10119–10127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lee, C.K.; Chan, H.K.; Choy, K.L.; Wu, Z. Swarm intelligence applied in green logistics: A literature review. Engineering Applications of Artificial Intelligence 2015, 37, 154–169. [Google Scholar] [CrossRef]
- Hu, Y.; Zheng, J.; Zou, J.; Yang, S.; Ou, J.; Wang, R. A dynamic multi-objective evolutionary algorithm based on intensity of environmental change. Information Sciences 2020, 523, 49–62. [Google Scholar] [CrossRef]
- Wang, C.; Pan, H.; Su, Y. A many-objective evolutionary algorithm with diversity-first based environmental selection. Swarm and evolutionary computation 2020, 53, 100641. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, H.; Rao, Z.; Ji, H. Hyperspectral imaging technology combined with deep forest model to identify frost-damaged rice seeds. Spectrochimica acta part A: molecular and biomolecular spectroscopy 2020, 229, 117973. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.; Xu, B.; Zhang, Y.; Guo, Y.; Yang, S. A similarity-based cooperative co-evolutionary algorithm for dynamic interval multiobjective optimization problems. IEEE Transactions on Evolutionary Computation 2019, 24, 142–156. [Google Scholar] [CrossRef]
- Yong, Z.; Dun-wei, G.; Wan-qiu, Z. Feature selection of unreliable data using an improved multi-objective PSO algorithm. Neurocomputing 2016, 171, 1281–1290. [Google Scholar] [CrossRef]
- Masoudi-Sobhanzadeh, Y.; Motieghader, H. World Competitive Contests (WCC) algorithm: A novel intelligent optimization algorithm for biological and non-biological problems. Informatics in Medicine Unlocked 2016, 3, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Kashan, A.H. League Championship Algorithm (LCA): An algorithm for global optimization inspired by sport championships. Applied Soft Computing 2014, 16, 171–200. [Google Scholar] [CrossRef]
- McCall, J. Genetic algorithms for modelling and optimisation. Journal of computational and Applied Mathematics 2005, 184, 205–222. [Google Scholar] [CrossRef]
- Shi, Y. Particle swarm optimization: developments, applications and resources. In Proceedings of the 2001 congress on evolutionary computation (IEEE Cat. No. 01TH8546) (Vol. 1, pp. 81–86). IEEE, 2001.
- Dorigo, M.; Bonabeau, E.; Theraulaz, G. Ant algorithms and stigmergy. Future generation computer systems 2000, 16, 851–871. [Google Scholar] [CrossRef]
- Atashpaz-Gargari, E.; Lucas, C. Imperialist competitive algorithm: an algorithm for optimization inspired by imperialistic competition. In 2007 IEEE congress on evolutionary computation (pp. 4661-4667). Ieee, 2007.
- Beigy, H.; Meybodi, M.R. Cellular learning automata with multiple learning automata in each cell and its applications. IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics) 2009, 40, 54–65. [Google Scholar] [CrossRef]
- Patel, V.K.; Savsani, V.J. Heat transfer search (HTS): a novel optimization algorithm. Information sciences 2015, 324, 217–246. [Google Scholar] [CrossRef]
- Ghaemi, M.; Feizi-Derakhshi, M.R. Feature selection using forest optimization algorithm. Pattern Recognition 2016, 60, 121–129. [Google Scholar] [CrossRef]
- Masoudi-Sobhanzadeh, Y.; Motieghader, H.; Masoudi-Nejad, A. FeatureSelect: a software for feature selection based on machine learning approaches. BMC bioinformatics 2019, 20, 1–17. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; pp. 1–3. [Google Scholar]
- Park, H.; Son, J.H. Machine learning techniques for THz imaging and time-domain spectroscopy. Sensors 2021, 21, 1186. [Google Scholar] [CrossRef]
- Meza Ramirez, C.A.; Greenop, M.; Ashton, L.; Rehman, I.U. Applications of machine learning in spectroscopy. Applied Spectroscopy Reviews 2021, 56, 733–763. [Google Scholar] [CrossRef]
- Kumar, A.; Alsadoon, A.; Prasad PW, C.; Abdullah, S.; Rashid, T.A.; Pham DT, H.; Nguyen TQ, V. Generative adversarial network (GAN) and enhanced root mean square error (ERMSE): deep learning for stock price movement prediction. Multimedia Tools and Applications 2022, 1–19. [Google Scholar] [CrossRef]
- Karunasingha DS, K. Root mean square error or mean absolute error? Use their ratio as well. Information Sciences 2022, 585, 609–629. [Google Scholar] [CrossRef]
- Menard, S. Coefficients of determination for multiple logistic regression analysis. The American Statistician 2000, 54, 17–24. [Google Scholar] [CrossRef]
- Dhakate, P.P.; Patil, S.; Rajeswari, K.; Abin, D. Preprocessing and Classification in WEKA using different classifiers. Inter J Eng Res Appl 2014, 4, 91–93. [Google Scholar]
- Ferreira, A.J.; Figueiredo, M.A. Efficient feature selection filters for high-dimensional data. Pattern recognition letters 2012, 33, 1794–1804. [Google Scholar] [CrossRef]
- Kumar, C.A.; Sooraj, M.P.; Ramakrishnan, S. A comparative performance evaluation of supervised feature selection algorithms on microarray datasets. Procedia computer science 2017, 115, 209–217. [Google Scholar] [CrossRef]
- Majd, A.; Sahebi, G.; Daneshtalab, M.; Plosila, J.; Lotfi, S.; Tenhunen, H. Parallel imperialist competitive algorithms. Concurrency and Computation: Practice and Experience 2018, 30, e4393. [Google Scholar] [CrossRef]
- Atashpendar, A.; Dorronsoro, B.; Danoy, G.; Bouvry, P. A scalable parallel cooperative coevolutionary PSO algorithm for multi-objective optimization. Journal of Parallel and Distributed Computing 2018, 112, 111–125. [Google Scholar] [CrossRef]
- Ramdania, D.R.; Irfan, M.; Alfarisi, F.; Nuraiman, D. Comparison of genetic algorithms and Particle Swarm Optimization (PSO) algorithms in course scheduling. In Journal of Physics: Conference Series (Vol. 1402, No. 2, p. 022079). IOP Publishing, 2019.
- Fu, J.; Yu, H.D.; Chen, Z.; Yun, Y.H. A review on hybrid strategy-based wavelength selection methods in analysis of near-infrared spectral data. Infrared Physics & Technology 2020, 104231. [Google Scholar]
- Yang, J.; Sun, L.; Xing, W.; Feng, G.; Bai, H.; Wang, J. Hyperspectral prediction of sugarbeet seed germination based on gauss kernel SVM. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 2021, 253, 119585. [Google Scholar] [CrossRef] [PubMed]
- Saputri DA, S.; Pahlawan MF, R.; Murti, B.M.; Masithoh, R.E. Vis/NIR spectroscopy for non-destructive method in detecting soybean seeds viability. In IOP Conference Series: Earth and Environmental Science (Vol. 1038, No. 1, p. 012043). IOP Publishing, 2022.
- Pahlawan MF, R.; Wati, R.K.; Masithoh, R.E. Development of a low-cost modular VIS/NIR spectroscopy for predicting soluble solid content of banana. In IOP Conference Series: Earth and Environmental Science (Vol. 644, No. 1, p. 012047). IOP Publishing, 2021.
- Wati, R.K.; Pahlawan MF, R.; Masithoh, R.E. Development of calibration model for pH content of intact tomatoes using a low-cost Vis/NIR spectroscopy. In IOP Conference Series: Earth and Environmental Science (Vol. 686, No. 1, p. 012049). IOP Publishing, 2021.
- Savi, A.; De Aguiar, L.M.; Tonial LM, S.; Lafay CB, B.; Assmann, T.S.; De Bortolli, M.A. Fast and Non-Destructive Determination of N, P, and K in Sorghum, Oat, and Corn Residue Using Near-Infrared Spectroscopy. J. Agric. Sci. 2019. [Google Scholar] [CrossRef]
- Yan, Y.L.; Chen, B.; Zhu, D. Near infrared spectroscopy-principles, technologies and applications. Beijing, China.
- Arora, R. Comparative analysis of classification algorithms on different datasets using WEKA. International Journal of Computer Applications 2012, 54. [Google Scholar] [CrossRef]
- Neo ER, K.; Yeo, Z.; Low JS, C.; Goodship, V.; Debattista, K. A review on chemometric techniques with infrared, Raman and laser-induced breakdown spectroscopy for sorting plastic waste in the recycling industry. Resources, Conservation and Recycling 2022, 180, 106217. [Google Scholar] [CrossRef]

| The studied index | Equation | References |
|---|---|---|
| Germination Energy | [10] | |
| Germination Value | [33] | |
| Germination Vigour | [34] | |
| Allometric Coefficien | [35] | |
| Daily Germination Speed | [36] | |
| Mean Daily Germination | [37] |
| Variety | Period | Number | Germination percentage | Germination Energy | Mean Daily Germination | Germination Value | Daily Germination Speed | Simple vitality index |
|---|---|---|---|---|---|---|---|---|
| NC-2 | 1 | 100 | 91 | 86 | 11.3750 | 250.2500 | 0.0879 | 7.0752 |
| NC-2 | 2 | 100 | 69 | 61 | 8.6250 | 146.6250 | 0.1159 | 3.7826 |
| NC-2 | 3 | 100 | 47 | 34 | 5.8750 | 64.6250 | 0.1702 | 0.9795 |
| Florispan | 1 | 100 | 94 | 92 | 11.7500 | 329.0000 | 0.0851 | 9.5836 |
| Florispan | 2 | 100 | 72 | 63 | 9.000 | 153.0000 | 0.1111 | 4.0761 |
| Florispan | 3 | 100 | 52 | 65 | 6.5000 | 84.5000 | 0.1538 | 2.0072 |
| AL | NOF | Wavelengths | ET | RMSE | CR(R2) |
|---|---|---|---|---|---|
| WCC | 14 | 704, 694, 775, 835,1025, 991, 906, 824, 852, 738, 795, 699, 963, 767 | 23.3603 | 0.0028 | 0.9870 |
| LCA | 10 | 748, 915, 783, 967, 887, 869, 801, 696, 744, 883 | 44.9389 | 0.0025 | 0.9872 |
| GA | 16 | 870, 799, 783, 636, 846, 785, 734, 737, 762, 954, 827, 913, 714, 810, 904, 725 | 3.4135 | 0.0028 | 0.9868 |
| PSO | 15 | 911, 784, 992, 713, 839, 726, 928, 840, 691, 791, 963, 832, 775, 737, 817 | 2.5181 | 0.0026 | 0.9870 |
| ACO | 16 | 780, 759, 868, 777, 814, 704, 804, 982, 952, 775, 1017, 934, 685, 905, 800, 657 | 31.1699 | 0.0027 | 0.9870 |
| ICA | 16 | 791, 844, 786, 731, 929, 1003, 798, 675, 1022, 774, 710, 888, 777, 978, 901, 697 | 2.3635 | 0.0027 | 0.9867 |
| LA | 15 | 935, 790, 768, 796, 776, 955, 732, 818, 883,694, 866, 1027, 783, 722, 824 | 33.8388 | 0.0025 | 0.9876 |
| HTS | 16 | 920, 828, 762, 804, 811, 503, 862, 837, 785, 779, 698, 846, 845, 957, 854, 633 | 14.3798 | 0.0028 | 0.9866 |
| FOA | 10 | 754, 825, 731, 778, 962, 902, 794, 738, 707, 856 | 17.1878 | 0.0025 | 0.9874 |
| DSOS | 16 | 915, 681, 672, 977, 815, 994, 956, 798, 939, 581, 522, 819, 690, 793, 760, 806 | 23.8145 | 0.0033 | 0.9854 |
| CUK | 12 | 874, 734, 706, 878, 1018, 775, 972, 742, 791, 843, 967, 723 | 33.3829 | 0.0027 | 0.9870 |
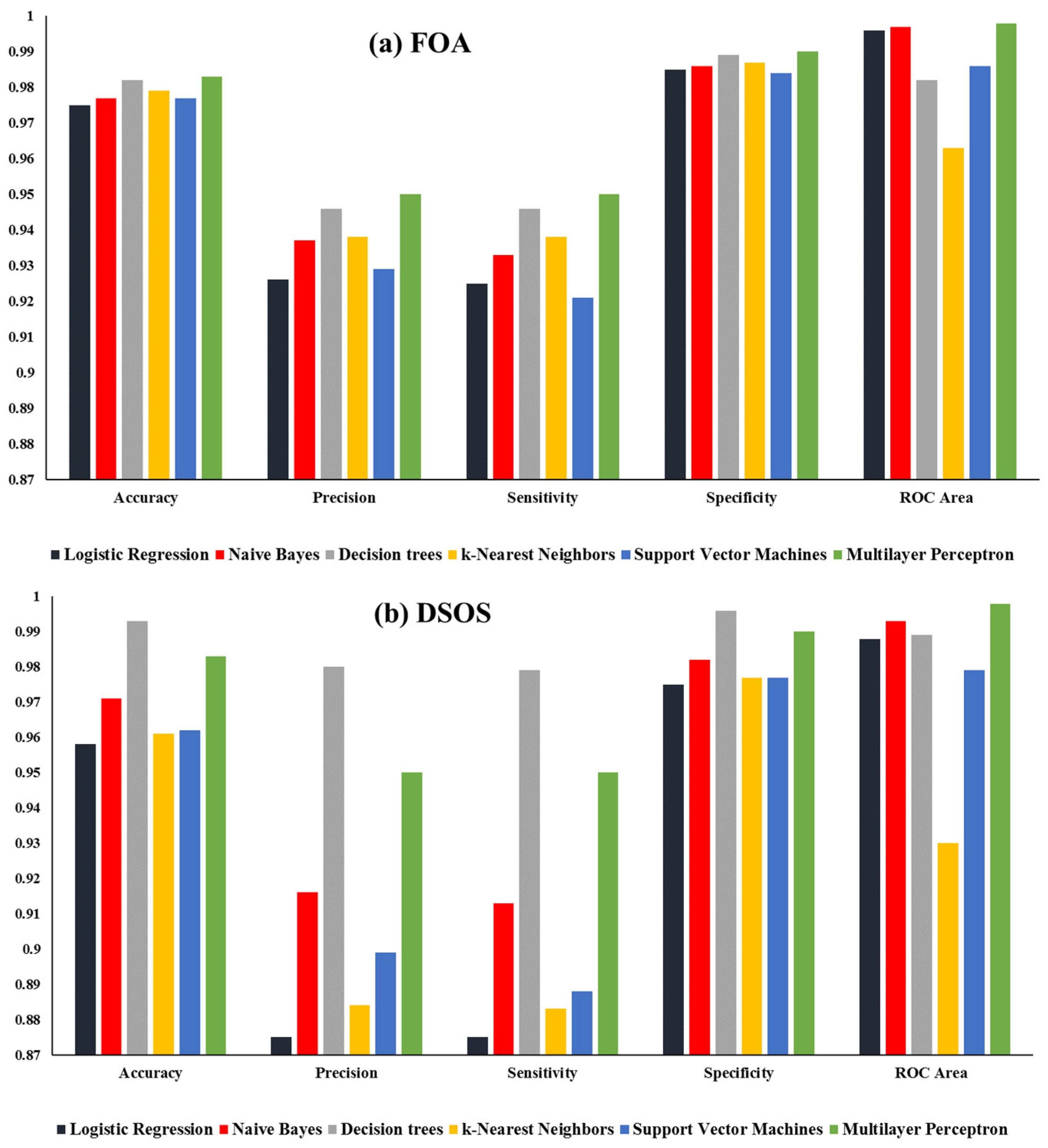
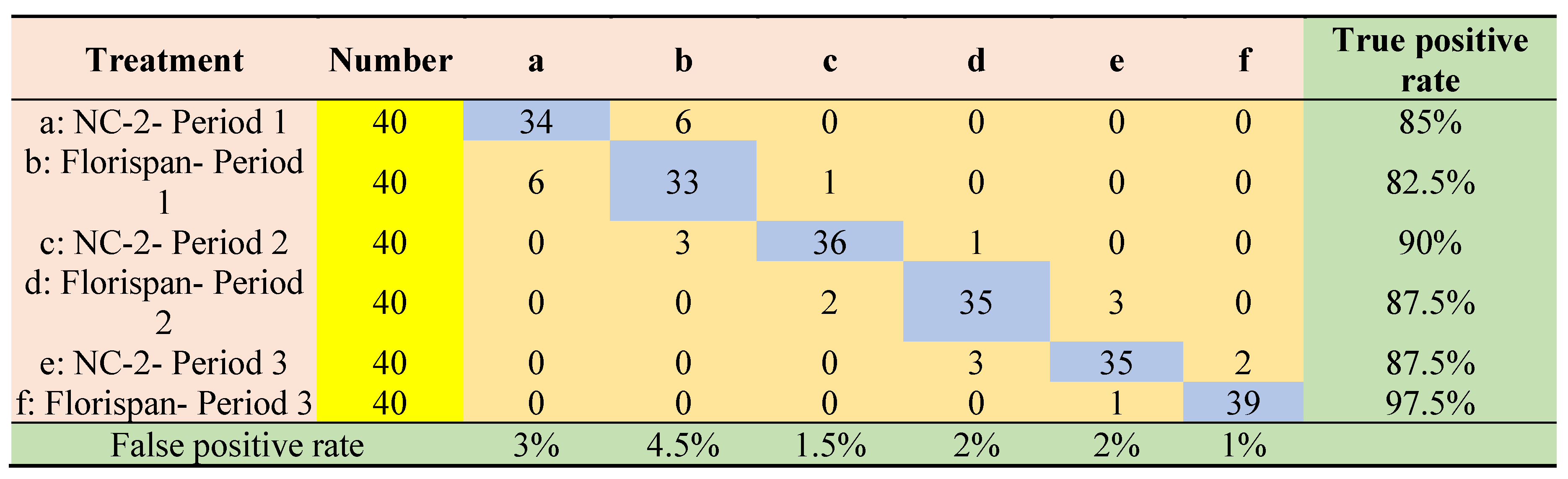
| Algorithm | classifiers | Accuracy | Precision | Sensitivity | Specificity | ROC Area |
|---|---|---|---|---|---|---|
| FOA | Logistic Regression | 0.9750 | 0.9260 | 0.9250 | 0.9850 | 0.9960 |
| Naive Bayes | 0.9770 | 0.9370 | 0.9330 | 0.9860 | 0.9970 | |
| Decision trees | 0.9820 | 0.9460 | 0.9460 | 0.9890 | 0.9820 | |
| k-Nearest Neighbors | 0.9790 | 0.9380 | 0.9380 | 0.9870 | 0.9630 | |
| Support Vector Machines | 0.9770 | 0.9290 | 0.9210 | 0.9840 | 0.9860 | |
| Multilayer Perceptron | 0.9830 | 0.9500 | 0.9500 | 0.9900 | 0.9980 | |
| DSOS | Logistic Regression | 0.9580 | 0.8750 | 0.8750 | 0.9750 | 0.9880 |
| Naive Bayes | 0.9710 | 0.9160 | 0.9130 | 0.9820 | 0.9930 | |
| Decision trees | 0.9930 | 0.9800 | 0.9790 | 0.9960 | 0.9890 | |
| k-Nearest Neighbors | 0.9610 | 0.8840 | 0.8830 | 0.9770 | 0.9300 | |
| Support Vector Machines | 0.9620 | 0.8990 | 0.8880 | 0.9770 | 0.9790 | |
| Multilayer Perceptron | 0.9830 | 0.9500 | 0.9500 | 0.9900 | 0.9980 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





