Submitted:
08 November 2023
Posted:
09 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
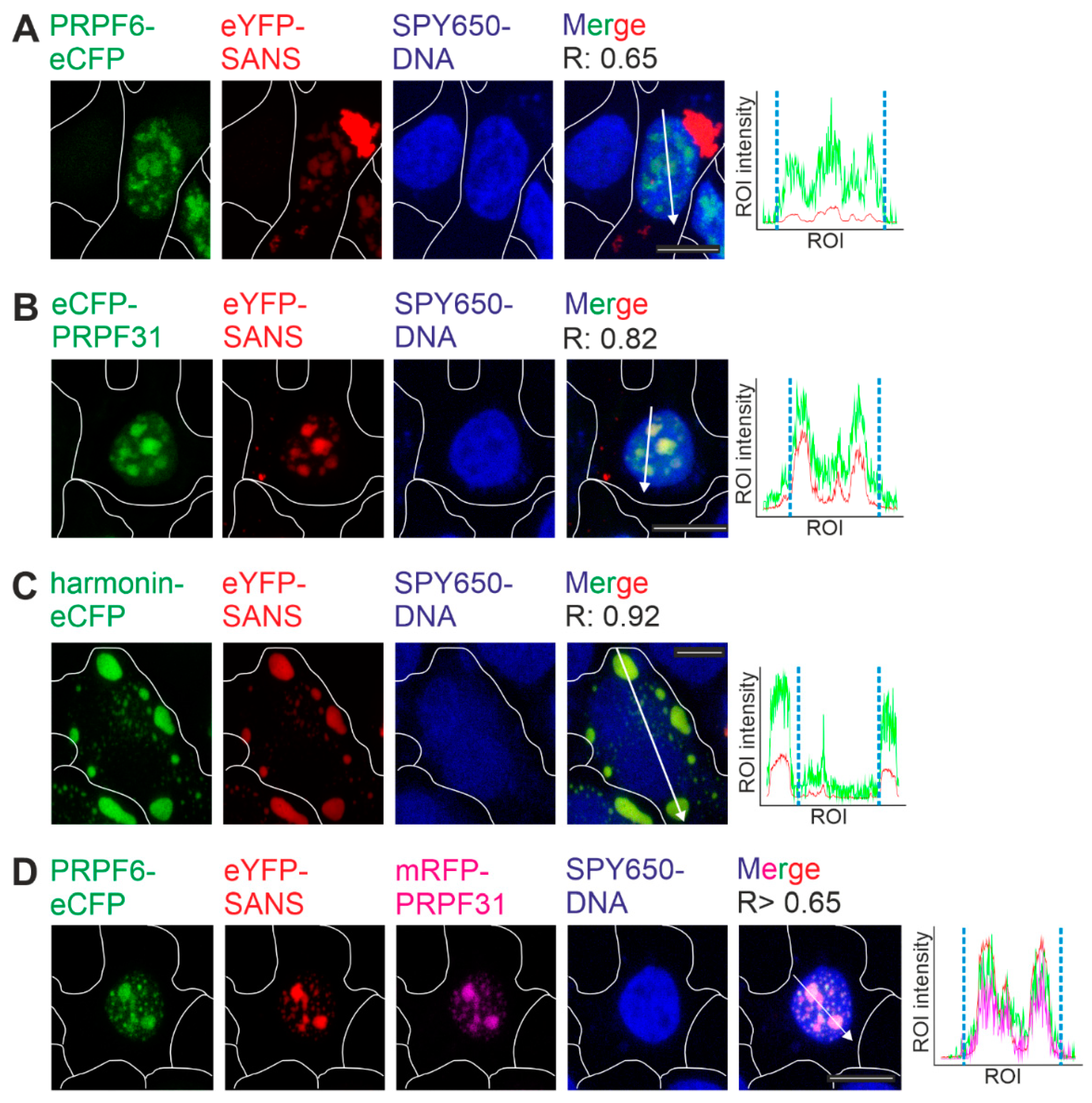
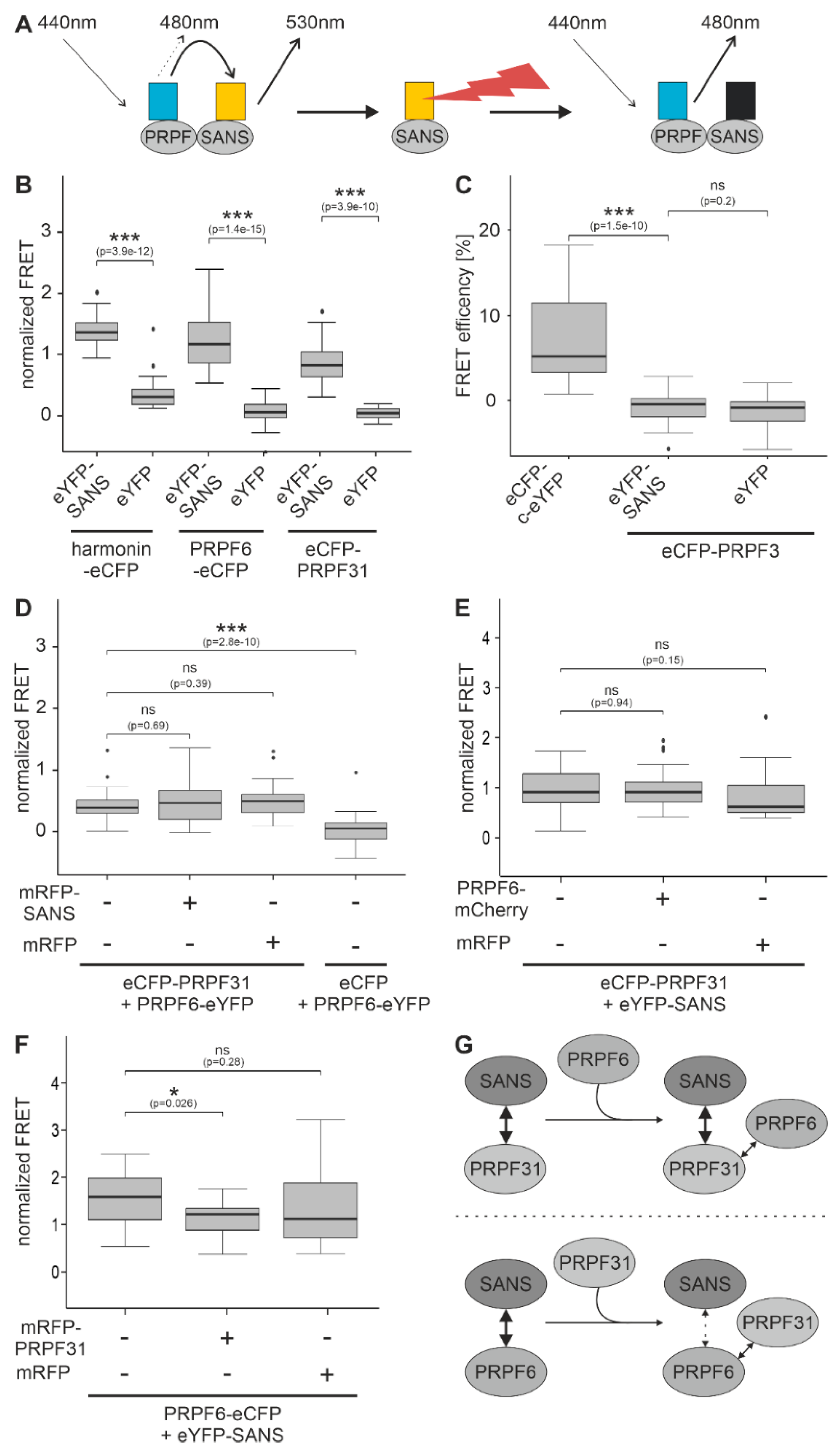
2.1. Binding of SANS to PRPF31 and PRPF6 in the nucleus revealed by FRET
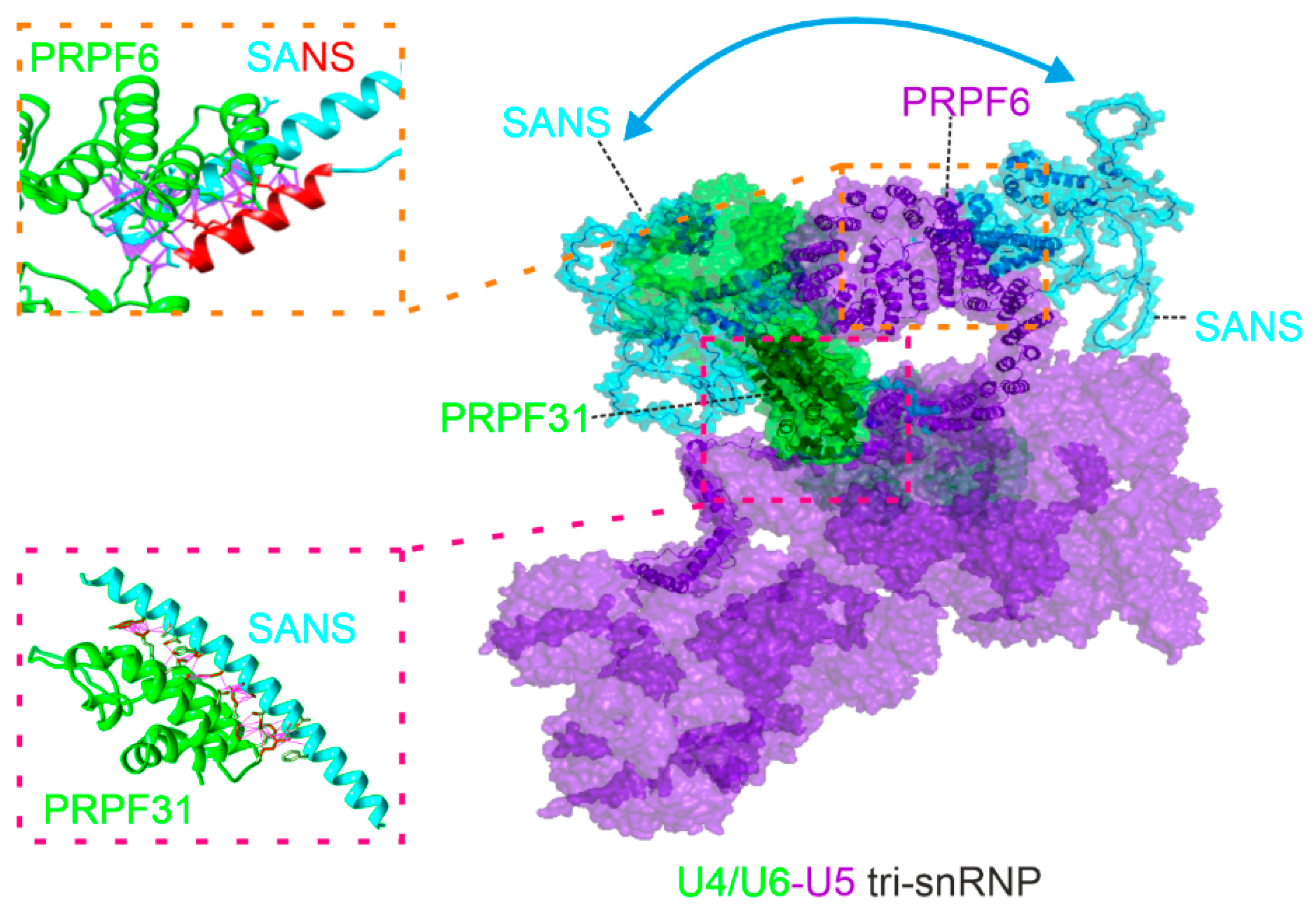
2.2. PRPF31 and PRPF6 interact with different regions in SANS CENTn domain
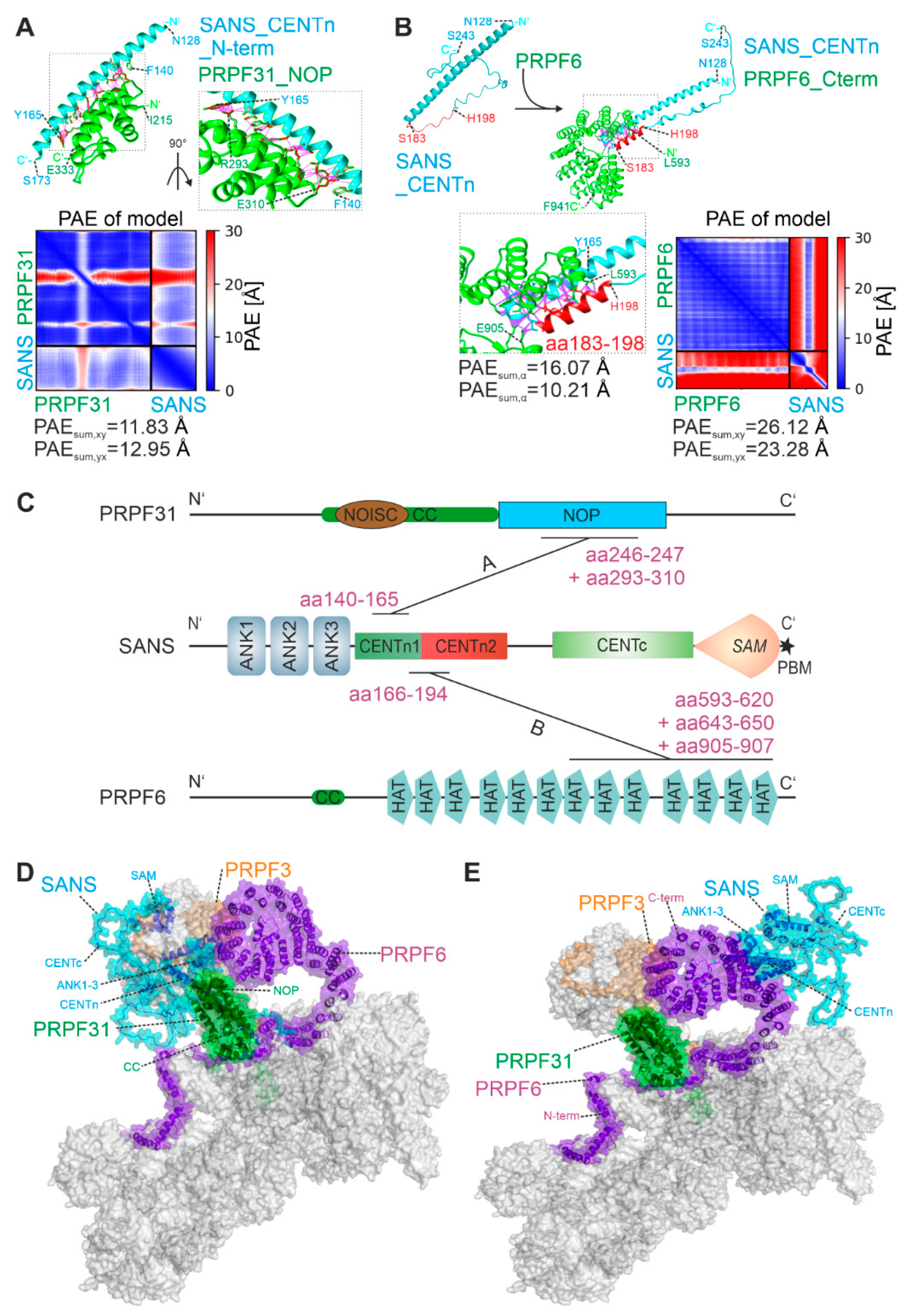
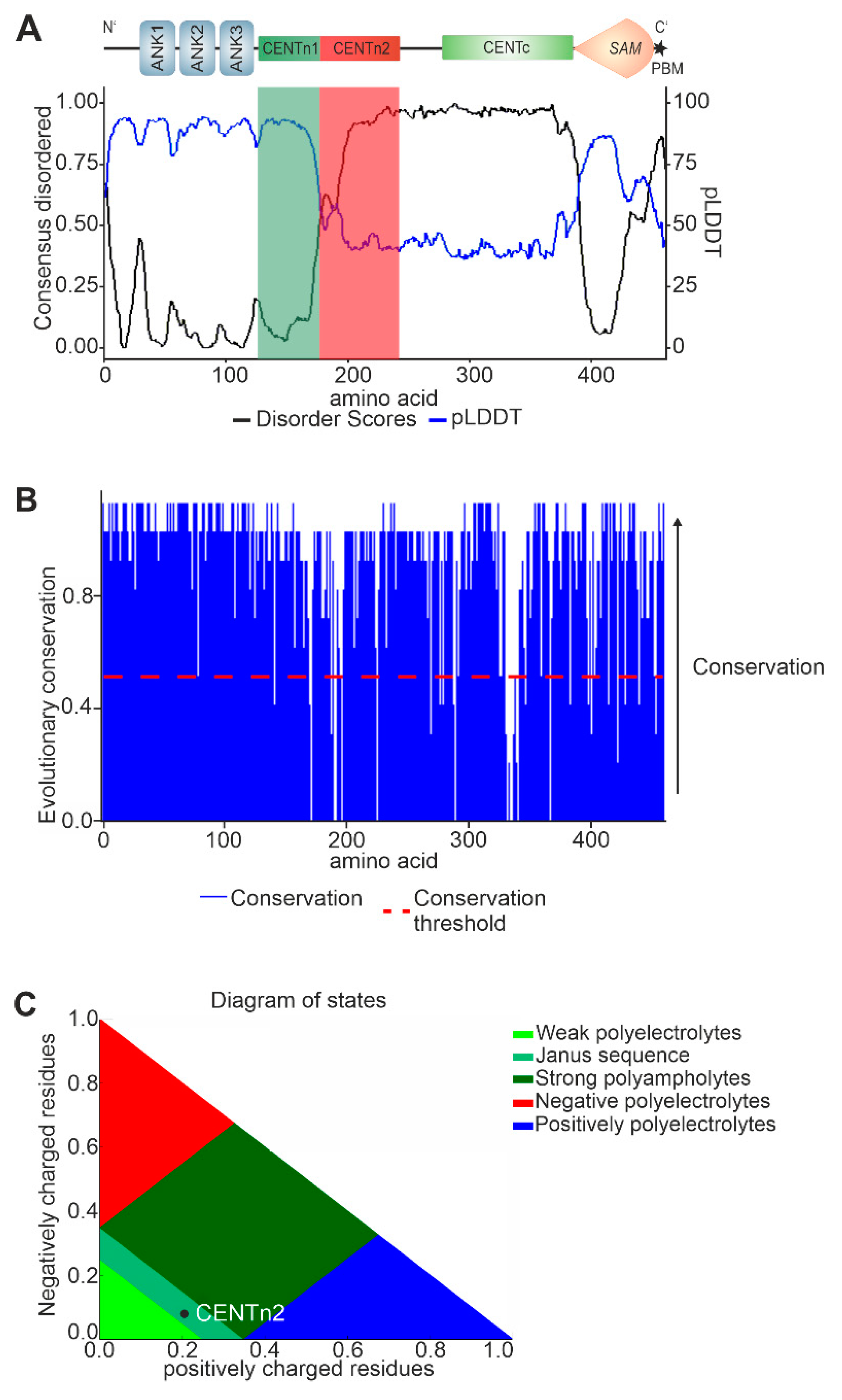
2.3. In silico analysis predicts evolutionary conserved multi-conformational intrinsically disordered regions (IRDs) for SANS
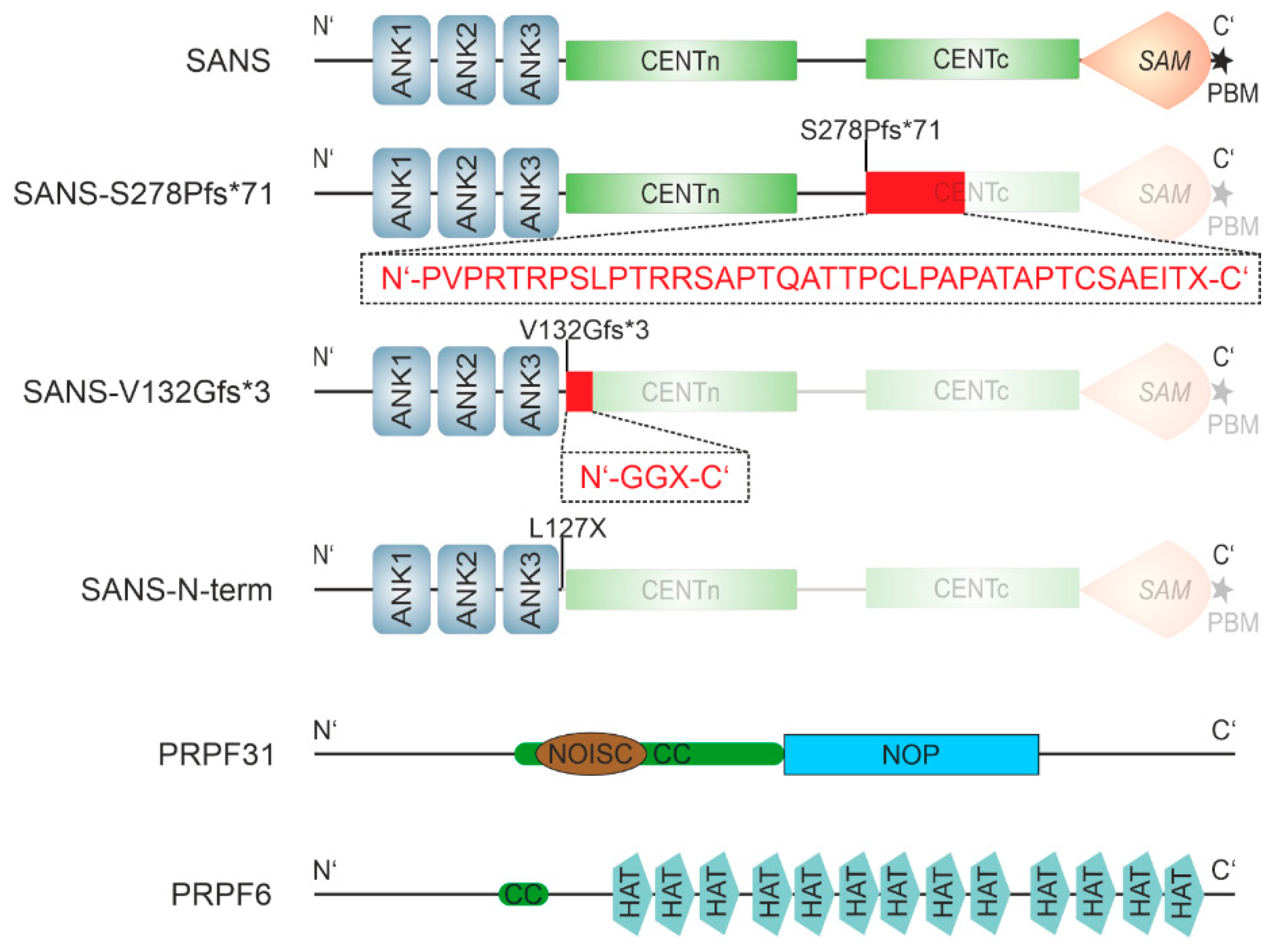
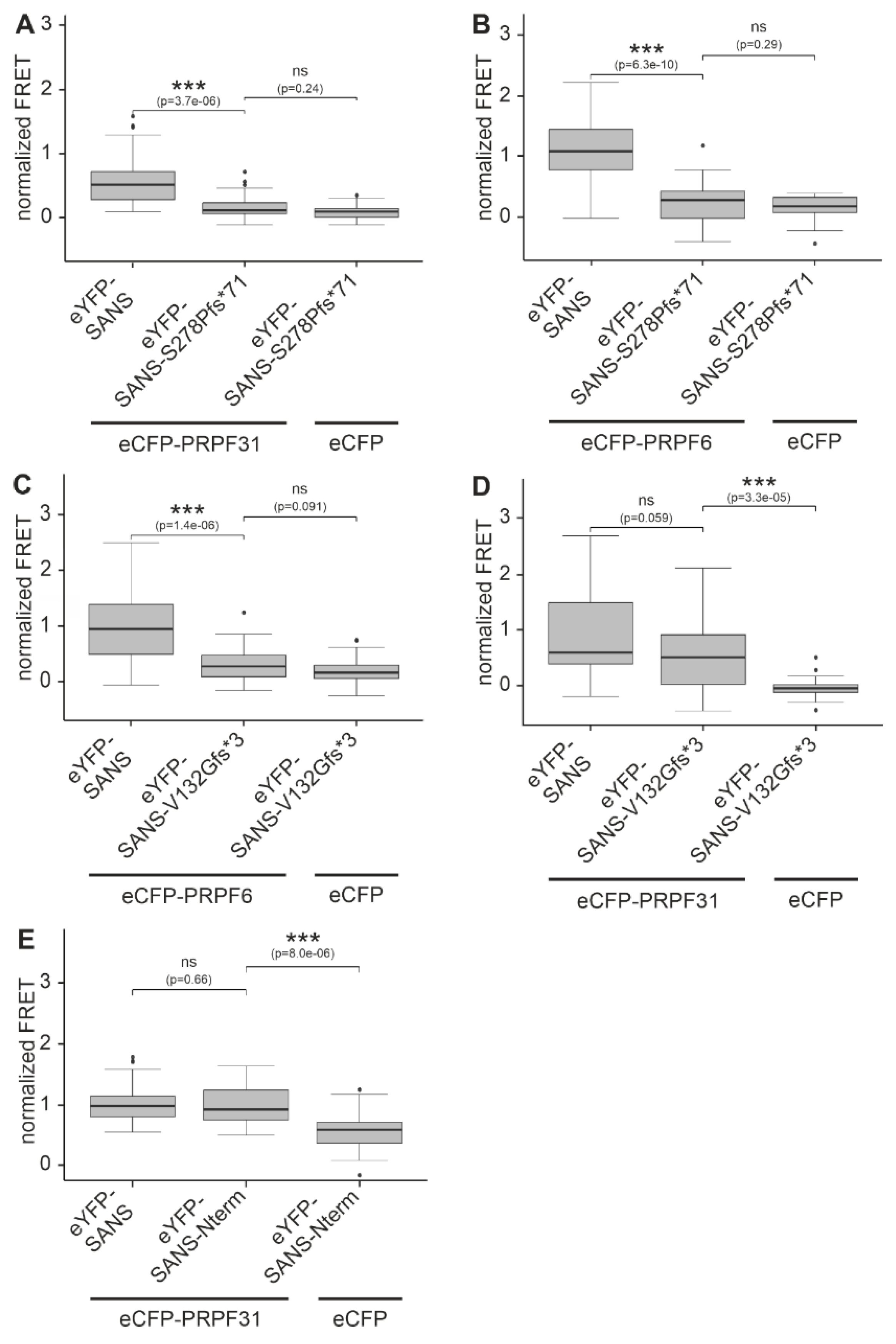
2.4. USH-causing pathogenic variants of SANS show altered interaction with PRPFs
3. Discussion
4. Materials and Methods
4.1. DNA constructs and primers
4.2. Cloning
4.3. Cell culture and cell lines
4.4. Fluorescence co-localization observation
4.5. Fluorescence resonance energy transfer (FRET) acceptor photobleaching assay
4.6. AlphaFold2-multimer
4.7. Evolutionary Conservation
4.8. Additional bioinformatic analyses
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Papasaikas, P.; Tejedor, J.R.; Vigevani, L.; Valcárcel, J. Functional Splicing Network Reveals Extensive Regulatory Potential of the Core Spliceosomal Machinery. Mol Cell 2015, 57, 7–22. [CrossRef]
- Will, C.L.; Lührmann, R. Spliceosome Structure and Function. Cold Spring Harb Perspect Biol 2011, 3, 1–2. [CrossRef]
- Yildirim, A.; Mozaffari-Jovin, S.; Wallisch, A.K.; Schäfer, J.; Ludwig, S.E.J.; Urlaub, H.; Lührmann, R.; Wolfrum, U. SANS (USH1G) Regulates Pre-MRNA Splicing by Mediating the Intra-Nuclear Transfer of Tri-SnRNP Complexes. Nucleic Acids Res 2021, 49, 5845–5866. [CrossRef]
- Matera, A.G.; Wang, Z. A Day in the Life of the Spliceosome. Nat Rev Mol Cell Biol 2014, 15, 108–121. [CrossRef]
- Fuster-Garcia, C.; Garcia-Bohorquez, B.; Rodriguez-Munoz, A.; Aller, E.; Jaijo, T.; Millan, J.M.; Garcia-Garcia, G. Usher Syndrome: Genetics of a Human Ciliopathy. Int J Mol Sci 2021, 22. [CrossRef]
- Velde, H.M.; Reurink, J.; Held, S.; Li, C.H.Z.; Yzer, S.; Oostrik, J.; Weeda, J.; Haer-Wigman, L.; Yntema, H.G.; Roosing, S.; et al. Usher Syndrome Type IV: Clinically and Molecularly Confirmed by Novel ARSG Variants. Hum Genet 2022, 141, 1723–1738. [CrossRef]
- Peter, V.G.; Quinodoz, M.; Sadio, S.; Held, S.; Rodrigues, M.; Soares, M.; Sousa, A.B.; Coutinho Santos, L.; Damme, M.; Rivolta, C. New Clinical and Molecular Evidence Linking Mutations in ARSG to Usher Syndrome Type IV. Hum Mutat 2021, 42, 261–271. [CrossRef]
- Weil, D.; El-Amraoui, A.; Masmoudi, S.; Mustapha, M.; Kikkawa, Y.; Laine, S.; Delmaghani, S.; Adato, A.; Nadifi, S.; Zina, Z. Ben; et al. Usher Syndrome Type I G (USH1G) Is Caused by Mutations in the Gene Encoding SANS, a Protein That Associates with the USH1C Protein, Harmonin. Hum Mol Genet 2003, 12, 463–471. [CrossRef]
- Sorusch, N.; Baub, K.; Plutniok, J.; Samanta, A.; Knapp, B.; Nagel-Wolfrum, K.; Wolfrum, U. Characterization of the Ternary Usher Syndrome SANS/Ush2a/Whirlin Protein Complex. Hum Mol Genet 2017, 26, 1157–1172. [CrossRef]
- Caberlotto, E.; Michel, V.; Foucher, I.; Bahloul, A.; Goodyear, R.J.; Pepermans, E.; Michalski, N.; Perfettini, I.; Alegria-Prévot, O.; Chardenoux, S.; et al. Usher Type 1G Protein sans Is a Critical Component of the Tip-Link Complex, a Structure Controlling Actin Polymerization in Stereocilia. Proc Natl Acad Sci U S A 2011, 108, 5825–5830. [CrossRef]
- He, Y.; Li, J.; Zhang, M. Myosin VII, USH1C, and ANKS4B or USH1G Together Form Condensed Molecular Assembly via Liquid-Liquid Phase Separation. Cell Rep 2019, 29, 974-986.e4. [CrossRef]
- Géléoc, G.G.S.; El-Amraoui, A. Disease Mechanisms and Gene Therapy for Usher Syndrome. Hear Res 2020, 394. [CrossRef]
- Maerker, T.; van Wijk, E.; Overlack, N.; Kersten, F.F.J.; Mcgee, J.; Goldmann, T.; Sehn, E.; Roepman, R.; Walsh, E.J.; Kremer, H.; et al. A Novel Usher Protein Network at the Periciliary Reloading Point between Molecular Transport Machineries in Vertebrate Photoreceptor Cells. Hum Mol Genet 2008, 17, 71–86. [CrossRef]
- Overlack, N.; Kilic, D.; Bauss, K.; Marker, T.; Kremer, H.; van Wijk, E.; Wolfrum, U.; Bauß, K.; Märker, T.; Kremer, H.; et al. Direct Interaction of the Usher Syndrome 1G Protein SANS and Myomegalin in the Retina. Biochim Biophys Acta 2011, 1813, 1883–1892. [CrossRef]
- Bauss, K.; Knapp, B.; Jores, P.; Roepman, R.; Kremer, H.; Wijk, E. V; Marker, T.; Wolfrum, U. Phosphorylation of the Usher Syndrome 1G Protein SANS Controls Magi2-Mediated Endocytosis. Hum Mol Genet 2014, 23, 3923–3942. [CrossRef]
- Papal, S.; Cortese, M.; Legendre, K.; Sorusch, N.; Dragavon, J.; Sahly, I.; Shorte, S.; Wolfrum, U.; Petit, C.; El-Amraoui, A. The Giant Spectrin BetaV Couples the Molecular Motors to Phototransduction and Usher Syndrome Type I Proteins along Their Trafficking Route. Hum Mol Genet 2013, 22, 3773–3788. [CrossRef]
- Sorusch, N.; Yildirim, A.; Knapp, B.; Janson, J.; Fleck, W.; Scharf, C.; Wolfrum, U. SANS (USH1G) Molecularly Links the Human Usher Syndrome Protein Network to the Intraflagellar Transport Module by Direct Binding to IFT-B Proteins. Front Cell Dev Biol 2019, 7, 216. [CrossRef]
- Sahly, I.; Dufour, E.; Schietroma, C.; Michel, V.; Bahloul, A.; Perfettini, I.; Pepermans, E.; Estivalet, A.; Carette, D.; Aghaie, A.; et al. Localization of Usher 1 Proteins to the Photoreceptor Calyceal Processes, Which Are Absent from Mice. J Cell Biol 2012, 199, 381–399. [CrossRef]
- May-Simera, H.; Nagel-Wolfrum, K.; Wolfrum, U. Cilia - The Sensory Antennae in the Eye. Prog Retin Eye Res 2017, 60, 144–180. [CrossRef]
- Cowan, C.S.; Renner, M.; De Gennaro, M.; Gross-Scherf, B.; Goldblum, D.; Hou, Y.; Munz, M.; Rodrigues, T.M.; Krol, J.; Szikra, T.; et al. Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution. Cell 2020, 182, 1623-1640 e34. [CrossRef]
- Adato, A.; Michel, V.; Kikkawa, Y.; Reiners, J.; Alagramam, K.N.; Weil, D.; Yonekawa, H.; Wolfrum, U.; El-Amraoui, A.; Petit, C. Interactions in the Network of Usher Syndrome Type 1 Proteins. Hum Mol Genet 2005, 14, 347–356. [CrossRef]
- Sorusch, N.; Wunderlich, K.; Bauá, K.; Nagel-Wolfrum, K.; Wolfrum, U. Usher Syndrome Protein Network Functions in the Retina and Their Relation to Other Retinal Ciliopathies. Adv Exp Med Biol 2014. [CrossRef]
- Overlack, N.; Maerker, T.; Latz, M.; Nagel-Wolfrum, K.; Wolfrum, U. SANS (USH1G) Expression in Developing and Mature Mammalian Retina. Vision Res 2008, 48, 400–412. [CrossRef]
- Evans, R.; O’neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein Complex Prediction with AlphaFold-Multimer. bioRxiv 2021, 2021.10.04.463034. [CrossRef]
- Emenecker, R.J.; Griffith, D.; Holehouse, A.S. Metapredict: A Fast, Accurate, and Easy-to-Use Predictor of Consensus Disorder and Structure. Biophys. J. 2021, 120, 4312–4319. [CrossRef]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ Ecosystem: An Open Platform for Biomedical Image Analysis. Mol Reprod Dev 2015, 82, 518–529. [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat Methods 2022, 19, 679–682. [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J Comput Chem 2004, 25, 1605–1612. [CrossRef]
- Nunez-Castilla, J.; Siltberg-Liberles, J. An Easy Protocol for Evolutionary Analysis of Intrinsically Disordered Proteins. In Methods mol biol; Humana Press Inc., 2020; Vol. 2141, pp. 147–177. [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [CrossRef]
- Positteam Download RStudio - Posit Available online: https://posit.co/downloads/ (accessed on 24 July 2023).
- Karpova, T.S.; Baumann, C.T.; He, L.; Wu, X.; Grammer, A.; Lipsky, P.; Hager, G.L.; McNally, J.G. Fluorescence Resonance Energy Transfer from Cyan to Yellow Fluorescent Protein Detected by Acceptor Photobleaching Using Confocal Microscopy and a Single Laser. J Microsc 2003, 209, 56–70. [CrossRef]
- Yan, J.; Pan, L.; Chen, X.; Wu, L.; Zhang, M. The Structure of the Harmonin/sans Complex Reveals an Unexpected Interaction Mode of the Two Usher Syndrome Proteins. Proc Natl Acad Sci U S A 2010, 107, 4040–4045. [CrossRef]
- Liu, S.; Li, P.; Dybkov, O.; Nottrott, S.; Hartmuth, K.; Lührmann, R.; Carlomagno, T.; Wahl, M.C. Binding of the Human Prp31 Nop Domain to a Composite RNA-Protein Platform in U4 SnRNP. Science 2007, 316, 115–120. [CrossRef]
- Charenton, C.; Wilkinson, M.E.; Nagai, K. Mechanism of 5’ Splice Site Transfer for Human Spliceosome Activation. Science 2019, 364, 362–367. [CrossRef]
- Colcombet-Cazenave, B.; Druart, K.; Bonnet, C.; Petit, C.; Sperandio, O.; Guglielmini, J.; Wolff, N. Phylogenetic Analysis of Harmonin Homology Domains. BMC Bioinformatics 2021, 22, 190. [CrossRef]
- Alderson, T.R.; Pritišanac, I.; Kolarić, E.; Moses, A.M.; Forman-Kay, J.D. Systematic Identification of Conditionally Folded Intrinsically Disordered Regions by AlphaFold2. bioRxiv 2023, 2022.02.18.481080. [CrossRef]
- Nguyen Ba, A.N.; Yeh, B.J.; Van Dyk, D.; Davidson, A.R.; Andrews, B.J.; Weiss, E.L.; Moses, A.M. Proteome-Wide Discovery of Evolutionary Conserved Sequences in Disordered Regions. Sci Signal 2012, 5. [CrossRef]
- Holehouse, A.S.; Das, R.K.; Ahad, J.N.; Richardson, M.O.G.; Pappu, R. V. CIDER: Resources to Analyze Sequence-Ensemble Relationships of Intrinsically Disordered Proteins. Biophys J 2017, 112, 16–21. [CrossRef]
- Das, R.K.; Ruff, K.M.; Pappu, R. V. Relating Sequence Encoded Information to Form and Function of Intrinsically Disordered Proteins. Curr Opin Struct Biol 2015, 32, 102–112. [CrossRef]
- Sheinerman, F.B.; Norel, R.; Honig, B. Electrostatic Aspects of Protein-Protein Interactions. Curr Opin Struct Biol 2000, 10, 153–159. [CrossRef]
- Hevekerl, H.; Spielmann, T.; Chmyrov, A.; Widengren, J. Förster Resonance Energy Transfer beyond 10 Nm: Exploiting the Triplet State Kinetics of Organic Fluorophores. J Phys Chem 2011, 115, 13360–13370. [CrossRef]
- Roszik, J.; Tóth, G.; Szöllosi, J.; Vereb, G. Validating Pharmacological Disruption of Protein-Protein Interactions by Acceptor Photobleaching FRET Imaging. Methods mol biol 2013, 986, 165–178. [CrossRef]
- Tancredi, T.; Carrà, G.; Guerrini, R.; Arduin, M.; Calò, G.; Regoli, D.; Salvadori, S.; Temussi, P.A. The Interaction of Highly Helical Structural Mutants with the NOP Receptor Discloses the Role of the Address Domain of Nociceptin/Orphanin FQ. Chem Eur J 2005, 11, 2061–2070. [CrossRef]
- Ghram, M.; Morris, G.; Culjkovic-Kraljacic, B.; Mars, J.; Gendron, P.; Skrabanek, L.; Revuelta, M.V.; Cerchietti, L.; Guzman, M.L.; Borden, K.L.B. The Eukaryotic Translation Initiation Factor EIF4E Reprograms Alternative Splicing. EMBO J 2023, 42. [CrossRef]
- Bertram, K.; Agafonov, D.E.; Dybkov, O.; Haselbach, D.; Leelaram, M.N.; Will, C.L.; Urlaub, H.; Kastner, B.; Lührmann, R.; Stark, H. Cryo-EM Structure of a Pre-Catalytic Human Spliceosome Primed for Activation. Cell 2017, 170, 701-713.e11. [CrossRef]
- Novotny, I.; Blazikova, M.; Stanek, D.; Herman, P.; Malinsky, J. In Vivo Kinetics of U4/U6.U5 Tri-SnRNP Formation in Cajal Bodies. Mol Biol Cell 2011, 22, 513–523. [CrossRef]
- Stanek, D.; Pridalova-Hnilicova, J.; Novotny, I.; Huranova, M.; Blazikova, M.; Wen, X.; Sapra, A.K.; Neugebauer, K.M. Spliceosomal Small Nuclear Ribonucleoprotein Particles Repeatedly Cycle through Cajal Bodies. Mol Biol Cell 2008, 19, 2534–2543. [CrossRef]
- Fokkema, I.F.A.C.; Taschner, P.E.M.; Schaafsma, G.C.P.; Celli, J.; Laros, J.F.J.; den Dunnen, J.T. LOVD v.2.0: The next Generation in Gene Variant Databases. Hum Mutat 2011, 32, 557–563. [CrossRef]
- Nagel-Wolfrum, K.; Fadl, B.R.; Becker, M.M.; Wunderlich, K.A.; Schäfer, J.; Sturm, D.; Fritze, J.; Gür, B.; Kaplan, L.; Andreani, T.; et al. Expression and Subcellular Localization of USH1C/Harmonin in Human Retina Provides Insights into Pathomechanisms and Therapy. Hum Mol Genet 2023, 32, 431–449. [CrossRef]
- Islam, Z.; Nagampalli, R.S.K.; Fatima, M.T.; Ashraf, G.M. New Paradigm in Ankyrin Repeats: Beyond Protein-Protein Interaction Module. Int J Biol Macromol 2018, 109, 1164–1173. [CrossRef]
- Tanackovic, G.; Ransijn, A.; Ayuso, C.; Harper, S.; Berson, E.L.; Rivolta, C. A Missense Mutation in PRPF6 Causes Impairment of Pre-MRNA Splicing and Autosomal-Dominant Retinitis Pigmentosa. Am. J. Hum. Genet. 2011, 88, 643–649. [CrossRef]
- Buskin, A.; Zhu, L.; Chichagova, V.; Basu, B.; Mozaffari-Jovin, S.; Dolan, D.; Droop, A.; Collin, J.; Bronstein, R.; Mehrotra, S.; et al. Disrupted Alternative Splicing for Genes Implicated in Splicing and Ciliogenesis Causes PRPF31 Retinitis Pigmentosa. Nat Commun 2018, 9. [CrossRef]
- Stenson, P.D.; Ball, E. V.; Mort, M.; Phillips, A.D.; Shiel, J.A.; Thomas, N.S.T.; Abeysinghe, S.; Krawczak, M.; Cooper, D.N. Human Gene Mutation Database (HGMD®): 2003 Update. Hum Mutat 2003, 21, 577–581. [CrossRef]
- Zelinger, L.; Swaroop, A. RNA Biology in Retinal Development and Disease. Trends Genet 2018, 34, 341–351. [CrossRef]
- Aisa-Marin, I.; Garcia-Arroyo, R.; Mirra, S.; Marfany, G. The Alter Retina: Alternative Splicing of Retinal Genes in Health and Disease. Int J Mol Sci 2021, 22. [CrossRef]
- Murphy, D.; Cieply, B.; Carstens, R.; Ramamurthy, V.; Stoilov, P. The Musashi 1 Controls the Splicing of Photoreceptor-Specific Exons in the Vertebrate Retina. PLoS Genet 2016, 12, e1006256. [CrossRef]
- Sundar, J.; Matalkah, F.; Jeong, B.; Stoilov, P.; Ramamurthy, V. The Musashi Proteins MSI1 and MSI2 Are Required for Photoreceptor Morphogenesis and Vision in Mice. J Biol Chem 2020, 296. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




