Submitted:
03 November 2023
Posted:
06 November 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Animals and cells
2.2. DNA isolation
2.3. Droplet digital PCR
2.3. Spleen and liver tissues from non-Auckland Island pigs
2.4. Tests for PERV-C
| Name | Sequence | Location (nucleotid number) | Accession number | Reference |
|---|---|---|---|---|
| PERV pol1-forward | CGACTGCCCCAAGGGTTCAA | 3568-3587 | HM159246 | Yang et al., 2015 [27] |
| PERV pol2-reverse | TCTCTCCTGCAAATCTGGGCC | 3803-3783 | ||
| PERV pol probe | /56FAM/CACGTACTGGAGGAGGGTCACCTG | 3678-3655 | ||
| Pig actin forward | TAACCGATCCTTTCAAGCATTT | Krüger et al., 2020 [23] | ||
| Pig actin reverse | TGGTTTCAAAGCTTGCATCATA | |||
| Pig actin probe | /5HEX/CGTGGGGATGCTTCCTGAGAAAG | |||
| Pig GAPDH forward Pig GAPDH reverse |
CCGCGATCTAATGTTCTCTTTC TTCACTCCGACCTTCACCAT | 3951-3970 4022-4001 |
NC_010447.5 (396823) | Krüger et al., 2020 [23] |
| Pig GAPDH probe | /5HEX/CAGCCGCGTCCCTGAGACAC | 3991-3972 | ||
| PCR1* PERV-C forward PERV-C reverse |
CTGACCTGGATTAGAACTGG ATGTTAGAGGATGGTCCTGG |
6606-6625 6867-6886 |
AM229312 |
Takeuchi et al. [25], Kaulitz et al. [26] |
| PCR6* PERV-C 2 forward PERV-C 2 reverse |
CCAGGACCACCAAATAATGG AAGTTTTGCCCCCATTTTAGT |
6435-6454 6924-6944 |
Kaulitz et al. [26] | |
| Real-time PCR* PERV-C 3 forward PERV-C 3 reverse PERV-C probe |
CCCCAACCCAAGGACCAG AAGTTTTGCCCCCATTTTAGT FAM-CTCTAACATAACTTCTGGATCAGACCC- BHQ1 |
6853-6870 6924-6944 6878-6904 |
2.5. Tests for additional porcine viruses
3. Results
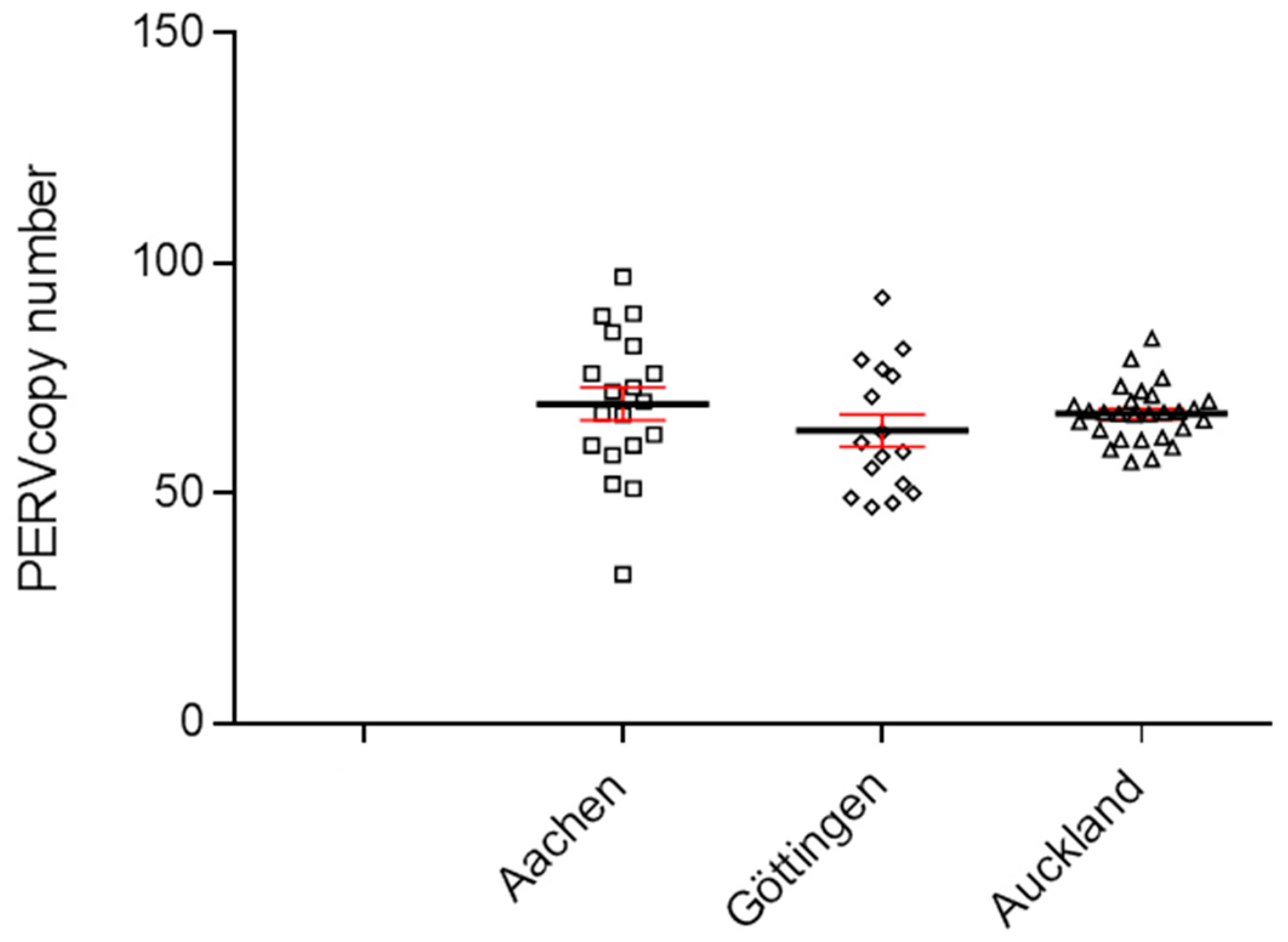
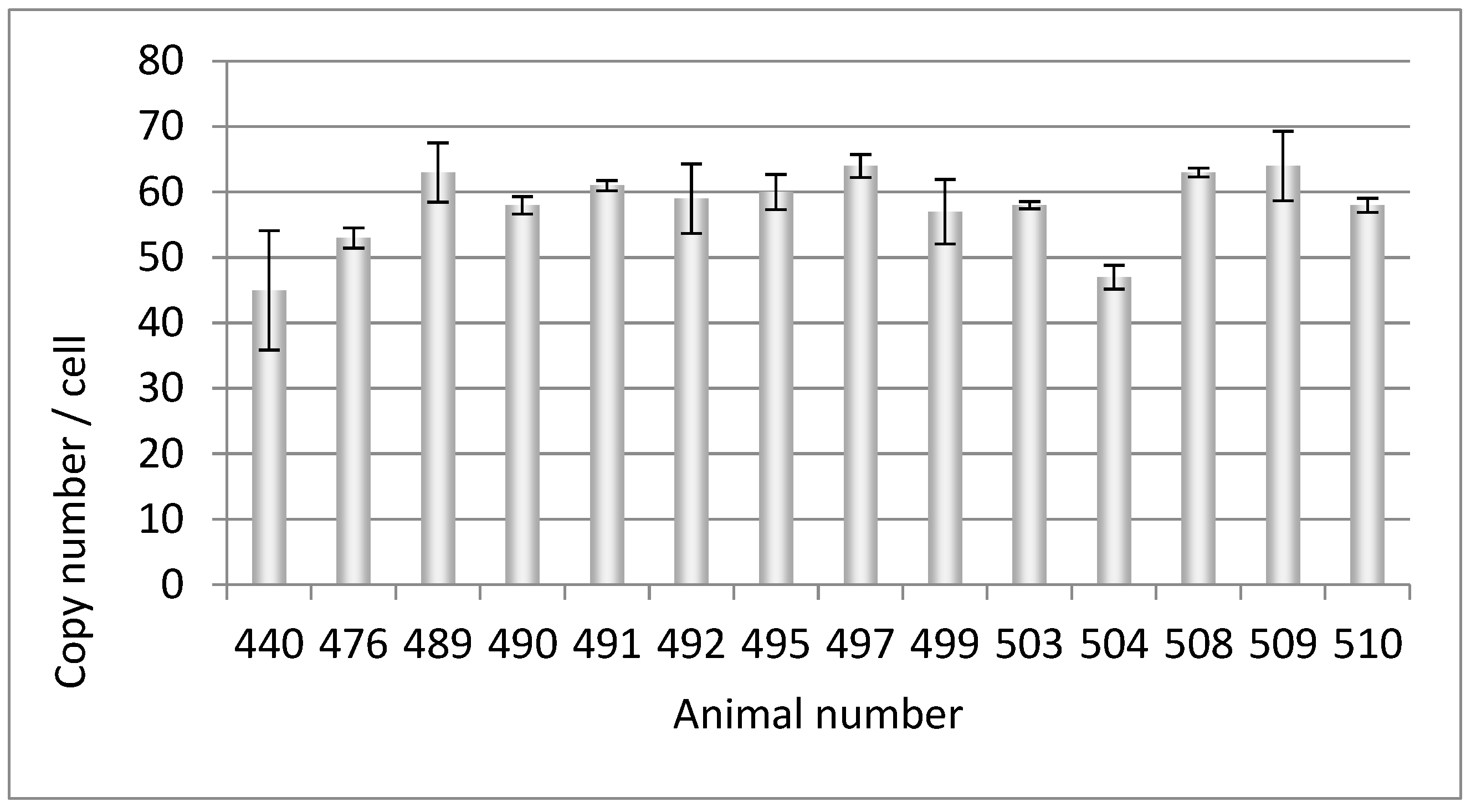
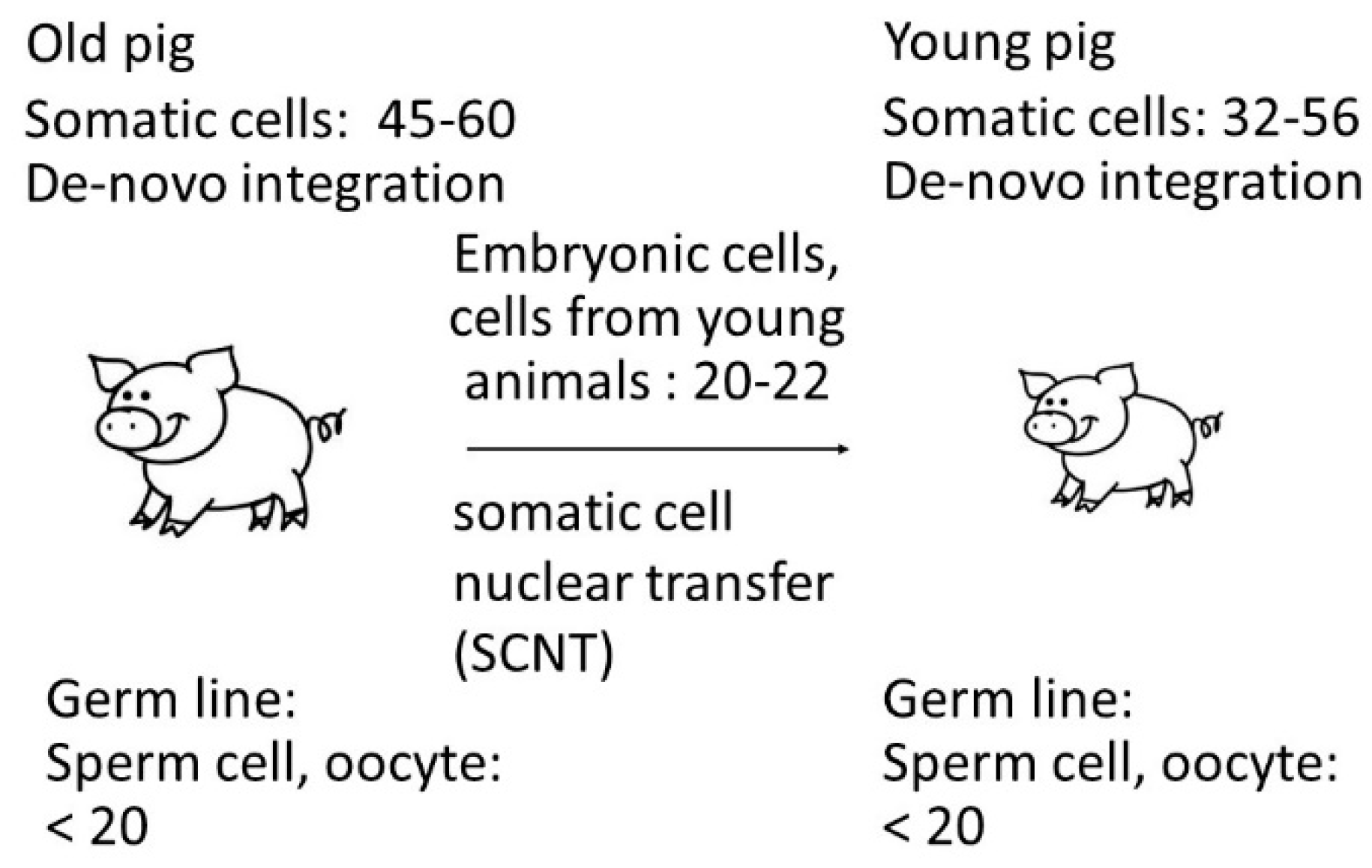
3.1. PERV copy number in the genome of adult Auckland Island pigs
3.2. Selection of PERV-C negative animals
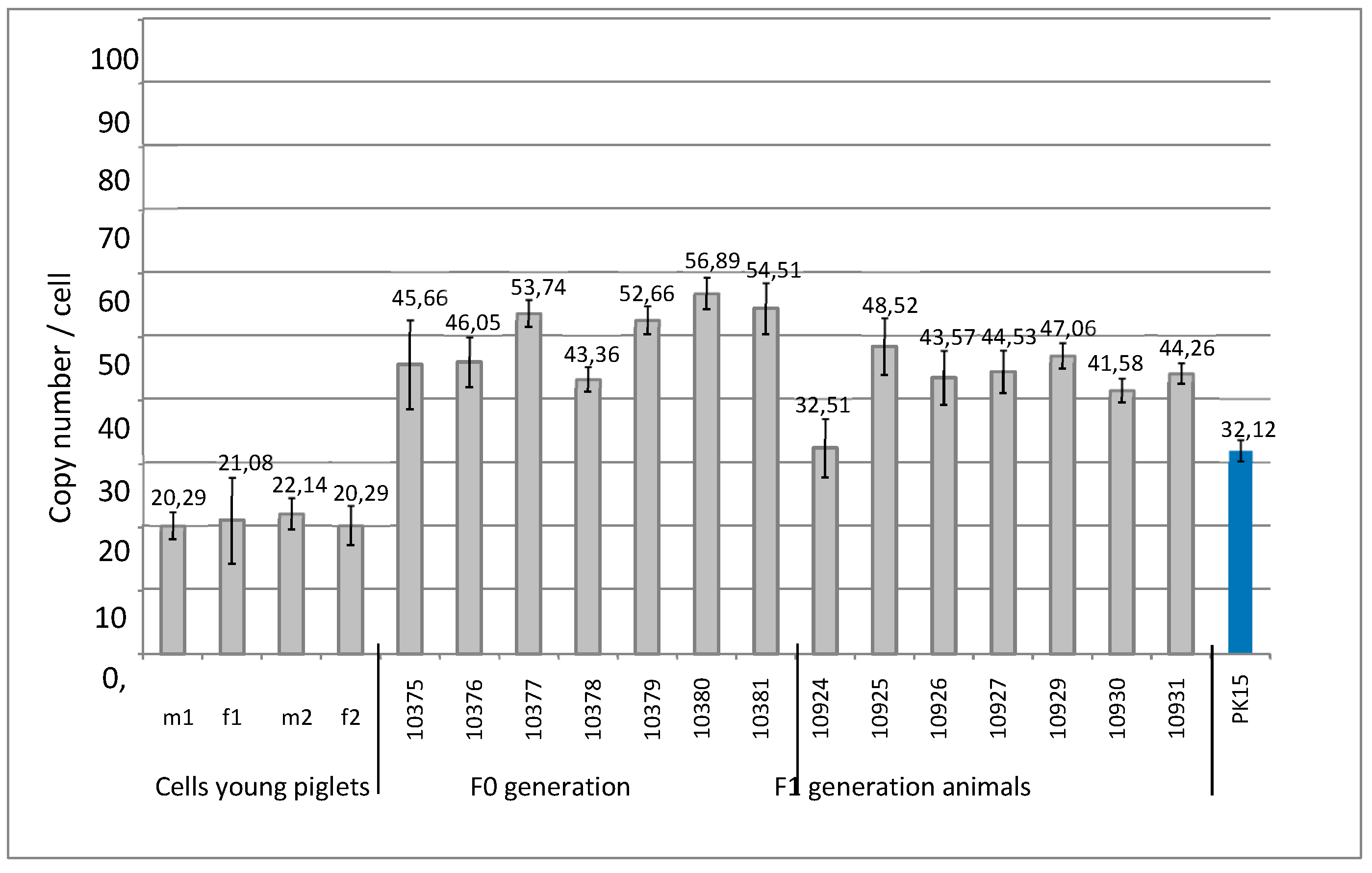
3.3. PERV copy number in the genome of cell lines from very young piglets
3.4. PERV copy number in the genome of Auckland Island pigs obtained by SCNT
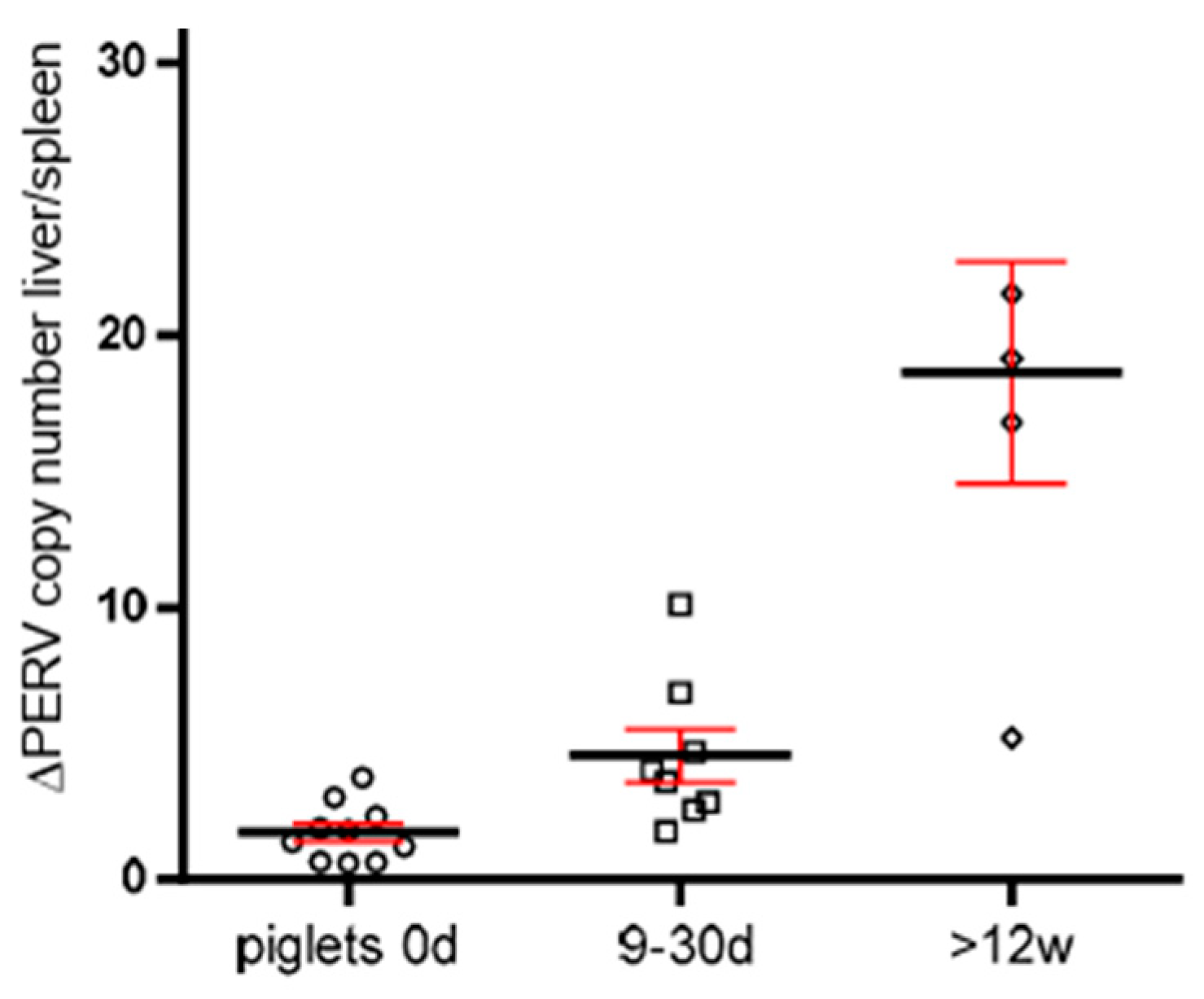
3.5. Increase of the PERV copy number with age in non-Auckland Island pigs
3.6. Further virological characterization of the Auckland Island pigs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gongora J, Garkavenko, O, Moran C. Origins of Kune Kune and Auckland Island pigs in New Zealand. 7th World Congress on Genetic Applied to Livestock Production, August 19-23, 2002, Montpellier, France.
- Robins JH, Matisoo-Smith, E Ross HA. The origins of the feral pigs on the Auckland Islands. J. Royal Society of New Zealand, 2003, 33 (2), 561-569.
- Fan B, Gongora J, Chen Y, Garkavenko O, Li Moran C. Population genetic variability and origin of Auckland Island feral pigs. J. Royal Society of New Zealand, 2005, 35(3), 279-285. [CrossRef]
- Garkavenko O, Muzina M, Muzina Z, Powels K, Elliott RB, Croxson MC. Monitoring for potentially xenozoonotic viruses in New Zealand pigs. J Med Virol. 2004, 72(2), 338-344. [CrossRef]
- Garkavenko O, Wynyard S, Nathu D, Simond D, Muzina M, Muzina Z, Scobie L, Hector RD, Croxson MC, Tan P, Elliott BR. Porcine endogenous retrovirus (PERV) and its transmission characteristics: a study of the New Zealand designated pathogen-free herd. Cell Transplant. 2008, 17(12), 1381-1388. [CrossRef]
- Garkavenko O, Dieckhoff B, Wynyard S, Denner J, Elliott RB, Tan PL, Croxson MC. Absence of transmission of potentially xenotic viruses in a prospective pig to primate islet xenotransplantation study. J Med Virol. 2008, 80(11), 2046-2052. [CrossRef]
- Wynyard S, Nathu D, Garkavenko O, Denner J, Elliott R. Microbiological safety of the first clinical pig islet xenotransplantation trial in New Zealand. Xenotransplantation. 2014, 21(4), 309-323. [CrossRef]
- Morozov VA, Wynyard S, Matsumoto S, Abalovich A, Denner J, Elliott R. No PERV transmission during a clinical trial of pig islet cell transplantation. Virus Res. 2017, 227, 34-40. [CrossRef]
- Denner J, Längin M, Reichart B, Krüger L, Fiebig U, Mokelke M, Radan J, Mayr T, Milusev A, Luther F, Sorvillo N, Rieben R, Brenner P, Walz C, Wolf E, Roshani B, Stahl-Hennig C, Abicht JM. Impact of porcine cytomegalovirus on long-term orthotopic cardiac xenotransplant survival. Sci Rep. 2020, 10(1):17531 . [CrossRef]
- Yamada K, Tasaki M, Sekijima M, Wilkinson RA, Villani V, Moran SG, Cormack TA, Hanekamp IM, Hawley RJ, Arn JS, Fishman JA, Shimizu A, Sachs DH. Porcine cytomegalovirus infection is associated with early rejection of kidney grafts in a pig to baboon xenotransplantation model. Transplantation. 2014, 98(4):411–418. [CrossRef]
- Sekijima M, Waki S, Sahara H, Tasaki M, Wilkinson RA, Villani V, Shimatsu Y, Nakano K, Matsunari H, Nagashima H, Fishman JA, Shimizu A, Yamada K. Results of life-supporting galactosyltransferase knockout kidneys in cynomolgus monkeys using two different sources of galactosyltransferase knockout Swine. Transplantation. 2014, 98(4):419-426. [CrossRef]
- Denner J. Reduction of the survival time of pig xenotransplants by porcine cytomegalovirus. Virol J. 2018, 15(1):171. [CrossRef]
- Griffith BP, Goerlich CE, Singh AK, Rothblatt M, Lau CL, Shah A, Lorber M, Grazioli A, Saharia KK, Hong SN, Joseph SM, Ayares D, Mohiuddin MM.Genetically Modified Porcine-to-Human Cardiac Xenotransplantation. N Engl J Med. 2022, 387(1):35-44. [CrossRef]
- Mohiuddin MM, Singh AK, Scobie L, Goerlich CE, Grazioli A, Saharia K, Crossan C, Burke A, Drachenberg C, Oguz C, Zhang T, Lewis B, Hershfeld A, Sentz F, Tatarov I, Mudd S, Braileanu G, Rice K, Paolini JF, Bondensgaard K, Vaught T, Kuravi K, Sorrells L, Dandro A, Ayares D, Lau C, Griffith BP. Graft dysfunction in compassionate use of genetically engineered pig-to-human cardiac xenotransplantation: a case report. Lancet. 2023, S0140-6736(23)00775-4. [CrossRef]
- Denner J, Specke V, Thiesen U, Karlas A, Kurth R. Genetic alterations of the long terminal repeat of an ecotropic porcine endogenous retrovirus during passage in human cells. Virology. 2003, 314(1):125-133. [CrossRef]
- Karlas A, Irgang M, Votteler J, Specke V, Ozel M, Kurth R, Denner J. Characterisation of a human cell-adapted porcine endogenous retrovirus PERV-A/C. Ann Transplant. 2010, 15(2):45-54.
- Harrison I, Takeuchi Y, Bartosch B, Stoye JP. Determinants of high titer in recombinant porcine endogenous retroviruses. J Virol. 2004, 78(24):13871-1389. [CrossRef]
- Wilson CA, Wong S, Muller J, Davidson CE, Rose TM, Burd P. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J Virol. 1998, 72(4):3082–3087. [CrossRef]
- Denner J, Schuurmann KJ. High prevalence of recombinant porcine endogenous retroviruses (PERV-A/Cs) in minipigs: a review on origin and presence. Viruses. 2021, 13:1869.
- Krüger L, Kristiansen Y, Reuber E, Möller L, Laue M, Reimer C, Denner J. A comprehensive strategy for screening for xenotransplantation-relevant viruses in a second isolated population of Göttingen Minipigs. Viruses. 2019, 12(1):38. [CrossRef]
- Halecker S, Krabben L, Kristiansen Y, Krüger L, Möller L, Becher D, Laue M, Kaufer B, Reimer C, Denner J. Rare isolation of human-tropic recombinant porcine endogenous retroviruses PERV-A/C from Göttingen minipigs. Virol J. 2022, 19(1):30.
- Pal N, Baker R, Schalk S, Scobie L, Tucker AW, Opriessnig T. Detection of porcine endogenous retrovirus (PERV) viremia in diseased versus healthy US pigs by qualitative and quantitative real-time RT-PCR. Transbound Emerg. Dis. 2011, 58, 344–351. [CrossRef]
- Krüger L, Stillfried M, Prinz C, Schröder V, Neubert LK, Denner J. Copy Number and Prevalence of Porcine Endogenous Retroviruses (PERVs) in German Wild Boars. Viruses. 2020, 12(4):419. [CrossRef]
- Fiebig U, Fischer K, Bähr A, Runge C, Schnieke A, Wolf E, Denner J. Porcine endogenous retroviruses: Quantification of the copy number in cell lines, pig breeds, and organs. Xenotransplantation. 2018, 25(4):e12445. [CrossRef]
- Takeuchi Y, Patience C, Magre S, Weiss RA, Banerjee PT, Le Tissier P, Stoye JP (1998) Host range and interference studies of three classes of pig endogenous retrovirus. J Virol 1998, 72:9986-9991.
- Kaulitz D, Mihica D, Adlhoch C, Semaan M, Denner J. Improved pig donor screening including newly identified variants of porcine endogenous retrovirus-C (PERV-C). Arch Virol. 2013, 158(2):341-348. [CrossRef]
- Yang, L.; Güell, M.; Niu, D.; George, H.; Lesha, E.; Grishin, D.; Aach, J.; Shrock, E.; Xu, W.; Poci, J.; et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science 2015, 350, 1101–1104. [CrossRef]
- Morozov VA, Plotzki E, Rotem A, Barkai U, Denner J. Extended microbiological characterization of Göttingen minipigs: porcine cytomegalovirus and other viruses. Xenotransplantation. 2016, 23(6), 490-496. [CrossRef]
- Morozov VA, Morozov AV, Denner J. New PCR diagnostic systems for the detection and quantification of porcine cytomegalovirus (PCMV). Arch Virol. 2016, 161(5), 1159-1168. [CrossRef]
- Heinze, J.; Plotzki, E.; Denner, J. Virus Safety of Xenotransplantation: Prevalence of Porcine Cicrovirus 2 (PCV2) in Pigs. Ann. Virol. Res. 2016, 2, 1023.
- Prinz, C.; Stillfried, M.; Neubert, L.K.; Denner, J. Detection of PCV3 in German wild boars. Virol. J. 2019, 16, 25. [CrossRef]
- Morozov, V.A.; Morozov, A.V.; Rotem, A.; Barkai, U.; Bornstein, S.; Denner, J. Extended Microbiological Characterization of Göttingen Minipigs in the Context of Xenotransplantation: Detection and Vertical Transmission of Hepatitis E Virus. PLoS ONE 2015, 10, e0139893. [CrossRef]
- Denner J. How Active Are Porcine Endogenous Retroviruses (PERVs)? Viruses. 2016, 8(8):215.
- Denner J. Porcine Lymphotropic Herpesviruses (PLHVs) and Xenotransplantation. Viruses 2021, 13(6), 1072.
- Wynyard S, Garkavenko O, Elliot R. Multiplex high resolution melting assay for estimation of Porcine Endogenous Retrovirus (PERV) relative gene dosage in pigs and detection of PERV infection in xenograft recipients. J Virol Methods. 2011, 175(1):95-100. [CrossRef]
- Mourad NI, Crossan C, Cruikshank V, Scobie L, Gianello P. Characterization of porcine endogenous retrovirus expression in neonatal and adult pig pancreatic islets. Xenotransplantation. 2017, 24(4). [CrossRef]
- Tacke SJ, Specke V, Denner J. Differences in release and determination of subtype of porcine endogenous retroviruses produced by stimulated normal pig blood cells. Intervirology. 2003, 46(1), 17-24. [CrossRef]
- Bartosch B, et al. Evidence and consequence of porcine endogenous retrovirus recombination. J. Virol. 2004; 78, 13880–13890. [CrossRef]
- Chen JQ, Zhang MP, Tong XK, Li JQ, Zhang Z, Huang F, Du HP, Zhou M, Ai HS, Huang LS. Scan of the endogenous retrovirus sequences across the swine genome and survey of their copy number variation and sequence diversity among various Chinese and Western pig breeds. Zool Res. 2022, 43(3), 423-441. [CrossRef]
- Le Tissier, P.; Stoye, J.P.; Takeuchi, Y.; Patience, C.; Weiss, R.A. Two sets of human-tropic pig retrovirus. Nature 1997,389, 681–682. [CrossRef]
- Patience, C.; Takeuchi, Y.; Weiss, R.A. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 1997, 3, 282–286. [CrossRef]
- Patience, C.; Switzer, W.M.; Takeuchi, Y.; Griffiths, D.J.; Goward, M.E.; Heneine, W.; Stoye, J.P.; Weiss, R.A. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 2001, 75, 2771–2775. [CrossRef]
- Liu, G.; Li, Z.; Pan, M.; Ge, M.; Wang, Y.; Gao, Y. Genetic prevalence of porcine endogenous retrovirus in Chinese experimental miniature pigs. Transplant. Proc. 2011, 43, 2762–2769. [CrossRef]
- Lee, D.; Lee, J.; Yoon, J.K.; Kim, N.Y.; Kim, G.W.; Park, C.; Oh, Y.K.; Kim, Y.B. Rapid determination of PERV copy number from porcine genomic DNA by real-time polymerase chain reaction. Anim. Biotechnol. 2011, 22, 175–180. [CrossRef]
- Yoon, J.K.; Choi, J.; Lee, H.J.; Cho, Y.; Gwon, Y.D.; Jang, Y.; Kim, S.; Choi, H.; Lee, J.H.; Kim, Y.B. Distribution of Porcine Endogenous Retrovirus in Different Organs of the Hybrid of a Landrace and a Jeju Domestic Pig in Korea. Transplant. Proc. 2015, 47, 2067–2071. [CrossRef]
- Zhang, P.; Yu, P.; Wang, W.; Zhang, L.; Li, S.; Bu, H. An effective method for the quantitative detection of porcine endogenous retrovirus in pig tissues. In Vitro Cell Dev. Biol. Anim. 2010, 46, 408–410. [CrossRef]
- Quereda, J.J.; Herrero-Medrano, J.M.; Abellaneda, J.M.; García-Nicolás, O.; Martínez-Alarcón, L.; Pallarés, F.J.; Ramírez, P.; Muñoz, A.; Ramis, G. Porcine endogenous retrovirus copy number in different pig breeds is not related to genetic diversity. Zoonoses Public Health 2012, 59, 401–407. [CrossRef]
- Mang, R.; Maas, J.; Chen, X.; Goudsmit, J.; van der Kuyl, A.C. Identification of a novel type C porcine endogenous retrovirus: Evidence that copy number of endogenous retroviruses increases during host inbreeding. J. Gen. Virol. 2001, 82, 1829–1834. [CrossRef]
- Lee, J.H.; Webb, G.C.; Allen, R.D.; Moran, C. Characterizing and mapping porcine endogenous retrovirusesin Westran pigs. J. Virol. 2002, 76, 5548–5556.
- Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Rogel-Gaillard C, Park C, Milan D, Megens HJ, Li S, Larkin DM, Kim H, Frantz LA, Caccamo M, Ahn H, Aken BL, Anselmo A, Anthon C, Auvil L, Badaoui B, Beattie CW, Bendixen C, Berman D, Blecha F, Blomberg J, Bolund L, Bosse M, Botti S, Bujie Z, Bystrom M, Capitanu B, Carvalho-Silva D, Chardon P, Chen C, Cheng R, Choi SH, Chow W, Clark RC, Clee C, Crooijmans RP, Dawson HD, Dehais P, De Sapio F, Dibbits B, Drou N, Du ZQ, Eversole K, Fadista J, Fairley S, Faraut T, Faulkner GJ, Fowler KE, Fredholm M, Fritz E, Gilbert JG, Giuffra E, Gorodkin J, Griffin DK, Harrow JL, Hayward A, Howe K, Hu ZL, Humphray SJ, Hunt T, Hornshøj H, Jeon JT, Jern P, Jones M, Jurka J, Kanamori H, Kapetanovic R, Kim J, Kim JH, Kim KW, Kim TH, Larson G, Lee K, Lee KT, Leggett R, Lewin HA, Li Y, Liu W, Loveland JE, Lu Y, Lunney JK, Ma J, Madsen O, Mann K, Matthews L, McLaren S, Morozumi T, Murtaugh MP, Narayan J, Nguyen DT, Ni P, Oh SJ, Onteru S, Panitz F, Park EW, Park HS, Pascal G, Paudel Y, Perez-Enciso M, Ramirez-Gonzalez R, Reecy JM, Rodriguez-Zas S, Rohrer GA, Rund L, Sang Y, Schachtschneider K, Schraiber JG, Schwartz J, Scobie L, Scott C, Searle S, Servin B, Southey BR, Sperber G, Stadler P, Sweedler JV, Tafer H, Thomsen B, Wali R, Wang J, Wang J, White S, Xu X, Yerle M, Zhang G, Zhang J, Zhang J, Zhao S, Rogers J, Churcher C, Schook LB. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 2012, 491, 393–398. [CrossRef]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation ofa droplet digital polymerase chain reaction format for DNA copy number quantification. Anal. Chem. 2012, 84, 1003–1011.
- Denner J. What does the PERV copy number tell us? Xenotransplantation. 2022, 2:e12732.
- Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011, 8:90. [CrossRef]
- Contreras-Galindo R, Kaplan MH, He S, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Kappes F, Dube D, Chan SM, Robinson D, Meng F, Dai M, Gitlin SD, Chinnaiyan AM, Omenn GS, Markovitz DM. HIV infection reveals widespread expansion of novel centromeric human endogenous retroviruses. Genome Res. 2013, 23(9), 1505-1513. [CrossRef]
- Denner J. Recombinant porcine endogenous retroviruses (PERV-A/C): a new risk for xenotransplantation? Adv. Virol. 2008, 153, 1421–1426.
- Dieckhoff B, Puhlmann J, Büscher K, Hafner-Marx A, Herbach N, Bannert N, Büttner M, Wanke R, Kurth R, Denner J. Expression of porcine endogenous retroviruses (PERVs) in melanomas of Munich miniature swine (MMS) Troll. Vet. Microbiol. 2007, 123, 53–68. [CrossRef]
- Bittmann I, Mihica D, Plesker R, Denner J. Expression of porcine endogenous retroviruses (PERV) in different organs of a pig. Virology. 2012, 433, 329–336. [CrossRef]
- Scobie L, Taylor S, Wood JC, Suling KM, Quinn G, Meikle S, Patience C, Schuurman HJ, Onions DE. Absence of replication-competent human-tropic porcine endogenous retroviruses in the germ line DNA of inbred miniature Swine. J. Virol. 2004, 78, 2502–2509. [CrossRef]
- Wood JC, Quinn G, Suling KM, Oldmixon BA, Van Tine BA, Cina R, Arn S, Huang CA, Scobie L, Onions DE, Sachs DH, Schuurman HJ, Fishman JA, Patience C. Identification of exogenous forms of human-tropic porcine endogenous retrovirus in miniature Swine. J. Virol. 2004, 78, 2494–2501. [CrossRef]
- Martin SI, Wilkinson R, Fishman JA. Genomic presence of recombinant porcine endogenous retrovirus in transmitting miniature swine. Virol. J. 2006, 3, 91. [CrossRef]
| Pig | Viruses* | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PERV copy number |
PERV-C | PCMV/PRV | HEV | HEV | HEV | PCV1/2 | PCV3 | PLHV-1/2 | PLHV-3 | |
| PCR | PCR | PCR | WB | ELISA | PCR | PCR | PCR | PCR | ||
| 440 | 45 | - | - | - | - | - | - | - | - | - |
| 476 | 53 | - | - | - | - | n.t. | - | - | - | + |
| 489 | 63 | + | - | - | - | - | - | - | - | - |
| 490 | 58 | + | - | - | - | n.t. | - | - | - | - |
| 491 | 61 | + | - | - | - | - | - | - | - | - |
| 492 | 59 | + | - | - | - | n.t. | - | - | - | - |
| 494 | n.t. | + | - | - | - | - | - | - | - | - |
| 495 | 60 | - | - | - | - | - | - | - | - | - |
| 497 | 64 | - | - | - | - | - | - | - | - | - |
| 499 | 57 | - | - | - | - | - | - | - | - | - |
| 503 | 58 | - | - | - | - | - | - | - | - | - |
| 504 | 47 | - | - | - | - | n.t. | - | - | - | - |
| 508 | 63 | + | - | - | - | n.t. | - | - | - | - |
| 509 | 64 | + | - | - | - | n.t. | - | - | - | - |
| 510 | 58 | + | - | - | - | n.t. | - | - | - | |
| f1 | 21 | - | - | n.t. | n.a. | n.a. | n.t. | - | n.t. | n.t. |
| f2 | 20 | - | - | n.t. | n.a. | n.a. | n.t. | - | n.t. | n.t. |
| m1 | 20 | - | - | n.t. | n.a. | n.a. | n.t. | - | n.t. | n.t. |
| m2 | 22 | - | - | n.t. | n.a. | n.a. | n.t. | - | n.t. | n.t. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





