1. Introduction
The human immune response against viruses is based on a coordinated activation of cellular and humoral components. To assess all these factors for an individual response against a specific virus infection requires complex laboratory approaches. The estimation of the T-cell mediated antiviral cellular response requires expensive and time-consuming laboratory methodologies [
1,
2,
3], although recently some simple methods, measuring the local Delayed Time Hypersensitivity (DTH) against virus antigens applied to the skin [
4], have also been advocated. Still, currently the best explored approach to assess the antiviral immune response is to measure the protective polyclonal antibodies generated after infection or vaccination. The key step of the SARS-CoV-2 virus infection is the entry of the virus into the epithelial cells through binding of the Spike protein to its ACE2 cellular receptor [
5,
6]. This binding depends on the interaction of the Spike Receptor Binding Domain (RBD) to the receptor; thus, the best-known virus neutralizing effects are evoked by antibodies binding to numerous epitopes of the RBD [
7,
8,
9,
10]. Correlation of RBD-based antibody assays with virus neutralization and with real-world protection efficiency against serious disease have been widely established [
11,
12,
13]. This is also well exemplified by the strong clinical protective effect of monoclonal antibodies developed against the RBD protein [
10,
14].
A major challenge in the estimate of the presence and efficiency of antiviral protective antibodies is the rapidly changing epitopes on the RBD in the case of emerging virus variants. In fact, monoclonal antibodies developed against the original, Wuhan-type virus [
15] became ineffective against the emerging Delta, and especially the Omicron virus variants [
16,
17]. Thus, a relatively simple diagnostic tool to assess the virus variant-dependent RBD binding of polyclonal antibodies generated in the human body after SARS-CoV-2 infection or immunization would be important in any further surge of this pandemic. New vaccines against emerging virus variants would require proper, RBD-specific assessment of their efficiency in large populations. Also, the waning presence of such polyclonal virus neutralizing antibodies is a major problem, especially in vulnerable individuals, due to their age, underlying disease conditions or inefficient immune response.
The aim of the present work was to develop a simple, rapid, high-throughput methodology to study the antibody-based immunity against emerging SARS-CoV-2 virus variants and demonstrate the correlation of this assay with commercially available, as well as direct virus neutralization assays. Here we also estimated the cross-reactivity of polyclonal antibodies against the Wuhan and Omicron virus variants in the sera of vaccinated donors and convalescent patients. The introduction should briefly place the study in a broad context and highlight why it is important. It should define the purpose of the work and its significance. The current state of the research field should be carefully reviewed, and key publications cited. Please highlight controversial and diverging hypotheses when necessary. Finally, briefly mention the main aim of the work and highlight the principal conclusions. As far as possible, please keep the introduction comprehensible to scientists outside your particular field of research. References should be numbered in order of appearance and indicated by a numeral or numerals in square brackets—e.g., [
1] or [
2,
3], or [
4,
5,
6]. See the end of the document for further details on references.
2. Materials and Methods
2.1. Recombinant RBD production
The RBD protein coding cassette with N-terminal His-tag as in the DNA constructs presented in Amanat et al. [
18], was cloned into p10 transposon vector to create stable cell lines with the SB100 Sleeping Beauty transposase. The RBD coding sequence was modified according to changes resulting in amino acid changes in the examined new Omicron SARS-CoV-2 variants Spike coding sequence. The modified RBD coding sequences were custom-made via gene synthesis and cloned into the plasmids used previously in the case of the Wuhan-RBD. The nucleotide sequences coding for the expressed proteins (see
Supplementary Materials) have been checked by sequencing in all cases (Wuhan-RBD, Omicron BA.1-RBD and Omicron BA.5-RBD).
For large-scale RBD protein expression we used stable expression in HEK cells, as described previously[
19], using the Sleeping-Beauty transposon-transposase system and the eGFP marker protein followed by an internal ribosome entry site (IRES2) that helps to sort cells with the desired protein expression and create stable clonal cell lines (see
Supplementary Materials and Suppl. Figures S1 and S2). The cell lines were grown in suspension, shaken (100 rpm) cultures in serum-free media (FreeStyle 293 Expression Medium, Gibco, Cat. 12338018) at 37°C, 5% CO
2. The RBD protein was isolated and purified by nickel ion affinity chromatography (ÄKTA pure
TM, GE Healthcare). Elution was performed by 200 mM imidazole, pH 7.4. Concentration and buffer exchange to PBS were performed by 30 kDa filters (Vivacell 100, Sartorious). The protein product size and purity were examined by SDS-polyacrylamide gel electrophoreses; concentration was estimated by UV-vis spectrophotometry at 280 nm. Samples were examined via Western blotting with anti-His (Sigma-Aldrich Cat.H1029) and in the case of the Wuhan-RBD anti-RBD antibody (Invitrogen Cat. MA5-38033). Anti-mouse HRP-conjugated secondary antibody (Invitrogen Cat. A16066) was used with ECL substrate for detection (BioRad Cat.1705061) (see
Suppl. Figure S2).
2.2. ELISA method
We have determined the specific recognition of the purified proteins in their native state by the anti-RBD monoclonal primary (Abcam ab273074 and Invitrogen 703959) and HRP-conjugated secondary antibody (Abcam ab6724) (see
Suppl. Figures S2 and S3). In order to calibrate the indirect ELISA allowing for detection of RBD-specific IgG antibodies in human sera, we have applied various concentrations of the purified RBD protein and pre-COVID-19 sera, as well as sera of non-vaccinated or fully vaccinated volunteer donors. We found that 0.4 µg RBD protein and 500x dilution of the sera gave well measurable interaction in all vaccinated donors, while minimum signal in the non-vaccinated donor (see
Suppl. Figure S3, and Figure S4). In this ELISA we used High Binding 96-well ELISA microplates (Corning Cat. 9018) and the tested amount of RBD in 100 µl PBS incubated overnight (16h) at 4 °C. The samples were blocked by 0.5% BSA/PBS for one hour at room temperature, then washed 3x in PBS-0.1%Tween 20. The commercially available anti-RBD monoclonal antibody (Abcam, cat. ab273074 and Invitrogen Cat.703959) was applied in 1:2000 dilution in 0.5% BSA/PBS for one hour at room temperature, washed 3 times with PBS-0.1% Tween 20. The secondary antibody (anti-rabbit HRP, Abcam, cat. ab6721) was applied in a dilution of 1:20000 in 0.5% BSA/PBS for 45 min, washed 3 times in PBS-0.1% Tween 20, then we used TMB substrate (Thermo Scientific cat. 34021) for quantification. The absorbance was read in a VictorX multilabel plate reader at 660 nm after 10 minutes, (A unit) was calculated by multiplying the absorbance by 1000.
In our indirect ELISA designed to measure SARS-CoV-2 Spike IgG titers from human sera, we used High Binding 96-well ELISA microplates (Corning Cat. 9018) coated with 0.5 µg RBD in 100 µl PBS per well. Coating was performed overnight (16 hours) at 4 °C. For blocking and the dilution of sera and antibodies we used 0.5% BSA/PBS (Bovine Serum Albumin Sigma-Aldrich Cat.A7030, PBS Gibco Cat.20012027). After 1 hour blocking at room temperature, we washed the plates 3 times with 0.1% Tween-20/PBS and incubated with 100 µl 1000x diluted sera in 0.5% BSA/PBS at room temperature for 1 hour, washed 3 times again and incubated for 45 minutes with HRP-conjugated anti-Human IgG secondary antibody (Invitrogen Cat.A18805). After washing 3 times, 100 µl TMB substrate solution (TMB Substrate Kit, Thermo ScientificTM Cat. 34021) per well was added, blue color reaction could be measured at 660nm, 5 minutes after adding the TMB substrate, the reaction was stopped by the addition of 100 µl 0.16 M H2SO4 solution per well. The absorbance at 450 nm was measured by a Perkin Elmer Victor X3 multilabel plate reader spectrophotometer, (A unit) was calculated by multiplying the absorbance by 1000.
Each serum sample was measured in two parallels, and the average of the two measurements was calculated. Control wells without serum in the case of all the three types of RBDs and controls with all used diluted serum samples with no coating antigen were used in all experiments. Sample preparation: fingertip blood samples were immediately centrifuged in polypropylene microcentrifuge tubes at 1,000x g for 5 minutes, and the serum was collected from the top into a sterile new tube. Samples were then frozen and stored at -20°C for short term and at -80 °C for long term. Clinical sera samples prepared in the hospital were stored at -80°C. Visualization of the results was performed using GraphPad Prism 8 Software.
2.3. Virus neutralization assay
To produce a VSV (vesicular stomatitis virus) pseudovirus carrying the SARS-CoV-2 spike protein, codon-optimized C-terminal 17 amino acid spike gene truncations (synthesized by Geneart, Regensburg) were cloned into the eukaryotic expression plasmid pSTZ. The cloning involved the insertion of the original Wuhan spike gene (Genbank MN908947.3) containing the D614G mutation (wild-type variant), as well as the spike Omicron BA.5 variant (Genbank UPN16705.1), resulting in the recombinant plasmids pCMVspike-wt and pCMVspike-BA5. Each of these plasmids was transfected into HEK293T cells. After 24 hours of transfection, the cells were inoculated with VSV-EGFP-ΔG-G (STZ Angewandte Biologische Chemie, Mannheim, Germany), a replication-deficient G-pseudotyped VSV pseudovirus encoding EGFP but lacking the genetic information for VSV-G. The inoculation lasted for one hour. Afterwards, the inoculum was removed, and the cells were washed with phosphate-buffered saline. Then, the cells were incubated in medium for 24 hours. Supernatants containing the S-pseudotyped VSV-EGFP-ΔG-S pseudovirus were harvested, clarified from cellular debris through centrifugation, and stored at -80°C. For neutralization experiments, VSV-EGFP-ΔG-S particles were preincubated for 1 hour at 37°C with varying concentrations of sera obtained from volunteer donors. The mixtures were then inoculated onto Calu-3 cells. Infection efficiency was determined at 18 hours post-inoculation by fluorescence measuring using a microplate fluorescence reader (ClarioStar, BMG Labtech, Ortenberg, Germany). The neutralization data were obtained from triplicate experiments. The inhibitory concentration 50 (IC50) value represents the reciprocal serum dilution that results in a 50% reduction in infection. The IC50 data were calculated using a non-linear regression model (agonist versus response, variable slope, four parameters) with GraphPad Prism 9.0 Software.
3. Results and Discussion
As shown in the
Suppl. Materials, Supplementary Figures S2–S4, we have confirmed that the antigenicity and the recognition by neutralizing antibodies of the purified RBD protein was preserved. Moreover, in 96 well plates, a single amount of the purified RBD (0.5 ug/well) and a single dilution (1,000x) of the human sera gave satisfactory results for all tested RBD variants to estimate the antibody levels, thus providing a one-step ELISA for anti-RBD polyclonal antibody determination.
3.1. Validation of the RBD-ELISA results with commercially available quantitative immunoassay and virus neutralization assay
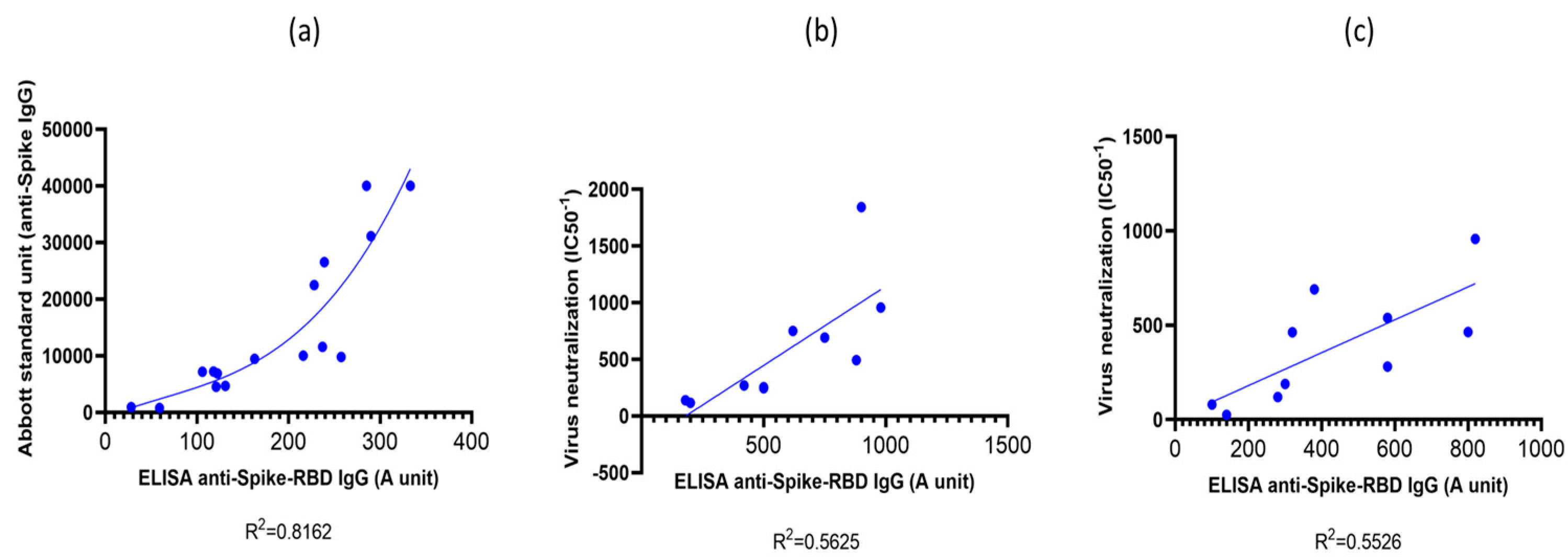
First, we validated our simple indirect RBD-ELISA assay by comparing it to the quantitative Abbott SARS-CoV-2 IgG immunoassay (Abbott Alinity-I immunology analyzer # 0-AB-ALINITYI by Abbott Diagnostics, SARS-CoV-2 anti-Spike IgG kit) calibrated to provide standard units (SU) for the circulating IgG against the Spike protein. This assay is commercially available, however, currently only to measure serum IgG against the Wuhan-type SARS-CoV-2 virus variant. As shown in
Figure 1(a), in vaccinated individuals we found a good correlation between the results of the two assays, especially at IgG titers above 300 SU levels. It has been shown in several publications that low anti-Spike IgG levels (below 2-300 SU – corresponding to 0.1-0.2 RBD-ELISA values), although indicate an antiviral immune response or successful immunization, provide only a low-level protection against real-world virus infection [
20,
21]. Thus, the range of antibody titers measured by the RBD-ELISA provide a good estimate of the antiviral humoral protection.
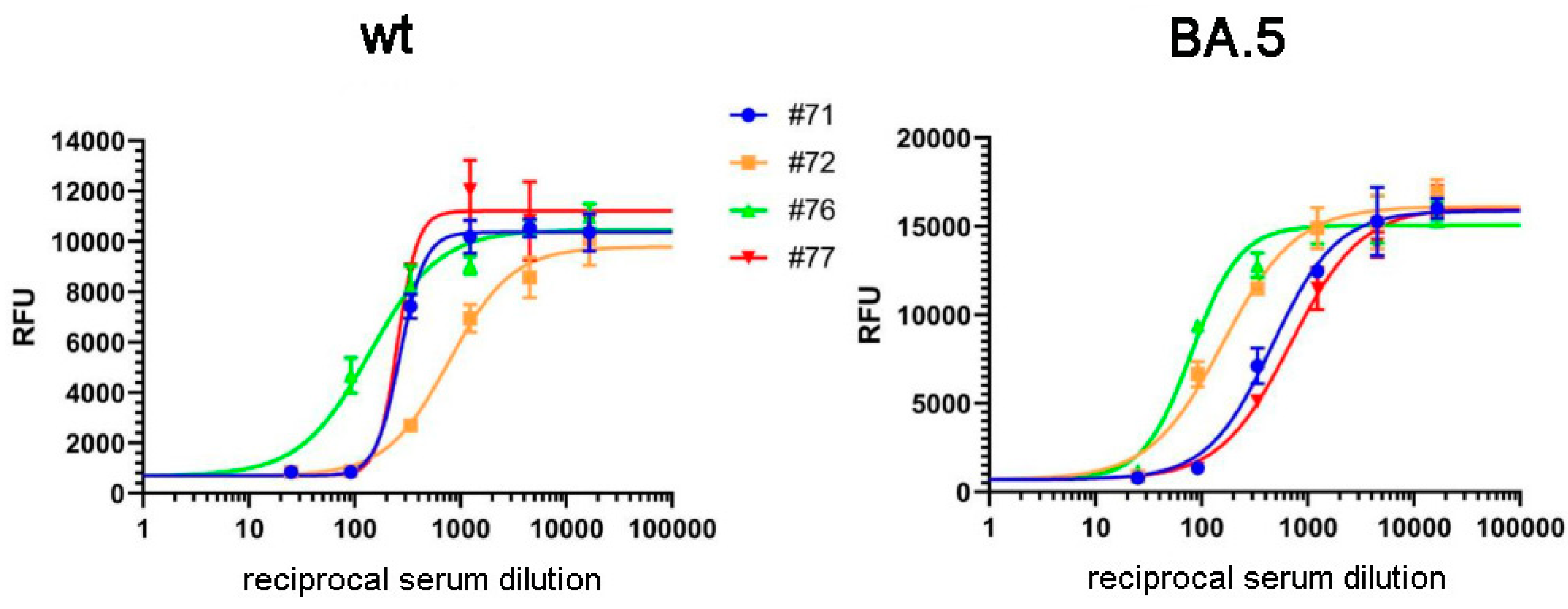
In the following experiments we validated the RBD-ELISA assay by comparing the measured anti-RBD serum IgG levels with direct virus neutralization efficiency (
Figure 2). In this case we could make this comparison both for the Wuhan-type and the Omicron BA.5 variants by using specific pseudovirus neutralization measurements. As shown in
Figure 1. (b) and
Figure 1. (c), these assays showed good correlation between the anti-RBD IgG levels, and the neutralization capacity of the sera examined, for both virus variants. This comparison showed that proper virus neutralization was provided only for IgG levels higher than 0.5 RBD-ELISA values, corresponding to about 300-600 SU in the commercial anti-Spike IgG assay (see
Figure 1 (a)). These data indicate again that the simple RBD-ELISA assay reported here provides a valuable measure of the potential humoral defense against the SARS-CoV-2 virus variants.
3.2. Results of RBD-ELISA for three virus variants in vaccinated donors and convalescent patients
3.2.1. Results obtained in vaccinated volunteer donors
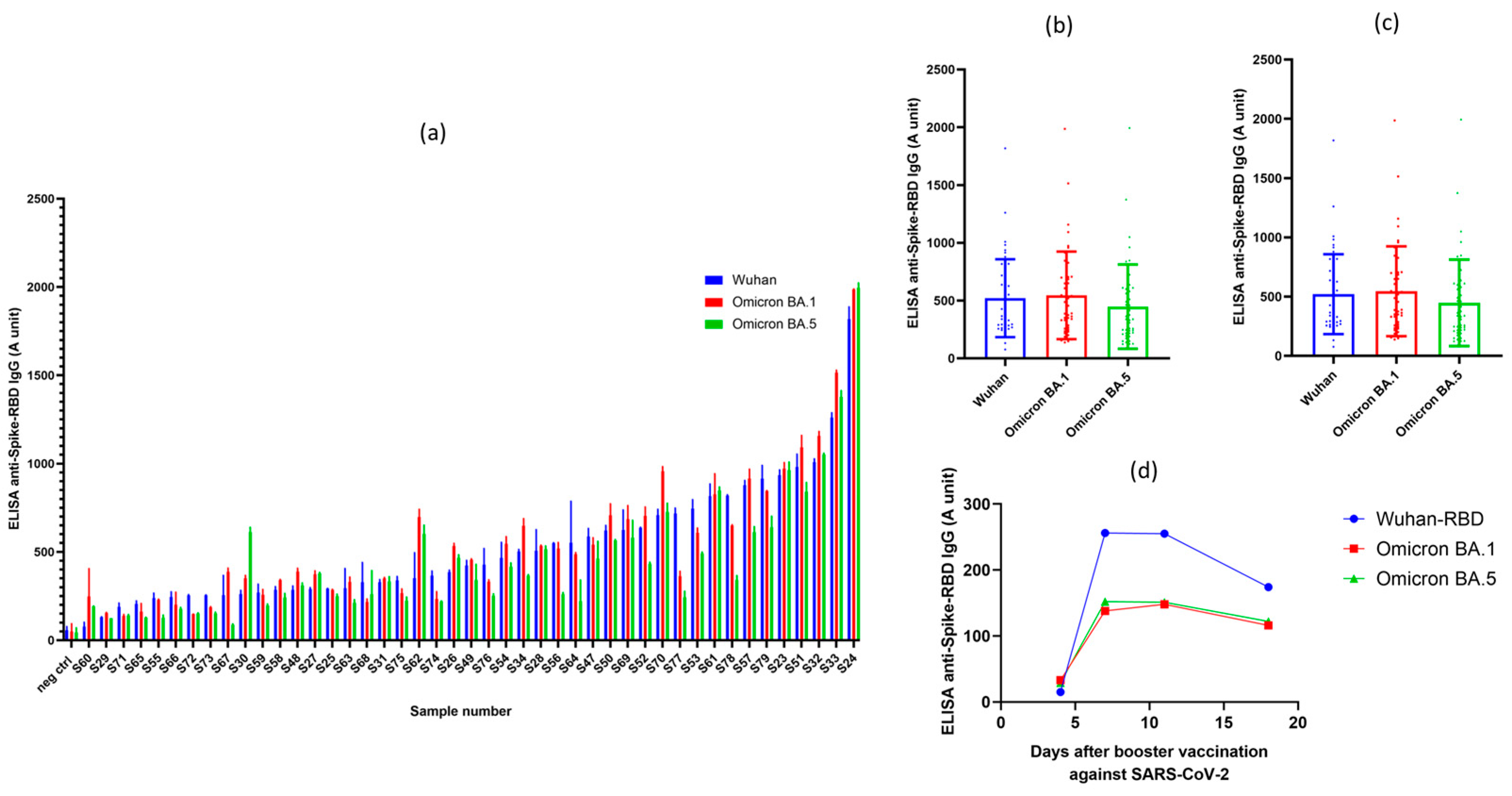
In the following experiments we have measured the anti-SARS-CoV-2 IgG levels by using the validated RBD-ELISA assay in 45 volunteers, in parallel for all the three virus variants (Wuhan, Omicron BA.1 and BA.5 variants). All individuals in this study obtained at least three vaccine shots, that is the monovalent two-dose BNT162b2 (Pfizer-BioNTech) (commercial name: Comirnaty (Pfizer-BioNTech)) or mRNA-1273 (Moderna-NIAID) (commercial name: Spikevax (Moderna-NIAID)) mRNA vaccine doses, and at least one booster shot (either homologous or heterologous forms of these vaccines). The time period after the last vaccination was between 3-12 months. These measurements were made easy by using only a few µL of sera from fingertip pricking, without the need of venous blood drawing.
As shown in
Figure 3(a), all fully vaccinated individuals had well measurable anti-RBD antibody titers, although the range of these titers was very wide, from 0.2-2.2 units. Interestingly, all vaccinated individuals, although at variable levels, also had antibodies against the Omicron variants. Since all these volunteers were vaccinated by the original Spikevax (Moderna-NIAID) or Comirnaty (Pfizer-BioNTech) vaccines, containing RNA coding for the Spike protein of the Wuhan variant, this was somewhat unexpected, although already noted in the relevant literature [
22]. While some of the volunteers may have been infected with the SARS-CoV-2 Omicron variant(s) in question, this is unlikely in this healthy volunteer cohort (see volunteer data).
Figure 3(b) shows collected data for the antibody titers against the three virus variants examined. Partly due to the relatively high scattering of the data, we found no statistical difference in the recognition of the RBD variants in the vaccinated individuals, although in many cases the Omicron BA.5 variant was less efficiently recognized in these volunteers.
Supplementary Figure S5 shows how in the case of the Omicron XBB.1.5 variant, this recognition decreased compared to the Wuhan RBD.
Figure 3(c) shows the effect of the time period after the last booster shot obtained in the vaccinated individuals. There is a significant decrease in the anti-RBD IgG levels after the longer time period following the last vaccination.
The one-step RBD-ELISA from finger-pricked serum samples against the virus variants allows to follow the specific IgG levels in a daily basis, without the burden of taking venous blood samples. Indeed, as shown in
Figure 3(d), after a booster shot the relatively low anti-RBD levels in this individual were greatly increased at about the 6th day following this re-vaccination. Interestingly, the Comirnaty (Pfizer-BioNTech) mRNA vaccine against the original SARS-CoV-2 Spike protein significantly increased the anti-RBD IgG levels against the Omicron variants as well. Based on these experiments we suggest that our simple ELISA method may be an important tool to assess the effects of multiple vaccinations in immunocompromised individuals, or with diseases affecting a proper immune response after vaccination.
3.2.2. Results obtained in convalescent patients after COVID-19 disease
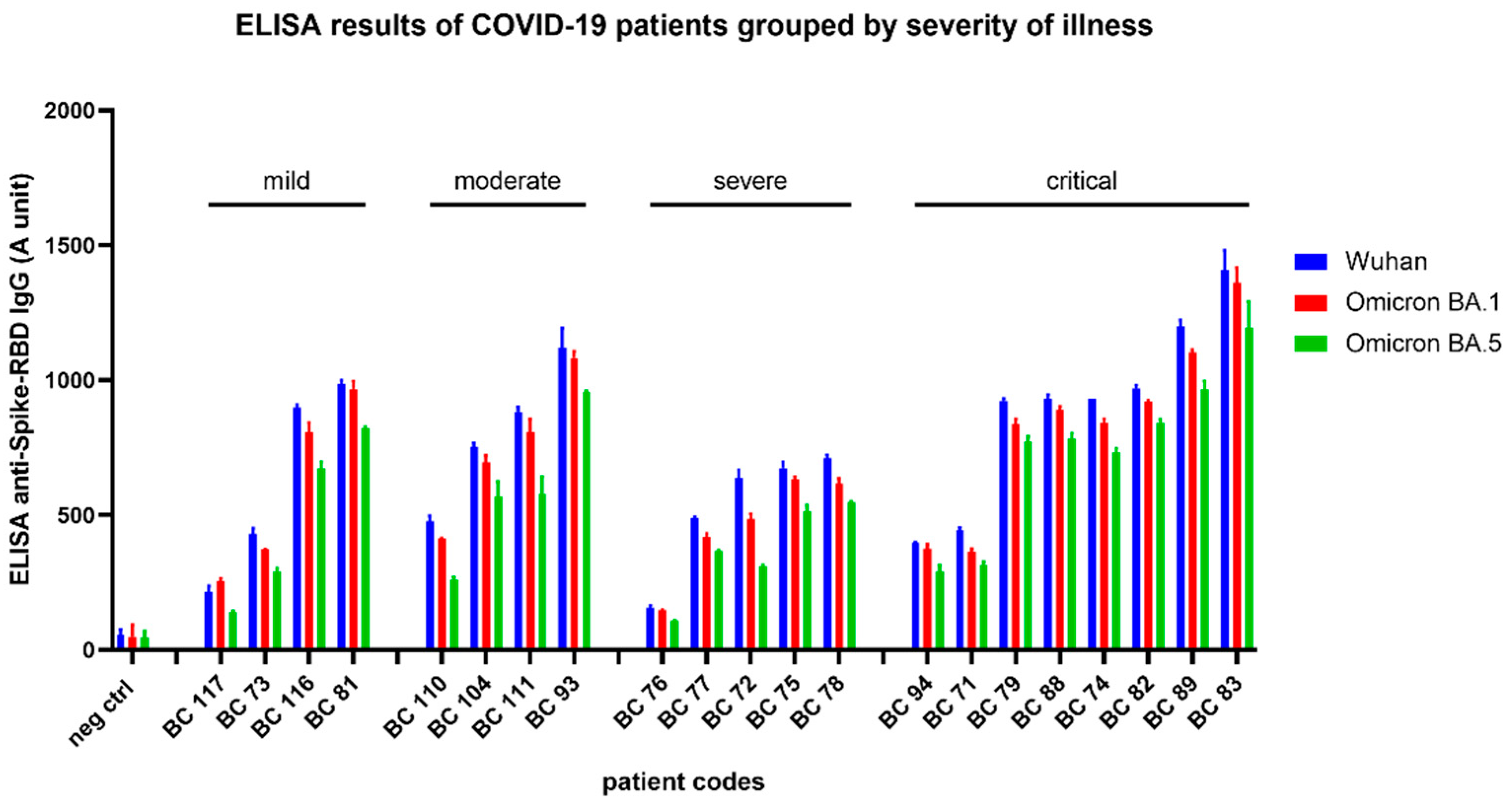
In this study we have analyzed the anti-RBD polyclonal IgG levels in 21 patients, treated in the National Korányi Institute of Pulmonology in Hungary, in the period of January 2020 – November 2021, with various manifestations of the COVID-19 disease: mild cases: Patient recovered at home, without hospital treatment; moderate: Patient recovered after hospital treatment, but not at the intensive care unit; severe: Patient recovered from illness after hospital treatment at the intensive care unit; critical: Patient recovered from illness after hospital treatment at the intensive care unit, with mechanical ventilation.
Collected sera after convalescence (within 7 days after recovery from illness) were stored at -80°C and analyzed by our RBD-ELISA for all the three SARS-CoV-2 variants. During the time these sera were collected, the Omicron virus variants were not present in Hungary. Consequently, these results indicate a cross-reactivity of the antibodies generated in the convalescent patients against the examined virus variants.
As shown in
Figure 4, in all these patients we found a significant level of anti-RBD polyclonal antibodies for all the three SARS-CoV-2 variants. Interestingly, the clinical severity (mild, moderate, severe, or critical level – see
Supplementary Figure S3) of the COVID-19 disease did not correlate with the antibody titers, that is lower and higher levels of anti-RBD antibodies were found in each group of the patients. Although there is a tendency for higher anti-RBD IgG levels in the patients having the COVID-19 disease at a critical level, the number of patients examined here is not enough to provide a quantitation of this potential difference.
5. Conclusions
As a summary, here we document the applicability of a simple, RBD-based ELISA to analyze the humoral immune response, that is anti-RBD IgG titers in vaccinated or convalescent patients. The sera obtained from small, finger-pricked blood samples allows a wide and easy sample collection, the assay uses a single dilution of the serum sample, and the assay can be performed in any commercially available ELISA developer and reader system. The simplified preparation of the RBD protein of emerging virus variants allows a rapid adjustment of the assay to any new surge of the disease.
An interesting finding of the present work is that, although to a highly variable extent, protective antibody production against the original Wuhan variant also coincides with the emergence of protective antibodies against two Omicron variants of the SARS-CoV-2 virus, both in vaccinated donors and in convalescent patients. In any further emerging SARS-CoV-2 variants such a rapid and simple assay may help to screen the actual humoral immune protection, which has been demonstrated to undergo changes with recent variants [
23]. We also demonstrated the application of this ELISA in the case of the recent Omicron XBB.1.5 variant (see
Suppl. Figure S5). We suggest that the presented and validated assay, easily modified to be applied in the case of newly emerging virus variants, may be helpful in a population-wide screening or a daily follow-up of potential antiviral humoral protection.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. S1: p10-RBD-IRES2-EGFP vector map; S2: RBD protein production; S3: ELISA with different serum dilutions grouped by variants; S4: RBD IgG ELISA results with different serum dilutions (Wuhan, BA.1 and BA.5 variants) grouped by dulition; S5: RBD (Wuhan and Omicron XBB.1.5 variant) IgG titers in sera samples from healthy volunteers and recovered COVID19 patients. Supplementary data: protein sequences of the produced RBD proteins (Wuhan, Omicron BA.1, BA.5, XBB.1.5).
Author Contributions
B. Sarkadi conceived, designed, and coordinated the study, conducted data interpretation, and wrote both the initial draft and the final version of the manuscript. O. Mozner performed experiments, conducted data collection, analysis, and interpretation, and contributed to the writing of the manuscript. J. Moldvay designed and coordinated the study, collected patient samples and data, and provided valuable clinical insights during the interpretation of the results. K.S. Szabo performed ELISA experiments, collected and analyzed the data. E. Hirsch, Cs. Feher, J. Domjan, D. Vasko, and Z. Ligeti were responsible for the production of the RBD protein, including the scale-up of cell culturing, purification, and analysis of the produced protein. They contributed to the interpretation of the results and reviewed the manuscript. M. Frey analyzed and interpreted the data regarding virus neutralization experiments and contributed to the writing of the manuscript. L. Puskas provided technical expertise, analyzed and interpreted the data regarding ELISA experiments, and contributed to the study design C. Stahl performed experiments and analyzed the data regarding virus neutralization experiments. D. Acs collected and analyzed the patient data. All authors have read and approved the final version of the manuscript.
Funding
B.S. was supported by the Post-Covid Research Program of the Hungarian Academy of Sciences (PC2022-7 /2022.); Project no. KDP-1017403 has been implemented with the support provided by the Ministry of Culture and Innovation of Hungary from the National Research, Development and Innovation Fund, financed under the KDP-2020 funding scheme (O.M.); Project no. RRF-2.3.1-21-2022-00015 has been implemented with the support provided by the European Union; The research has been implemented with the support provided by the Ministry of Culture and Innovation from the National Research, Development and Innovation Fund, financed under the [K-143039 and PD-142301] funding scheme.
Institutional Review Board Statement
This project has been carried out by the ethical permission of 24004-7/2021/EihO, issued on May 7, 2021, by the NNK Hungary.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We greatly appreciate the help of Prof. F. Krammer, for providing the original pCAGGS plasmid for SARS-CoV-2 Spike-RBD sequence. We also appreciate the help of Kelen Kórház (Dr. Zsuzsanna Kövesd) for helping to collect volunteer blood samples, and all the volunteers in the RCNS to provide blood samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mathew, D.; Giles, J.R.; Baxter, A.E.; Oldridge, D.A.; Greenplate, A.R.; Wu, J.E.; Alanio, C.; Kuri-Cervantes, L.; Pampena, M.B.; D’Andrea, K.; et al. Deep Immune Profiling of COVID-19 Patients Reveals Distinct Immunotypes with Therapeutic Implications. Science 2020, 369. [CrossRef]
- Cohen, K.W.; Linderman, S.L.; Moodie, Z.; Czartoski, J.; Lai, L.; Mantus, G.; Norwood, C.; Nyhoff, L.E.; Edara, V.V.; Floyd, K.; et al. Longitudinal Analysis Shows Durable and Broad Immune Memory after SARS-CoV-2 Infection with Persisting Antibody Responses and Memory B and T Cells. Cell Rep Med 2021, 2. [CrossRef]
- Sekine, T.; Perez-Potti, A.; Rivera-Ballesteros, O.; Strålin, K.; Gorin, J.B.; Olsson, A.; Llewellyn-Lacey, S.; Kamal, H.; Bogdanovic, G.; Muschiol, S.; et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell 2020, 183. [CrossRef]
- Díaz-Espada, F.; Matheu, V.; Barrios, Y. A Review of Hypersensitivity Methods to Detect Immune Responses to SARS-CoV-2. In Methods in Microbiology; 2022; Vol. 50.
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. 2020. [CrossRef]
- Hassan, A.O.; Case, J.B.; Winkler, E.S.; Thackray, L.B.; Kafai, N.M.; Bailey, A.L.; McCune, B.T.; Fox, J.M.; Chen, R.E.; Alsoussi, W.B.; et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell 2020, 182. [CrossRef]
- Zost, S.J.; Gilchuk, P.; Case, J.B.; Binshtein, E.; Chen, R.E.; Nkolola, J.P.; Schäfer, A.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; et al. Potently Neutralizing and Protective Human Antibodies against SARS-CoV-2. Nature 2020, 584. [CrossRef]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.M.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 Neutralizing Antibody Structures Inform Therapeutic Strategies. Nature 2020, 588. [CrossRef]
- Guo, Y.; Huang, L.; Zhang, G.; Yao, Y.; Zhou, H.; Shen, S.; Shen, B.; Li, B.; Li, X.; Zhang, Q.; et al. A SARS-CoV-2 Neutralizing Antibody with Extensive Spike Binding Coverage and Modified for Optimal Therapeutic Outcomes. [CrossRef]
- Ni, L.; Ye, F.; Cheng, M.L.; Feng, Y.; Deng, Y.Q.; Zhao, H.; Wei, P.; Ge, J.; Gou, M.; Li, X.; et al. Detection of SARS-CoV-2-Specific Humoral and Cellular Immunity in COVID-19 Convalescent Individuals. Immunity 2020, 52, 971-977.e3. [CrossRef]
- Garcia-Beltran, W.F.; Lam, E.C.; Astudillo, M.G.; Yang, D.; Miller, T.E.; Feldman, J.; Hauser, B.M.; Caradonna, T.M.; Clayton, K.L.; Nitido, A.D.; et al. COVID-19-Neutralizing Antibodies Predict Disease Severity and Survival. Cell 2021, 184. [CrossRef]
- Liu, C.; Zhang, J.; Zeng, Y.; Huang, C.; Chen, F.; Cao, Y.; Wu, S.; Wei, D.; Lin, Z.; Zhang, Y.; et al. Effectiveness of SARS-CoV-2-Inactivated Vaccine and the Correlation to Neutralizing Antibodies: A Test-Negative Case–Control Study. J Med Virol 2023, 95, e28280. [CrossRef]
- Gupta, A.; Gonzalez-Rojas, Y.; Juarez, E.; Crespo Casal, M.; Moya, J.; Falci, D.R.; Sarkis, E.; Solis, J.; Zheng, H.; Scott, N.; et al. Early Treatment for Covid-19 with SARS-CoV-2 Neutralizing Antibody Sotrovimab. New England Journal of Medicine 2021, 385. [CrossRef]
- Gottlieb, R.L.; Nirula, A.; Chen, P.; Boscia, J.; Heller, B.; Morris, J.; Huhn, G.; Cardona, J.; Mocherla, B.; Stosor, V.; et al. Effect of Bamlanivimab as Monotherapy or in Combination with Etesevimab on Viral Load in Patients with Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA - Journal of the American Medical Association 2021, 325. [CrossRef]
- Correction to Lancet Infect Dis 2022; Published Online Nov 18. Https://Doi.Org/10.1016/S1473-3099(22)00733-2 (The Lancet Infectious Diseases (2023) 23(1) (22–23), (S1473309922007332), (10.1016/S1473-3099(22)00733-2)). Lancet Infect Dis 2023, 23.
- Tada, T.; Zhou, H.; Dcosta, B.M.; Samanovic, M.I.; Chivukula, V.; Herati, R.S.; Hubbard, S.R.; Mulligan, M.J.; Landau, N.R. Increased Resistance of SARS-CoV-2 Omicron Variant to Neutralization by Vaccine-Elicited and Therapeutic Antibodies. EBioMedicine 2022, 78. [CrossRef]
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.O.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J.; et al. A Serological Assay to Detect SARS-CoV-2 Seroconversion in Humans. Nat Med 2020, 26, 1033–1036. [CrossRef]
- Zámbó, B.; Mózner, O.; Bartos, Z.; Török, G.; Várady, G.; Telbisz, Á.; Homolya, L.; Orbán, T.I.; Sarkadi, B. Cellular Expression and Function of Naturally Occurring Variants of the Human ABCG2 Multidrug Transporter. Cellular and Molecular Life Sciences 2020, 77, 365–378. [CrossRef]
- Barda, N.; Canetti, M.; Gilboa, M.; Asraf, K.; Indenboim, V.; Weiss-Ottolenghi, Y.; Amit, S.; Zubli, D.; Doolman, R.; Mendelson, E.; et al. The Association Between Prebooster Vaccination Antibody Levels and the Risk of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection. Clinical Infectious Diseases 2022. [CrossRef]
- Khoury, D.S.; Schlub, T.E.; Cromer, D.; Steain, M.; Fong, Y.; Gilbert, P.B.; Subbarao, K.; Triccas, J.A.; Kent, S.J.; Davenport, M.P. Correlates of Protection, Thresholds of Protection, and Immunobridging among Persons with SARS-CoV-2 Infection. Emerg Infect Dis 2023, 29. [CrossRef]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O’Connell, A.-M.; et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. New England Journal of Medicine 2022, 386. [CrossRef]
- Miller, J.; Hachmann, N.P.; Collier, A.Y.; Lasrado, N.; Mazurek, C.R.; Patio, R.C.; Powers, O.; Surve, N.; Theiler, J.; Korber, B.; et al. Substantial Neutralization Escape by SARS-CoV-2 Omicron Variants BQ.1.1 and XBB.1. New England Journal of Medicine 2023, 388. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).