Submitted:
26 October 2023
Posted:
27 October 2023
You are already at the latest version
Abstract
Keywords:
1. INTRODCUTION
2. MATERIAL AND METHODS
2.1. Bacterial growth conditions
2.2. Preliminary screening of bacteriocins producers
2.2.1. Agar Well Diffusion method
2.2.2. Antagonistic activity of crude bacteriocins
2.3. Evaluating bacteriocins extraction methods
2.3.1. Convectional bacteriocins extraction
2.3.2. Ammonium sulphate precipitation.
2.4. Bacteriocin extraction using solvents
2.4.1. Solvent extraction
2.4.2. Ethyl acetate extraction
2.4.3. Butanol Extraction
2.4.4. Petroleum ether extraction
2.4.5. Chloroform extraction.
2.5. Physic-chemical characterization of bacteriocins
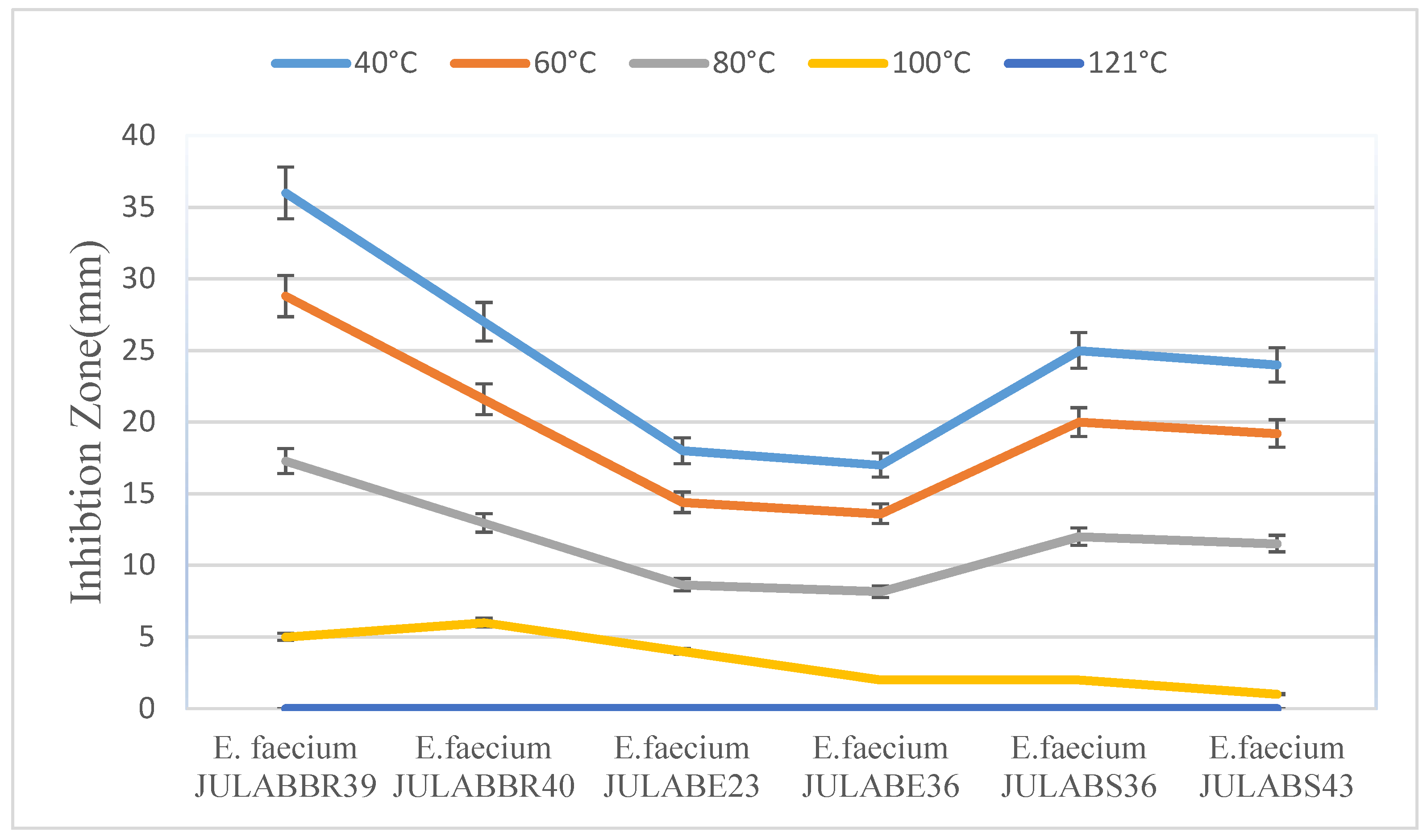
2.5.1. Effect of Temperature
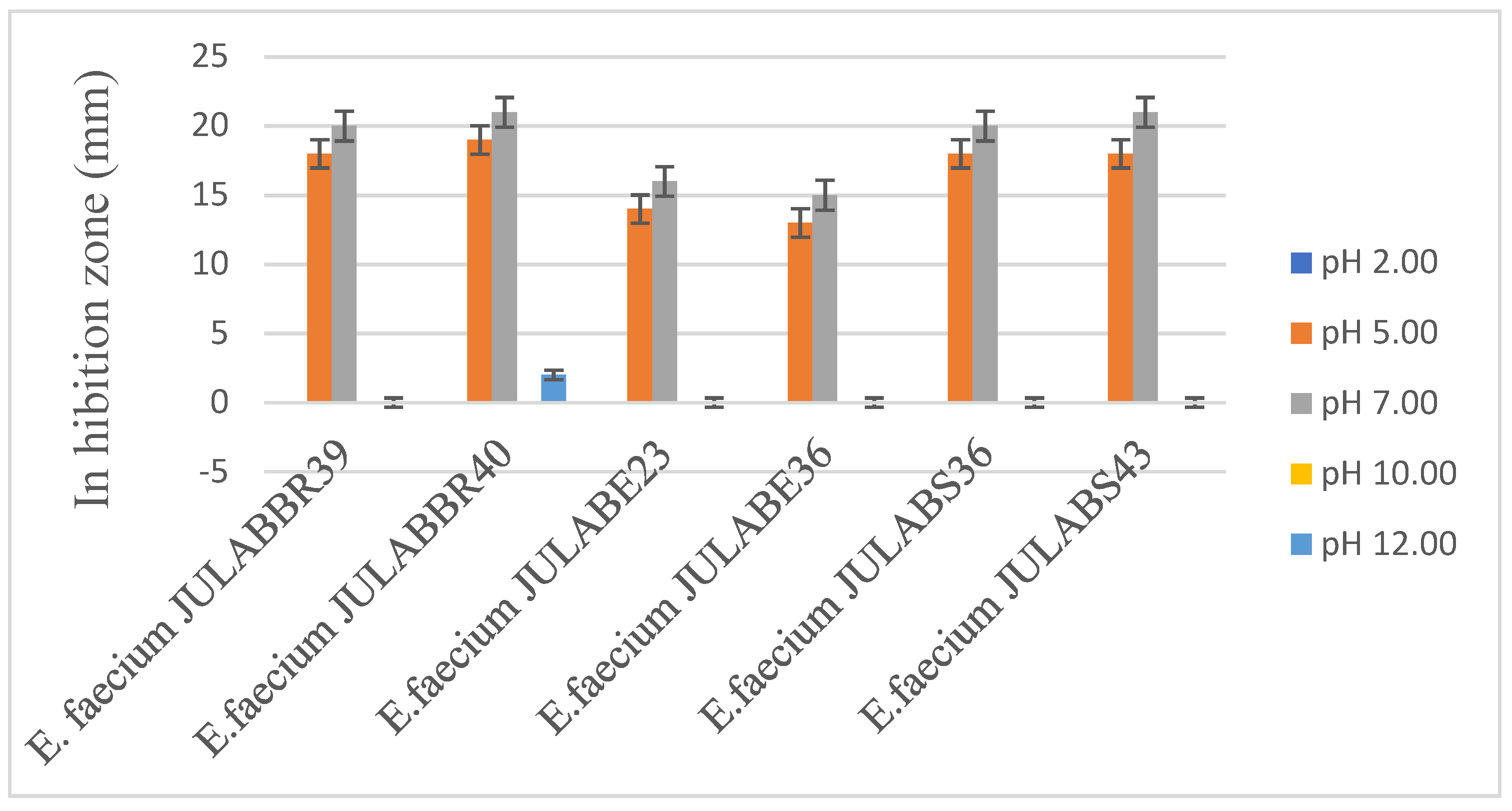
2.5.1.1. Effect of pH
2.5.1.2. Effect of enzymes
2.5.1.3. Effect of solvents
2.5.1.4. Effect of surfactants
2.6. Optimization for enhanced bacteriocins production
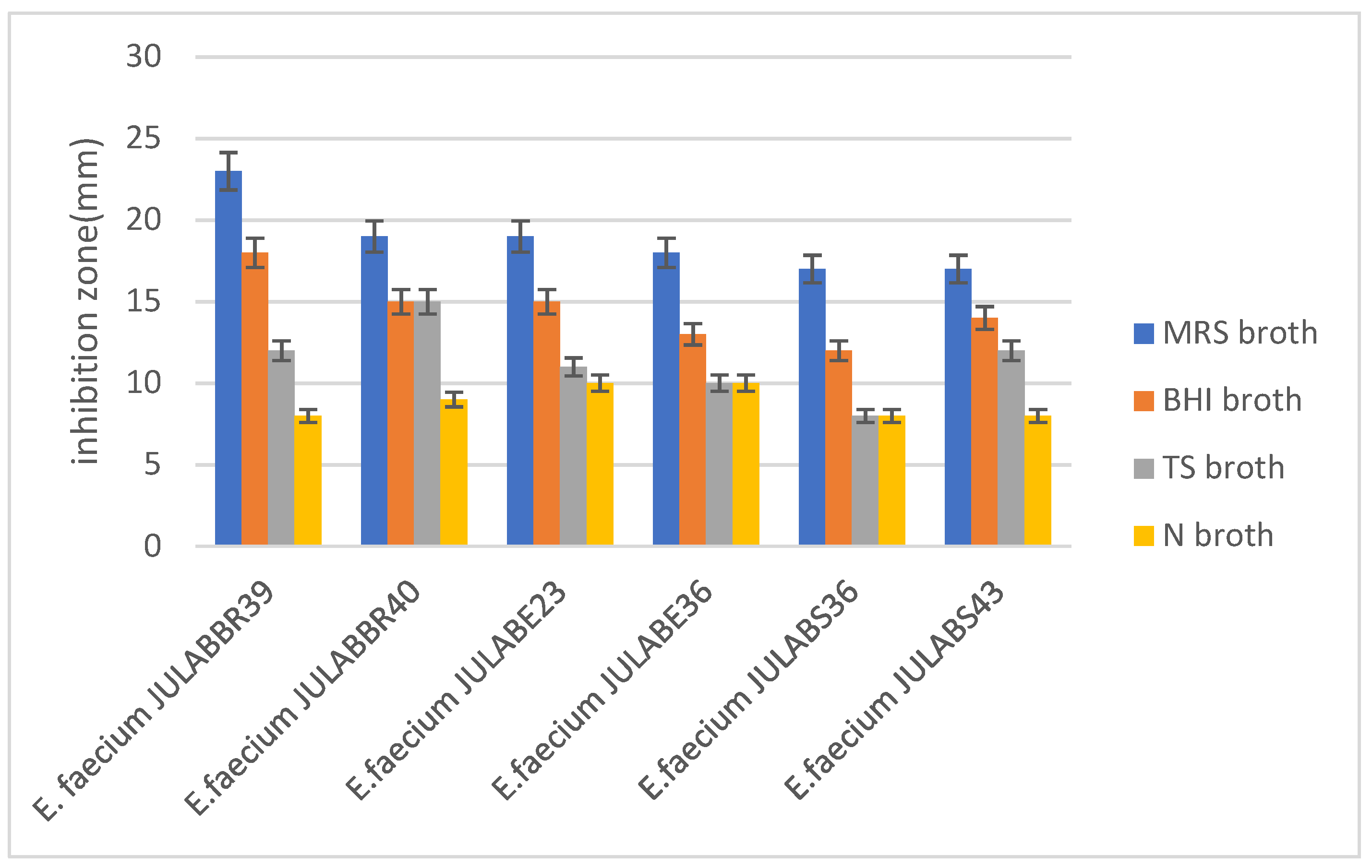
2.6.1. Media selection for bacteriocins optimization
2.6.2. Effect of carbon sources on bacteriocins production
2.6.3. Effect of nitrogen sources bacteriocins production
2.6.4. Effect of Incubation Period on Bacteriocins Production
2.7. Data analysis
3. Results
3.1. Evaluating bacteriocins producing potential
3.2. Antimicrobial activity of bacteriocins crude extract
3.2.1. Determination of antimicrobial substance
3.2.2. Preparation of bacteriocin crude extract
3.3. Characterization of the partially purified supernatants
3.3.1. Effects of Temperature
3.3.2. Effect of pH on the inhibitory activity
3.3.3. Effect of enzymes
3.3.4. Effects of surfactants and additives of the antimicrobial activity of bacteriocins
3.4. Bacteriocins production
3.5. Effects of carbon sources on antimicrobial activity of Enterococcus faecium
3.6. Effects of Nitrogen sources on antimicrobial activity of Enterococcus faecium
4. Discussion
Authors Contributions
Funding
Availability of data and materials
Ethics approval and consent to participate
Consent for publication
Acknowledgment
Competing interests
Abbreviation
References
- Abanoz, H. S., & Kunduhoglu, B. (2018). Antimicrobial Activity of a Bacteriocin Produced byEnterococcus faecalis KT11 against Some Pathogens andAntibiotic-Resistant Bacteria. Korean Journal for Food Science of Animal Resources, 38(5) 1064. [CrossRef]
- Ahmed, D., Fatima, K., Saeed, R., & Masih, R. (2016). Isolation and identification of bioactive compounds from chloroform fraction of methanolic extract of Carissa opaca roots. Natural Product Research, 30(17), 2012–2016. [CrossRef]
- Akhtar, M. S., Panwar, J., & Yun, Y. S. (2013). Biogenic synthesis of metallic nanoparticles by plant extracts. ACS Sustainable Chemistry and Engineering, 1(6). [CrossRef]
- Amit, S. K., Uddin, M. M., Rahman, R., Islam, S. M. R., & Khan, M. S. (2017). A review on mechanisms and commercial aspects of food preservation and processing. Agriculture and Food Security, 6(1), 1–22. [CrossRef]
- Ammor, S., Dufour, E., Zagorec, M., Chaillou, S., & Chevallier, I. (2005). Characterization and selection of Lactobacillus sakei strains isolated from traditional dry sausage for their potential use as starter cultures. Food Microbiology, 22(6). [CrossRef]
- Balla, E., Dicks, L. M. T., Du Toit, M., Van Der Merwe, M. J., & Holzapfel, W. H. (2000). Characterization and cloning of the genes encoding enterocin 1071A and enterocin 1071B, two antimicrobial peptides produced by Enterococcus faecalis BFE 1071. Applied and Environmental Microbiology, 66(4), 1298–1304. [CrossRef]
- Barale, S. S., Ghane, S. G., & Sonawane, K. D. (2022a). Purification and characterization of antibacterial surfactin isoforms produced by Bacillus velezensis SK. AMB Express, 12(1), 1–20. [CrossRef]
- Barale, S. S., Ghane, S. G., & Sonawane, K. D. (2022b). Purification and characterization of antibacterial surfactin isoforms produced by Bacillus velezensis SK. AMB Express, 12(1), 1–20. [CrossRef]
- Ben Braïek, O., Ghomrassi, H., Cremonesi, P., Morandi, S., Fleury, Y., Le Chevalier, P., Hani, K., Bel Hadj, O., & Ghrairi, T. (2017). Isolation and characterisation of an enterocin P-producing Enterococcus lactis strain from a fresh shrimp (Penaeus vannamei). Antonie van Leeuwenhoek, International Journal of General and Molecular Microbiology, 110(6), 771–786. [CrossRef] [PubMed]
- Burianek, L. L., & Yousef, A. E. (2000). Solvent extraction of bacteriocins from liquid cultures. Letters in Applied Microbiology, 31(3), 193–197. [CrossRef] [PubMed]
- Cocolin, L., Foschino, R., Comi, G., & Grazia Fortina, M. (2007). Description of the bacteriocins produced by two strains of Enterococcus faecium isolated from Italian goat milk. Food Microbiology, 24(7–8), 752–758. [CrossRef]
- do Nascimento, M. da S., Moreno, I., & Kuaye, A. Y. (2010). Antimicrobial activity of Enterococcus faecium FAIR-E 198 against gram-positive pathogens. Brazilian Journal of Microbiology, 41(1), 74–81. [CrossRef]
- Drider, D., Belguesmia, Y., Choiset, Y., Prévost, H., Dalgalarrondo, M., & Chobert, J. M. (2010). Partial purification and characterization of the mode of action of enterocin S37: A bacteriocin produced by enterococcus faecalis S37 isolated from poultry feces. Journal of Environmental and Public Health, 2010. [CrossRef]
- Du, L., Liu, F., Zhao, P., Zhao, T., & Doyle, M. P. (2017). Characterization of Enterococcus durans 152 bacteriocins and their inhibition of Listeria monocytogenes in ham. Food Microbiology, 68, 97–103. [CrossRef]
- Ellis, M. V. (2001). Harmful supervision, a cause for alarm: Comment on Gray et al. (2001) and Nelson and Friedlander (2001). In Journal of Counseling Psychology (Vol. 48, Issue 4). [CrossRef]
- Elyass, M. E., Mahdi, A. A., & Attitalla, I. H. (2015). Characterization and Evaluation of Antimicrobial Activity of Bacteriocins from Lactobacillus Curvatus and Pediococcus Pentosaceus. Infectious & Non Infectious Diseases, 1(1), 1–7. [CrossRef]
- Erb, D. K., Steidel, C. C., Shapley, A. E., Pettini, M., Reddy, N. A., & Adelberger, K. L. (2006). Hα Observations of a Large Sample of Galaxies at z ∼ 2: Implications for Star Formation in High-Redshift Galaxies. The Astrophysical Journal, 647(1). [CrossRef]
- Franzetti, L., Pompei, M., Scarpellini, M., & Galli, A. (2004). Phenotypic and genotypic characterization of Enterococcus spp. of different origins. Current Microbiology, 49(4), 255–260. [CrossRef]
- Freitas, T. K. F. S., Oliveira, V. M., de Souza, M. T. F., Geraldino, H. C. L., Almeida, V. C., Fávaro, S. L., & Garcia, J. C. (2015). Optimization of coagulation-flocculation process for treatment of industrial textile wastewater using okra (A. esculentus) mucilage as natural coagulant. Industrial Crops and Products, 76, 538–544. [CrossRef]
- Fugaban, J. I. I., Bucheli, J. E. V., Holzapfel, W. H., & Todorov, S. D. (2021). Characterization of partially purified bacteriocins produced by enterococcus faecium strains isolated from soybean paste active against listeria spp. and vancomycin-resistant enterococci. Microorganisms, 9(5), 1085. https://www.mdpi.com/2076-2607/9/5/1085/htm.
- Gaaloul, N., ben Braiek, O., Hani, K., Volski, A., Chikindas, M. L., & Ghrairi, T. (2015). Isolation and characterization of large spectrum and multiple bacteriocin-producing Enterococcus faecium strain from raw bovine milk. Journal of Applied Microbiology, 118(2), 343–355. [CrossRef] [PubMed]
- Ghrairi, T., Frere, J., Berjeaud, J. M., & Manai, M. (2008). Purification and characterisation of bacteriocins produced by Enterococcus faecium from Tunisian rigouta cheese. Food Control, 19(2), 162–169. [CrossRef]
- Goh, H. F., & Philip, K. (2015). Purification and Characterization of Bacteriocin Produced by Weissella confusa A3 of Dairy Origin. PLOS ONE, 10(10), e0140434. [CrossRef]
- Gutiérrez-Cortés, C., Suarez, H., Buitrago, G., Nero, L. A., & Todorov, S. D. (2018). Characterization of bacteriocins produced by strains of Pediococcus pentosaceus isolated from Minas cheese. Annals of Microbiology, 68(6), 383–398. https://annalsmicrobiology.biomedcentral.com/articles/10.1007/s13213-018-1345-z.
- Hosseini, S. V., Arlindo, S., Böhme, K., Fernández-No, C., Calo-Mata, P., & Barros-Velázquez, J. (2009). Molecular and probiotic characterization of bacteriocin-producing Enterococcus faecium strains isolated from nonfermented animal foods. Journal of Applied Microbiology, 107(4), 1392–1403. [CrossRef]
- Isleroglu, H., Yildirim, Z., Tokatli, M., Oncul, N., & Yildirim, M. (2012). Partial characterisation of enterocin KP produced by Enterococcus faecalis KP, a cheese isolate. International Journal of Dairy Technology, 65(1), 90–97. [CrossRef]
- Ivy, J., Fugaban, I., Enrique Vazquez Bucheli, J., Park, J.Y., Suh, H.D., Jung, E. S., de Melo Franco, B.D.M., Ivanova, I.V., Holzapfel, W.H., Todorov, S.D., Paulo, S., Correspondence, B., & Dimitrov Todorov, S. (2022). Antimicrobial properties of Pediococcus acidilactici and Pediococcus pentosaceus isolated from silage. Journal of Applied Microbiology, 132(1), 311–330. [CrossRef]
- Kang, J. H., & Lee, M. S. (2005). Characterization of a bacteriocin produced by Enterococcus faecium GM-1 isolated from an infant. Journal of Applied Microbiology, 98(5), 1169–1176. [CrossRef]
- Kelleher, D. J., Karaoglu, D., Mandon, E. C., & Gilmore, R. (2003). Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Molecular Cell, 12(1). [CrossRef]
- Kumar, M., Tiwari, S. K., & Srivastava, S. (2010). Purification and characterization of enterocin LR/6, a bacteriocin from enterococcus faecium LR/6. Applied Biochemistry and Biotechnology, 160(1), 40–49. [CrossRef]
- Kumar, V., Sheoran, P., Gupta, A., Yadav, J. P., & Tiwari, S. K. (2016). Antibacterial property of bacteriocin produced by Lactobacillus plantarum LD4 isolated from a fermented food. Annals of Microbiology, 66(4), 1431–1440.
- Molognoni, L., Daguer, H., Motta, G. E., Merlo, T. C., & Lindner, J. D. D. (2019). Interactions of preservatives in meat processing: Formation of carcinogenic compounds, analytical methods, and inhibitory agents. In Food Research International (Vol. 125). [CrossRef]
- Moreira, S. M., Picinin, C. T., do Amarante, C. C., Pilatti, L. A., & Pedroso, B. (2020). Simulation of the new proposed method by CAPES for the Qualis 2017-2020 classification of the Brazilian archives of biology and technology. Brazilian Archives of Biology and Technology, 63. [CrossRef]
- Muhammad Shahid Riaz Rajoka a, Hafiza Mahreen Mehwish c, Muhammad Siddiq b, Zhao Haobin a, Jing Zhu a, Li Yan a, Dongyan Shao a, Xiaoguang Xu a, J. S., Riaz Rajoka, M. S., Mehwish, H. M., Siddiq, M., Haobin, Z., Zhu, J., Yan, L., Shao, D., Xu, X., & Shi, J. (2017). Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. 84, 271–280. [CrossRef]
- O’Sullivan, L., Ross, R. P., & Hill, C. (2002). Potential of bacteriocin-producing lactic acid bacteria for improvements in food safety and quality. In Biochimie (Vol. 84, Issues 5–6). [CrossRef]
- Ogunbanwo, S. T., Sanni, A. I., & Onilude, A. A. (2003). Characterization of bacteriocin produced by Lactobacillus plantarum F1 and Lactobacillus brevis OG1. African Journal of Biotechnology, 2(8). [CrossRef]
- Ogunbanwo, S. T., Sanni, A. I., & Onilude, A. A. (2004). Effect of bacteriocinogenic Lactobacillus spp. on the shelf life of fufu, a traditional fermented cassava product. World Journal of Microbiology and Biotechnology, 20(1), 57–63. [CrossRef]
- Ohmomo, S., Murata, S., Katayama, N., Nitisinprasart, S., Kobayashi, M., Nakajima, T., Yajima, M., & Nakanishi, K. (2000). Purification and some characteristics of enterocin ON-157, a bacteriocin produced by Enterococcus faecium NIAI 157. Journal of Applied Microbiology, 88(1), 81–89. [CrossRef]
- Parada, J., & Aguilera, J. M. (2007). Food microstructure affects the bioavailability of several nutrients. Journal of Food Science, 72(2). [CrossRef]
- Perumal, V., & Venkatesan, A. (2017). Antimicrobial, cytotoxic effect and purification of bacteriocin from vancomycin susceptible Enterococcus faecalis and its safety evaluation for probiotization. LWT, 78, 303–310. [CrossRef]
- Phumisantiphong, U., Siripanichgon, K., Reamtong, O., & Diraphat, P. (2017). A novel bacteriocin from Enterococcus faecalis 478 exhibits a potent activity against vancomycin-resistant enterococci. PLoS ONE, 12(10). [CrossRef]
- Quintela-Baluja, M., Jobling, K., Graham, D. W., Tabraiz, S., Shamurad, B., Alnakip, M., Böhme, K., Barros-Velázquez, J., Carrera, M., & Calo-Mata, P. (2022). Rapid Proteomic Characterization of Bacteriocin-Producing Enterococcus faecium Strains from Foodstuffs. [CrossRef]
- Rajeswari, T., Venugopal, A., Viswanathan, C., Kishmu, L., Venil, C. K., & Kumar, S. (2010). Antibacterial activity of honey against Staphylococcus aureus from infected wounds. Pharmacologyonline.Silae.It, 1, 537–541. https://pharmacologyonline.silae.it/files/newsletter/2010/vol1/59.Kumar.pdf.
- Rehaiem, A., Martínez, B., Manai, M., & Rodríguez, A. (2010). Production of enterocin A by Enterococcus faecium MMRA isolated from ‘Rayeb’, a traditional Tunisian dairy beverage. Journal of Applied Microbiology, 108(5), 1685–1693. [CrossRef]
- Rushdy, A. A., & Gomaa, E. Z. (2013). Antimicrobial compounds produced by probiotic Lactobacillus brevis isolated from dairy products. Annals of Microbiology, 63(1). [CrossRef]
- Sanlibaba, P., Güçer, Y., & Şanlıbaba, P. (2015). ANTIMICROBIAL ACTIVITY OF LACTIC ACID BACTERIA. Agriculture & Food ISSN, 3. www.scientific-publications.net.
- Santos, R., Oliva-Teles, A., Saavedra, M., Enes, P., & Serra, C. (2018). Bacillus spp. as source of Natural Antimicrobial Compounds to control aquaculture bacterial fish pathogens. Frontiers in Marine Science, 5. [CrossRef]
- Sarkar, S. L., Hossain, M. I., Monika, S. A., Sanyal, S. K., Roy, P. C., Hossain, M. A., & Jahid, I. K. (2020). Probiotic potential of pediococcus acidilactici and enterococcus faecium isolated from indigenous yogurt and raw goat milk. Microbiology and Biotechnology Letters, 48(3). [CrossRef]
- Sasidharan, S., Chen, Y., Saravanan, D., Sundram, K. M., & Yoga Latha, L. (2011). Extraction, Isolation and Characterization of Bioactive Compounds from Plants’ Extracts. African Journal of Traditional, Complementary, and Alternative Medicines, 8(1), 1. [CrossRef]
- Silva, C. C. G. C. C. G., Silva, S. P. M. S. C. G., & Ribeiro, S. C. (2018). Application of Bacteriocins and Protective Cultures in Dairy Food Preservation. 9(APR), 594. www.frontiersin.org.
- Sivapalasingam, S., Friedman, C. R., Cohen, L., & Tauxe, R. V. (2004). Fresh produce: A growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. In Journal of Food Protection (Vol. 67, Issue 10). [CrossRef]
- Todorov, S. D. (2009). Bacteriocinas de Lactobacillus plantarum - produção, organização genética e modo de ação. Brazilian Journal of Microbiology, 40(2), 209–221. [CrossRef]
- Vimont, A., Fernandez, B., Hammami, R., Ababsa, A., Daba, H., & Fliss, I. (2017). Bacteriocin-Producing Enterococcus faecium LCW 44: A High Potential Probiotic Candidate from Raw Camel Milk. Frontiers in Microbiology, 8(MAY). [CrossRef]
- Weaver, D. B., & Lawton, L. J. (2007). Twenty years on: The state of contemporary ecotourism research. Tourism Management, 28(5). [CrossRef]
- Weine, E. R., & Kim, N. S. (2019). Systematic distortions in clinicians’ memories for client cases: Increasing causal coherence. Journal of Experimental Psychology: Learning Memory and Cognition, 45(2). [CrossRef]
- Wingfield, P. T. (2001). Wingfield, P. T. (2001). Protein Precipitation Using Ammonium Sulfate. Current Protocols in Protein Science / Editorial Board, John E. Coligan... [et Al.], APPENDIX 3, AppendixPrecipitation Using Ammonium Sulfate. [CrossRef]
- Wray, N. R., Ripke, S., Mattheisen, M., Trzaskowski, M., Byrne, E. M., Abdellaoui, A., Adams, M. J., Agerbo, E., Air, T. M., Andlauer, T. M. F., Bacanu, S. A., Bækvad-Hansen, M., Beekman, A. F. T., Bigdeli, T. B., Binder, E. B., Blackwood, D. R. H., Bryois, J., Buttenschøn, H. N., Bybjerg-Grauholm, J., … Sullivan, P. F. (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nature Genetics, 50(5). [CrossRef]
- Xie, Y., Peng, Q., Ji, Y., Xie, A., Yang, L., Mu, S., Li, Z., He, T., Xiao, Y., Zhao, J., & Zhang, Q. (2021). Isolation and Identification of Antibacterial Bioactive Compounds From Bacillus megaterium L2. Frontiers in Microbiology, 12, 662. [CrossRef]
- Xu, H., Jeong, H. S., Lee, H. Y., & Ahn, J. (2009). Assessment of cell surface properties and adhesion potential of selected probiotic strains. Letters in Applied Microbiology, 49(4). [CrossRef]
- Yamada, R., Sato, M., Kawabata, M., Nakatsuka, H., Nakamura, K., & Takashima, S. (1983). Hepatic artery embolization in 120 patients with unresectable hepatoma. Radiology, 148(2). [CrossRef]
- Yang, E., Fan, L., Jiang, Y., Doucette, C., & Fillmore, S. (2012). Antimicrobial activity of bacteriocin-producing lactic acid bacteria isolated from cheeses and yogurts. AMB Express, 2(1), 1–12. [CrossRef]
- Yang, J., Ji, Y., Park, H., Lee, J., Park, S., Yeo, S., Shin, H., & Holzapfel, W. H. (2014). International Journal of Food Microbiology Selection of functional lactic acid bacteria as starter cultures for the fermentation of Korean leek ( Allium tuberosum Rottler ex Sprengel.). International Journal of Food Microbiology, 191, 164–171. [CrossRef] [PubMed]
- Zhang, J., Yang, Y., Yang, H., Bu, Y., Yi, H., Zhang, L., Han, X., & Ai, L. (2018). Purification and partial characterization of bacteriocin Lac-B23, a novel bacteriocin production by lactobacillus plantarumJ23, isolated from Chinese traditional fermented milk. Frontiers in Microbiology, 9(OCT). [CrossRef]
- Zommiti, M., Bouffartigues, E., Maillot, O., Barreau, M., Szunerits, S., Sebei, K., Feuilloley, M., Connil, N., & Ferchichi, M. (2018). In vitroassessment of the probiotic properties and bacteriocinogenic potential of pediococcus pentosaceusMZF16 isolated from artisanal tunisian meat "dried ossban. Frontiers in Microbiology, 9(NOV), 2607. [CrossRef]
- Zommiti, M., Cambronel, M., Maillot, O., Barreau, M., Sebei, K., Feuilloley, M., Ferchichi, M., & Connil, N. (2018). Evaluation of probiotic properties and safety of Enterococcus faecium isolated from artisanal Tunisian Meat ‘Dried Ossban’. Frontiers in Microbiology, 9(AUG). [CrossRef]

| Sources of isolates | Designation/code | Identity |
| EPHI* | ATCC@25922 | Escherichia. Coli |
| EPHI | ATCC@25923 | Staphylococcus aureus |
| EPHI | ATCC@7644 | Listeria monocytogens |
| EPHI | ATCC@27853 | Pseudomonas auroginoasa |
| EPHI | ATCC@13311 | Salmonella Typhimurium |
| EPHI | ATCC@ 14053 | Candida albicans |
| Borde | JULAB-Br40 | Enterococcus faecium |
| Borde | JULAB-Br39 | Enterococcus faecium |
| Ergo | JULAB-E23 | Enterococcus faecium |
| Ergo | JULAB-E36 | Enterococcus faecium |
| Cabbage-Shamita | JULAB-S43 | Enterococcus faecium |
| Cabbage-Shamita | JULAB-S36 | Enterococcus faecium |
| S/N | LAB isolates |
E.coli ATCC®25922 |
S.aurues ATCC®25923 |
L.monocytogenes ATCC®7644 |
P.auroginosa ATCC®27853 |
S. Tphimurium ATCC®13311 |
C.albicans ATCC®14053 |
| 1) | JULABBE23 | 13.50±0.71d | 16.00±1.41bc | 35.31±0.44a | 20.50±0.71bc | 18.88±0.18abc | 16.13±0.18d |
| 2) | JULABBr39 | 20.50±0.71a | 17.50±0.71ab | 20.26±0.37de | 21.25±0.35a | 20.17±0.71a | 15.50±0.71ef |
| 3) | JULABBR40 | 17.75±0.35b | 17.50±0.71ab | 28.32±0.45bc | 16.13±0.18c | 15.13±0.18c | 24.38±0.53cd |
| 4) | JULABE36 | 18.50±0.71b | 18.00±0.00a | 28.25±0.35bc | 13.25±0.35de | 20.17±0.71a | 21.50±0.71c |
| 5) | JULABS36 | 20.25±0.35a | 17.50±0.71ab | 25.25±0.35c | 16.13±0.18c | 19.13±0.18ab | 19.26±0.37de |
| 6) | JULABS43 | 17.93±0.11b | 15.50±0.7ca | 20.13±0.18de | 14.13±0.18de | 18.13±0.18abc | 30.26±0.37a |
| Tetracycline | 18.75±1.06b | 17.50±0.71ab | 18.38±0.53d | 14.13±0.18de | 20.13±0.18a | 25.26±0.37cd |
| Diameter of inhibition zone (mm) | ||||
| S/N | LAB isolates | E.coli ATCC@25922 | S.aurues ATCC@25923 | L.monocytogenes ATCC@7644 |
| 1) | JULABE36 | 18.5±0.00ab | 18.55±0.00b | 17.00±0.00c |
| 2) | JULABE23 | 20.5±0.18a | 18.4±0.18b | 18.22±0.18c |
| 3) | JULABS43 | 14.75±0.35c | 17.05±0.71bc | 24.00±0.71bc |
| 4) | JULABS36 | 14.5±0.18c | 17.5±0.18bc | 25.25±0.37bc |
| 5) | JULABBR40 | 18.93±0.18ab | 18.00±0.18b | 27.00±0.53b |
| 6) | JULABBR39 | 19.00±0.71a | 20.22±0.18a | 36.00±0.18a |
|
E.coli ATCC@25922 |
S.aurues ATCC@25923 |
L.monocytogenes ATCC@7644 | |||||||||||
| S/N | LAB isolates | 40% | 50% | 60% | 80% | 40% | 50% | 60% | 80% | 40% | 50% | 60% | 80% |
| 1.00 | JULABBR39 | – | – | * | **** | – | – | * | **** | – | – | * | **** |
| 2.00 | JULABBR40 | – | – | * | **** | – | – | * | **** | – | – | * | **** |
| 3.00 | JULABE23 | – | – | * | **** | – | – | * | **** | – | – | * | **** |
| 4.00 | JULABE36 | – | – | * | **** | – | – | * | **** | – | – | * | **** |
| 5.00 | JULABS36 | – | – | * | **** | – | – | * | **** | – | – | * | **** |
| 6.00 | JULABS43 | – | – | * | **** | – | – | * | **** | – | – | * | **** |
| S/N | LAB isolates | Chloroform | Iso-propaly | Ethyl Acetate | Petroleum ether | Methanol | Butanol | Ethanol |
| 1) | JULABBR39 | **** | **** | *** | ** | ** | * | * |
| 2) | JULABBR40 | **** | **** | *** | ** | ** | * | * |
| 3) | JULABE23 | **** | *** | *** | ** | * | * | * |
| 4) | JULABE36 | **** | *** | *** | ** | * | * | – |
| 5) | JULABS36 | **** | *** | *** | ** | * | * | – |
| 6) | JULABS43 | **** | *** | *** | ** | * | * | * |
| Enterococcus faecium strains | Glucose | Sucrose | Maltose | Fructose | Lactose | MRS broth |
| JULABBR39 | 31.33±0.41a | 29.67±1.70a | 25.00±0.82ab | 18.00±0.82c | 16.00±0.82b | 15.67±1.70abc |
| JULABBR40 | 30.67±0.47ab | 28.00±0.82a | 27.33±0.47a | 28.00±0.00a | 17.33±0.94a | 17.00±0.82a |
| JULABE23 | 29.00±0.81ab | 25.00±0.83ab | 25.67±0.47ab | 25.00±0.85b | 15.00±0.82b | 13.67±2.36bc |
| JULABE36 | 25.00±0.82b | 22.00±0.85abc | 21.00±0.82c | 18.00±0.00c | 13.00±1.41bc | 10.67±0.94d |
| JULABS36 | 21.00±0.82bc | 18.90±0.97b | 15.00±0.74d | 13.00±0.82de | 15.33±1.89b | 16.33±1.25ab |
| JULABS43 | 17.50±0.40c | 18.33±0.48b | 20.33±0.52c | 14.83±0.24d | 12.33±0.47bd | 15.00±0.82abc |
| S/N | LAB isolates | Pepsin (5mg/1ml) | Trypsin (5mg/1ml) |
Proteinase K (5mg/1ml) |
α-amylase | Enzyme-free cell supernatants |
| 1 | JULABBR39 | - | - | - | + | + |
| 2 | JULABBR40 | - | - | - | + | + |
| 3 | JULABE23 | - | - | - | + | + |
| 4 | JULABE36 | - | - | - | + | + |
| 5 | JULABS36 | - | - | - | + | + |
| 6 | JULABS43 | - | - | - | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





