Submitted:
24 October 2023
Posted:
25 October 2023
You are already at the latest version
Abstract
Keywords:
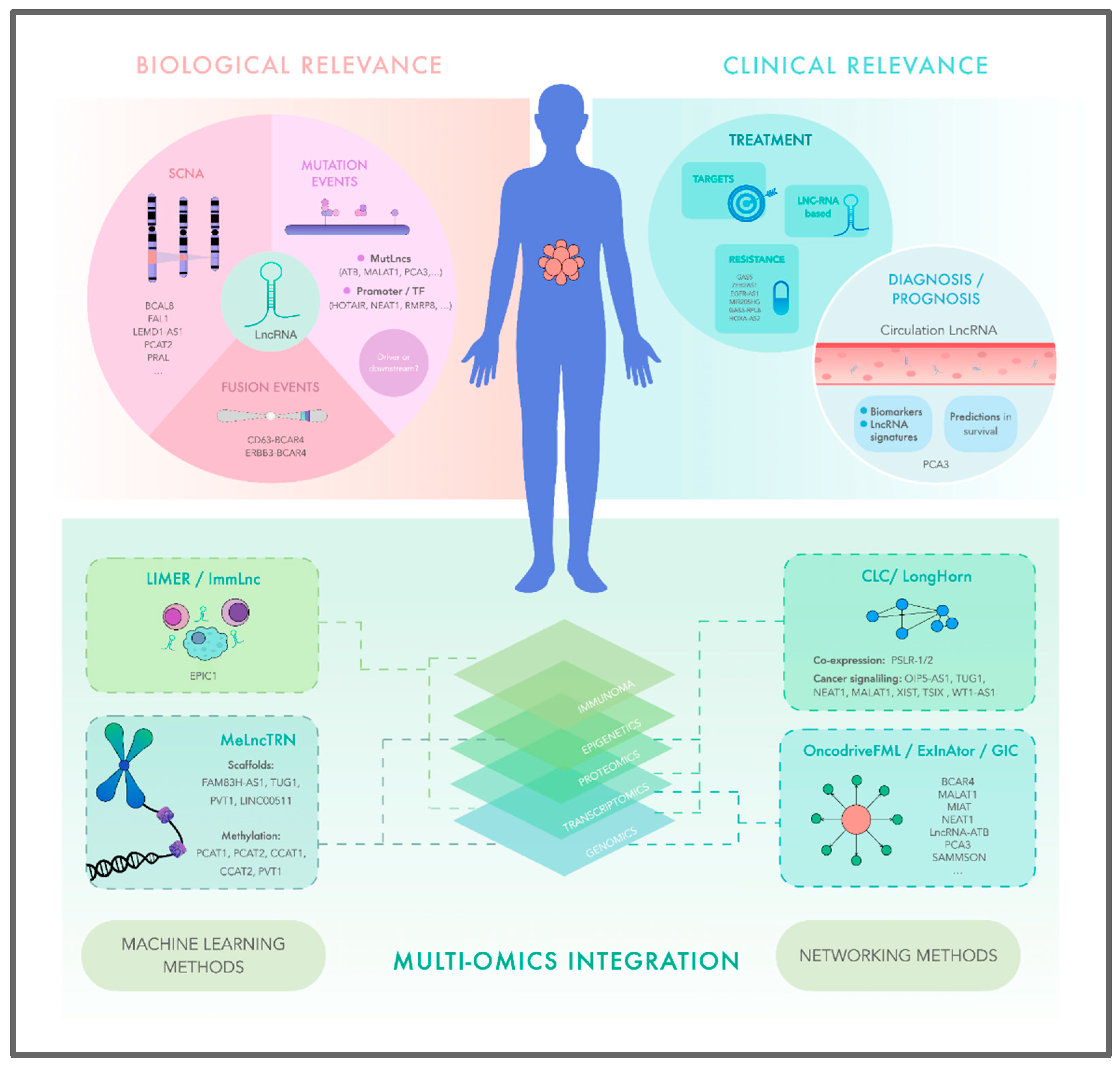
1. INTRODUCTION
2. THE REVOLUTION OF NON-CODING TRANSCRIPTOME IN CANCER STUDIES
3. LncRNAS ASSOCIATED TO CANCER DRIVER SOMATIC MUTATIONS PROFILES
4. APPROACHES TO PRIORITIZE CANCER DRIVER LNCRNAS USING SOMATIC MUTATIONS PROFILES
5. COPY-NUMBER VARIATIONS IN GENOMIC REGIONS ENCODING LNCRNAS
| LncRNA | Potential association based on biological or clinical data | Relation with other Omics features | Reference |
|---|---|---|---|
| ESR1, TRPS1, ERG, RUNX1, SNHG16, HOTAIR | cancer development and progression | somatic mutations on lncRNA TF binding site | [45] |
| RMRP8, NEAT1 | breast cancer (BRCA) | somatic mutations on lncRNA promoter | [44] |
| ENSG0000021403, ENSG00000261650, ENSG00000281406, G001643 | Relapse in colorectal cancer (CRC) | Mutations accumulated in lncRNA loci | [49] |
| LINC00460, AC156455.1, AC015977.2, ‘PRDM16-dt’, AL139351.1, AL035661.1, LINC01606 | Poor overall survival risk in renal cell carcinoma (RCC) | High Somatic mutation-associated lncRNAs signature |
[50] |
| LINC00460, LINC01234 | Poor overall survival risk in clear cell renal carcinoma (CCRC) | High Somatic mutation-associated lncRNAs signature |
[51] |
| FAM30A, CACNA1C-AS1, LINC02595 | Poor overall survival risk in AML | High Somatic mutation-associated lncRNAs signature |
[52] |
| AC007996.1, AC009237.14, AP003555.1, AL590483.1 | Poor overall survival risk in CRC | High Somatic mutation-associated lncRNAs signature |
[53] |
|
ZNF503-AS1, AL353747.2, AC129492.1, AP003555.1, AC009237.14 |
Poor overall survival risk in CRC | High Somatic mutation-associated lncRNAs signature |
[54] |
| AC107464.2, MIR100HG, AP001527.2 | Poor overall survival risk in cervical cancer (CESC) | High Somatic mutation-associated lncRNAs signature |
[55] |
| AC002511.2, LINC00501, LINC02055, LINC02714, LINC01508, LOC105371967, RP11_96A15.1, RP11_305F18.1, RP11_342M1.3, RP11_432J24.3, U95743.1 | Poor overall survival risk in hepatocellular carcinoma (HCC) | High Somatic mutation-associated lncRNAs signature |
[56] |
| C116351.1, ZFPM2-AS1, AC145343.1, MIR210HG | Poor overall survival risk in hepatocellular carcinoma (HCC) | High Somatic mutation-associated lncRNAs signature |
[58] |
| FAL1 | Oncogenic lncRNA | SCNA related lncRNA | [73] |
| BCAL8 | Poor overall survival risk BRCA | SCNA related lncRNAs | [74] |
| RUSC1-AS1, LINC01990, LINC01411, LINC02099, H19, LINC00452, ADPGK-AS1, C1QTNF1-AS1 | Poor overall survival risk in CESC | SCNA related lncRNAs | [77] |
| PRAL | Tumor suppressor | SCNA related lncRNA | [78] |
| LOC101927604, LOC105377267, CASC15, LINC-PINT, CLDN10-AS1, C14orf132, LMF1, LINC00675, CCDC144NL-AS1, LOC284454 | low disease-free survival in CRC | SCNA related lncRNA | [79] |
| RP11-571M6.8 | immunosuppressive function in GBM | SCNA related lncRNA | [80] |
| RP11-1020A11.1 | Poor overall survival risk in bladder carcinoma | SCNA related lncRNA | [80] |
| CTD-2256P15.2 | Treatment with methyl ethyl ketone (MEK) inhibitors prediction in LUAD | SCNA related lncRNA | [80] |
| LINC00896, MCM8-AS1, LINC01251, LNX1-AS1, GPRC5D-AS1, CTD-2350J17.1, LINC01133, LINC01121, and AC073130.1 | Poor overall survival risk in LUSC | SCNA related lncRNA | [81] |
| RP11-241F15.10 | Tumor suppressor in osteosarcoma | SCNA related lncRNA | [82] |
| ALAL-1 | Related to lower levels of immune infiltration LUSC | SCNA related lncRNA | [83] |
| LOC339803, F11-AS1, PCAT2 TMEM220-AS1 | Poor overall survival risk in HCC | SCNA related lncRNA | [85] |
| ENSG00000261582 | Poor overall survival risk in LUAD and CESC | SCNA related lncRNA | [89] |
| PCAN-R1 (Ensembl ID ENSG00000228288), PCAN-R2 (Ensembl ID ENSG00000231806) | Oncogenic in PRAD related to Poor overall survival risk | SCNA related lncRNA | [89] |
| LOC101927151, LINC00861, LEMD1-AS1 | Poor overall survival risk in OV | SCNA related lncRNA | [90] |
6. THE ADVENT OF MACHINE LEARNING TO DEEPEN THE FUNCTIONAL AND BIOLOGICAL ROLES OF LNCRNAS IN CANCER
7. MULTI-OMIC NETWORKS APPROACHES REVEAL LNCRNA BIOLOGICAL RELEVANCE ON CANCER BIOLOGY
7.1. Describing new LncRNAs drivers in cancer through multi-omic integration
8. THERAPEUTIC HARNESSING OF LNCRNAS THROUGH MULTI-OMIC ONCOLOGY: PERSPECTIVE OF THE FUTURE APPLICATION OF LNCRNAS
9. CONCLUSIONS
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Francies, H.E.; McDermott, U.; Garnett, M.J. Genomics-Guided Pre-Clinical Development of Cancer Therapies. Nature Cancer 2020, 1, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular Profiling for Precision Cancer Therapies. Genome Med. 2020, 12, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Perakakis, N.; Yazdani, A.; Karniadakis, G.E.; Mantzoros, C. Omics, Big Data and Machine Learning as Tools to Propel Understanding of Biological Mechanisms and to Discover Novel Diagnostics and Therapeutics. Metabolism 2018, 87, A1–A9. [Google Scholar] [CrossRef]
- Vlachavas, E.I.; Bohn, J.; Ückert, F.; Nürnberg, S. A Detailed Catalogue of Multi-Omics Methodologies for Identification of Putative Biomarkers and Causal Molecular Networks in Translational Cancer Research. Int. J. Mol. Sci. 2021, 22, 2822. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.-T.; Huang, X.; Huang, S.; Xie, S.-J.; Xiao, Z.-D.; Zhang, H. RNA Sequencing: New Technologies and Applications in Cancer Research. J. Hematol. Oncol. 2020, 13, 1–16. [Google Scholar] [CrossRef]
- Adelman, K.; Egan, E. Non-Coding RNA: More Uses for Genomic Junk. Nature 2017, 543, 183–185. [Google Scholar] [CrossRef]
- Consortium, T.E.P. The ENCODE Project Consortium An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The Landscape of Long Noncoding RNAs in the Human Transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Zhang, X.; Meyerson, M. Illuminating the Noncoding Genome in Cancer. Nature Cancer 2020, 1, 864–872. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Matsui, M.; Corey, D.R. Non-Coding RNAs as Drug Targets. Nat. Rev. Drug Discov. 2017, 16, 167–179. [Google Scholar] [CrossRef]
- Amodio, N.; Stamato, M.A.; Juli, G.; Morelli, E.; Fulciniti, M.; Manzoni, M.; Taiana, E.; Agnelli, L.; Cantafio, M.E.G.; Romeo, E.; et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO Inhibits Gene Expression of Proteasome Subunits and Triggers Anti-Multiple Myeloma Activity. Leukemia 2018, 32, 1948–1957. [Google Scholar] [CrossRef]
- Tang, Q.; Zheng, F.; Liu, Z.; Wu, J.; Chai, X.; He, C.; Li, L.; Hann, S.S. Novel Reciprocal Interaction of lncRNA HOTAIR and miR-214-3p Contribute to the Solamargine-Inhibited PDPK1 Gene Expression in Human Lung Cancer. J. Cell. Mol. Med. 2019, 23, 7749–7761. [Google Scholar] [CrossRef]
- Cieślik, M.; Chinnaiyan, A.M. Cancer Transcriptome Profiling at the Juncture of Clinical Translation. Nat. Rev. Genet. 2018, 19, 93–109. [Google Scholar] [CrossRef]
- Zhou, M.; Hu, L.; Zhang, Z.; Wu, N.; Sun, J.; Su, J. Recurrence-Associated Long Non-Coding RNA Signature for Determining the Risk of Recurrence in Patients with Colon Cancer. Mol. Ther. Nucleic Acids 2018, 12, 518–529. [Google Scholar] [CrossRef]
- Yuan, L.; Zhao, J.; Sun, T.; Shen, Z. A Machine Learning Framework That Integrates Multi-Omics Data Predicts Cancer-Related LncRNAs. BMC Bioinformatics 2021, 22, 1–18. [Google Scholar] [CrossRef]
- Heo, Y.J.; Hwa, C.; Lee, G.-H.; Park, J.-M.; An, J.-Y. Integrative Multi-Omics Approaches in Cancer Research: From Biological Networks to Clinical Subtypes. Mol. Cells 2021, 44, 433–443. [Google Scholar] [CrossRef]
- Qian, Y.; Shi, L.; Luo, Z. Long Non-Coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 2020, 7, 612393. [Google Scholar] [CrossRef]
- Yan, C.; Zhang, Z.; Bao, S.; Hou, P.; Zhou, M.; Xu, C.; Sun, J. Computational Methods and Applications for Identifying Disease-Associated lncRNAs as Potential Biomarkers and Therapeutic Targets. Mol. Ther. Nucleic Acids 2020, 21, 156–171. [Google Scholar] [CrossRef]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. LncRNADisease: A Database for Long-Non-Coding RNA-Associated Diseases. Nucleic Acids Res. 2012, 41, D983–D986. [Google Scholar] [CrossRef]
- Volders, P.-J.; Anckaert, J.; Verheggen, K.; Nuytens, J.; Martens, L.; Mestdagh, P.; Vandesompele, J. LNCipedia 5: Towards a Reference Set of Human Long Non-Coding RNAs. Nucleic Acids Res. 2019, 47, D135–D139. [Google Scholar] [CrossRef]
- Sun, Y.; Ma, L. New Insights into Long Non-Coding RNA in Cancer and Metastasis. Cancers 2019, 11, 216. [Google Scholar] [CrossRef]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A Master Regulator of Chromatin Dynamics and Cancer. Biochim. Biophys. Acta 2015, 1856, 151–164. [Google Scholar] [CrossRef]
- Ferreira, L.B.; Palumbo, A.; de Mello, K.D.; Sternberg, C.; Caetano, M.S.; de Oliveira, F.L.; Neves, A.F.; Nasciutti, L.E.; Goulart, L.R.; Gimba, E.R.P. PCA3 Noncoding RNA Is Involved in the Control of Prostate-Cancer Cell Survival and Modulates Androgen Receptor Signaling. BMC Cancer 2012, 12, 507. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a Novel Noncoding RNA, and Thymosin beta4 Predict Metastasis and Survival in Early-Stage Non-Small Cell Lung Cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Okugawa, Y.; Toiyama, Y.; Hur, K.; Toden, S.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Kusunoki, M.; Boland, C.R.; et al. Metastasis-Associated Long Non-Coding RNA Drives Gastric Cancer Development and Promotes Peritoneal Metastasis. Carcinogenesis 2014, 35, 2731–2739. [Google Scholar] [CrossRef]
- Zheng, H.-T.; Shi, D.-B.; Wang, Y.-W.; Li, X.-X.; Xu, Y.; Tripathi, P.; Gu, W.-L.; Cai, G.-X.; Cai, S.-J. High Expression of lncRNA MALAT1 Suggests a Biomarker of Poor Prognosis in Colorectal Cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 3174–3181. [Google Scholar]
- Lu, R.; Zhang, J.; Zhang, W.; Huang, Y.; Wang, N.; Zhang, Q.; Qu, S. Circulating HOTAIR Expression Predicts the Clinical Response to Neoadjuvant Chemotherapy in Patients with Breast Cancer. Cancer Biomark. 2018, 22, 249–256. [Google Scholar] [CrossRef]
- Yang, Z.; Zhou, L.; Wu, L.-M.; Lai, M.-C.; Xie, H.-Y.; Zhang, F.; Zheng, S.-S. Overexpression of Long Non-Coding RNA HOTAIR Predicts Tumor Recurrence in Hepatocellular Carcinoma Patients Following Liver Transplantation. Ann. Surg. Oncol. 2011, 18, 1243–1250. [Google Scholar] [CrossRef]
- Lemos, A.E.G.; Matos, A. da R.; Ferreira, L.B.; Gimba, E.R.P. The Long Non-Coding RNA: An Update of Its Functions and Clinical Applications as a Biomarker in Prostate Cancer. Oncotarget 2019, 10, 6589–6603. [Google Scholar] [CrossRef]
- Pal, R.P.; Maitra, N.U.; Mellon, J.K.; Khan, M.A. Defining Prostate Cancer Risk before Prostate Biopsy. Urol. Oncol. 2013, 31, 1408–1418. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic Instability—An Evolving Hallmark of Cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational Heterogeneity in Cancer and the Search for New Cancer-Associated Genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef]
- Korf, B.R.; Rehm, H.L. New Approaches to Molecular Diagnosis. JAMA 2013, 309, 1511–1521. [Google Scholar] [CrossRef]
- Piraino, S.W.; Furney, S.J. Beyond the Exome: The Role of Non-Coding Somatic Mutations in Cancer. Ann. Oncol. 2016, 27, 240–248. [Google Scholar] [CrossRef]
- Rheinbay, E.; Nielsen, M.M.; Abascal, F.; Wala, J.A.; Shapira, O.; Tiao, G.; Hornshøj, H.; Hess, J.M.; Juul, R.I.; Lin, Z.; et al. Analyses of Non-Coding Somatic Drivers in 2,658 Cancer Whole Genomes. Nature 2020, 578, 102–111. [Google Scholar] [CrossRef]
- Rheinbay, E.; Parasuraman, P.; Grimsby, J.; Tiao, G.; Engreitz, J.M.; Kim, J.; Lawrence, M.S.; Taylor-Weiner, A.; Rodriguez-Cuevas, S.; Rosenberg, M.; et al. Recurrent and Functional Regulatory Mutations in Breast Cancer. Nature 2017, 547, 55–60. [Google Scholar] [CrossRef]
- Fujimoto, A.; Furuta, M.; Totoki, Y.; Tsunoda, T.; Kato, M.; Shiraishi, Y.; Tanaka, H.; Taniguchi, H.; Kawakami, Y.; Ueno, M.; et al. Whole-Genome Mutational Landscape and Characterization of Noncoding and Structural Mutations in Liver Cancer. Nat. Genet. 2016, 48, 500–509. [Google Scholar] [CrossRef]
- Gasic, V.; Karan-Djurasevic, T.; Pavlovic, D.; Zukic, B.; Pavlovic, S.; Tosic, N. Diagnostic and Therapeutic Implications of Long Non-Coding RNAs in Leukemia. Life 2022, 12, 1770. [Google Scholar] [CrossRef]
- Puente, X.S.; Beà, S.; Valdés-Mas, R.; Villamor, N.; Gutiérrez-Abril, J.; Martín-Subero, J.I.; Munar, M.; Rubio-Pérez, C.; Jares, P.; Aymerich, M.; et al. Non-Coding Recurrent Mutations in Chronic Lymphocytic Leukaemia. Nature 2015, 526, 519–524. [Google Scholar] [CrossRef]
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Landscape of Somatic Mutations in 560 Breast Cancer Whole-Genome Sequences. Nature 2016, 534, 47–54. [Google Scholar] [CrossRef]
- Gao, Y.; Li, X.; Zhi, H.; Zhang, Y.; Wang, P.; Wang, Y.; Shang, S.; Fang, Y.; Shen, W.; Ning, S.; et al. Comprehensive Characterization of Somatic Mutations Impacting lncRNA Expression for Pan-Cancer. Mol. Ther. Nucleic Acids 2019, 18, 66–79. [Google Scholar] [CrossRef]
- Rezaie, N.; Bayati, M.; Hamidi, M.; Tahaei, M.S.; Khorasani, S.; Lovell, N.H.; Breen, J.; Rabiee, H.R.; Alinejad-Rokny, H. Somatic Point Mutations Are Enriched in Non-Coding RNAs with Possible Regulatory Function in Breast Cancer. Commun Biol 2022, 5, 556. [Google Scholar] [CrossRef]
- Esposito, R.; Lanzós, A.; Polidori, T.; Guillen-Ramirez, H.; Merlin, B.M.; Mela, L.; Zoni, E.; Büchi, I.; Hovhannisyan, L.; McCluggage, F.; et al. Tumour Mutations in Long Noncoding RNAs Enhance Cell Fitness. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, P.; Guo, Q.; Hao, Y.; Qi, Y.; Xin, M.; Zhang, Y.; Cui, B.; Wang, P. Oncogenic Landscape of Somatic Mutations Perturbing Pan-Cancer lncRNA-ceRNA Regulation. Front Cell Dev Biol 2021, 9, 658346. [Google Scholar] [CrossRef] [PubMed]
- Iraola-Guzmán, S.; Brunet-Vega, A.; Pegueroles, C.; Saus, E.; Hovhannisyan, H.; Casalots, A.; Pericay, C.; Gabaldón, T. Target Enrichment Enables the Discovery of lncRNAs with Somatic Mutations or Altered Expression in Paraffin-Embedded Colorectal Cancer Samples. Cancers 2020, 12, 2844. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Liu, X.; Lu, L.; Liu, G. Identification of a Somatic Mutation-Derived Long Non-Coding RNA Signatures of Genomic Instability in Renal Cell Carcinoma. Front. Oncol. 2021, 11, 728181. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yan, K.; Wang, L.; Bi, J. Genome Instability-Related Long Non-Coding RNA in Clear Renal Cell Carcinoma Determined Using Computational Biology. BMC Cancer 2021, 21, 727. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Guo, T. Somatic Mutation-Associated Risk Index Based on lncRNA Expression for Predicting Prognosis in Acute Myeloid Leukemia. Hematology 2022, 27, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Yun, D.; Yang, Z. Identification of a Four-lncRNA Prognostic Signature for Colon Cancer Based on Genome Instability. J. Oncol. 2021, 2021, 7408893. [Google Scholar] [CrossRef]
- Yin, T.; Zhao, D.; Yao, S. Identification of a Genome Instability-Associated LncRNA Signature for Prognosis Prediction in Colon Cancer. Front. Genet. 2021, 12, 679150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ding, N.; He, Y.; Tao, C.; Liang, Z.; Xin, W.; Zhang, Q.; Wang, F. Bioinformatic Identification of Genomic Instability-Associated lncRNAs Signatures for Improving the Clinical Outcome of Cervical Cancer by a Prognostic Model. Sci. Rep. 2021, 11, 20929. [Google Scholar] [CrossRef]
- Jin, C.; Zhao, J.-S.; Huang, X.-Q.; Yang, X.-Z.; Niu, F.-Y.; Lin, J.-R.; Ma, L.; Shi, Y.-X.; Li, X.-S.; Jiang, P.; et al. A Somatic Mutation-Derived LncRNA Signatures of Genomic Instability Predicts the Prognosis and Tumor Microenvironment Immune Characters in Hepatocellular Carcinoma. Hepatol. Int. 2022, 16, 1220–1233. [Google Scholar] [CrossRef]
- Brady, S.W.; Gout, A.M.; Zhang, J. Therapeutic and Prognostic Insights from the Analysis of Cancer Mutational Signatures. Trends Genet. 2021. [Google Scholar] [CrossRef]
- Wu, J.; Ren, X.; Wang, N.; Zhou, R.; Chen, M.; Cai, Y.; Lin, S.; Zhang, H.; Xie, X.; Dang, C.; et al. A Mutation-Related Long Noncoding RNA Signature of Genome Instability Predicts Immune Infiltration and Hepatocellular Carcinoma Prognosis. Front. Genet. 2021, 12, 779554. [Google Scholar] [CrossRef]
- Gibney, G.T.; Weiner, L.M.; Atkins, M.B. Predictive Biomarkers for Checkpoint Inhibitor-Based Immunotherapy. Lancet Oncol. 2016, 17, e542–e551. [Google Scholar] [CrossRef]
- Ding, C.; Shan, Z.; Li, M.; Xia, Y.; Jin, Z. Exploration of the Associations of lncRNA Expression Patterns with Tumor Mutation Burden and Prognosis in Colon Cancer. Onco. Targets. Ther. 2021, 14, 2893–2909. [Google Scholar] [CrossRef]
- Lee, J.J.-K.; Park, S.; Park, H.; Kim, S.; Lee, J.; Lee, J.; Youk, J.; Yi, K.; An, Y.; Park, I.K.; et al. Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma. Cell 2019, 177, 1842–1857.e21. [Google Scholar] [CrossRef] [PubMed]
- Bae, K.; Kim, J.H.; Jung, H.; Kong, S.-Y.; Kim, Y.-H.; Kim, S.; Lee, G.K.; Lee, J.S.; Lee, J.J.-K.; Ju, Y.S.; et al. A Fusion of CD63-BCAR4 Identified in Lung Adenocarcinoma Promotes Tumorigenicity and Metastasis. Br. J. Cancer 2021, 124, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Jordan, E.J.; Kim, H.R.; Arcila, M.E.; Barron, D.; Chakravarty, D.; Gao, J.; Chang, M.T.; Ni, A.; Kundra, R.; Jonsson, P.; et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov. 2017, 7, 596–609. [Google Scholar] [CrossRef] [PubMed]
- Koivunen, J.P.; Mermel, C.; Zejnullahu, K.; Murphy, C.; Lifshits, E.; Holmes, A.J.; Choi, H.G.; Kim, J.; Chiang, D.; Thomas, R.; et al. EML4-ALK Fusion Gene and Efficacy of an ALK Kinase Inhibitor in Lung Cancer. Clin. Cancer Res. 2008, 14, 4275–4283. [Google Scholar] [CrossRef]
- Wang, C.; Yin, R.; Dai, J.; Gu, Y.; Cui, S.; Ma, H.; Zhang, Z.; Huang, J.; Qin, N.; Jiang, T.; et al. Whole-Genome Sequencing Reveals Genomic Signatures Associated with the Inflammatory Microenvironments in Chinese NSCLC Patients. Nat. Commun. 2018, 9, 2054. [Google Scholar] [CrossRef]
- Godinho, M.F.E.; Sieuwerts, A.M.; Look, M.P.; Meijer, D.; Foekens, J.A.; Dorssers, L.C.J.; van Agthoven, T. Relevance of BCAR4 in Tamoxifen Resistance and Tumour Aggressiveness of Human Breast Cancer. Br. J. Cancer 2010, 103, 1284–1291. [Google Scholar] [CrossRef]
- Guichard, C.; Amaddeo, G.; Imbeaud, S.; Ladeiro, Y.; Pelletier, L.; Maad, I.B.; Calderaro, J.; Bioulac-Sage, P.; Letexier, M.; Degos, F.; et al. Integrated Analysis of Somatic Mutations and Focal Copy-Number Changes Identifies Key Genes and Pathways in Hepatocellular Carcinoma. Nat. Genet. 2012, 44, 694–698. [Google Scholar] [CrossRef]
- Shao, X.; Lv, N.; Liao, J.; Long, J.; Xue, R.; Ai, N.; Xu, D.; Fan, X. Copy Number Variation Is Highly Correlated with Differential Gene Expression: A Pan-Cancer Study. BMC Med. Genet. 2019, 20, 175. [Google Scholar] [CrossRef]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhang, C.-Z.; Wala, J.; Mermel, C.H.; et al. Pan-Cancer Patterns of Somatic Copy Number Alteration. Nature Genetics 2013, 45, 1134–1140. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, L.; Kucherlapati, M.; Chen, F.; Hadjipanayis, A.; Pantazi, A.; Bristow, C.A.; Lee, E.A.; Mahadeshwar, H.S.; Tang, J.; et al. A Pan-Cancer Compendium of Genes Deregulated by Somatic Genomic Rearrangement across More Than 1,400 Cases. Cell Rep. 2018, 24, 515–527. [Google Scholar] [CrossRef]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The Landscape of Somatic Copy-Number Alteration across Human Cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Ping, Y.; Zhou, Y.; Hu, J.; Pang, L.; Xu, C.; Xiao, Y. Dissecting the Functional Mechanisms of Somatic Copy-Number Alterations Based on Dysregulated ceRNA Networks across Cancers. Molecular Therapy - Nucleic Acids 2020, 21, 464–479. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Feng, Y.; Zhang, D.; Zhao, S.D.; Hu, Z.; Greshock, J.; Zhang, Y.; Yang, L.; Zhong, X.; Wang, L.-P.; et al. A Functional Genomic Approach Identifies FAL1 as an Oncogenic Long Noncoding RNA That Associates with BMI1 and Represses p21 Expression in Cancer. Cancer Cell 2014, 26, 344–357. [Google Scholar] [CrossRef]
- Yan, X.; Hu, Z.; Feng, Y.; Hu, X.; Yuan, J.; Zhao, S.D.; Zhang, Y.; Yang, L.; Shan, W.; He, Q.; et al. Comprehensive Genomic Characterization of Long Non-Coding RNAs across Human Cancers. Cancer Cell 2015, 28, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-L.; Li, B.; Piccolo, S.R.; Zhang, X.-Q.; Li, J.-H.; Zhou, H.; Yang, J.-H.; Qu, L.-H. Integrative Analysis Reveals Clinical Phenotypes and Oncogenic Potentials of Long Non-Coding RNAs across 15 Cancer Types. Oncotarget 2016, 7, 35044–35055. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of Transcription in Human Cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Zhong, Q.; Lu, M.; Yuan, W.; Cui, Y.; Ouyang, H.; Fan, Y.; Wang, Z.; Wu, C.; Qiao, J.; Hang, J. Eight-lncRNA Signature of Cervical Cancer Were Identified by Integrating DNA Methylation, Copy Number Variation and Transcriptome Data. J. Transl. Med. 2021, 19, 58. [Google Scholar] [CrossRef]
- Zhou, C.-C.; Yang, F.; Yuan, S.-X.; Ma, J.-Z.; Liu, F.; Yuan, J.-H.; Bi, F.-R.; Lin, K.-Y.; Yin, J.-H.; Cao, G.-W.; et al. Systemic Genome Screening Identifies the Outcome Associated Focal Loss of Long Noncoding RNA PRAL in Hepatocellular Carcinoma. Hepatology 2016, 63, 850–863. [Google Scholar] [CrossRef]
- Liu, H.; Gu, X.; Wang, G.; Huang, Y.; Ju, S.; Huang, J.; Wang, X. Copy Number Variations Primed lncRNAs Deregulation Contribute to Poor Prognosis in Colorectal Cancer. Aging 2019, 11, 6089–6108. [Google Scholar] [CrossRef]
- Zhang, Y.; Liao, G.; Bai, J.; Zhang, X.; Xu, L.; Deng, C.; Yan, M.; Xie, A.; Luo, T.; Long, Z.; et al. Identifying Cancer Driver lncRNAs Bridged by Functional Effectors through Integrating Multi-Omics Data in Human Cancers. Mol. Ther. Nucleic Acids 2019, 17, 362–373. [Google Scholar] [CrossRef]
- Ning, J.; Wang, F.; Zhu, K.; Li, B.; Shu, Q.; Liu, W. Characterizing the Copy Number Variation of Non-Coding RNAs Reveals Potential Therapeutic Targets and Prognostic Markers of LUSC. Front. Genet. 2021, 12, 779155. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Xiao, L.; Li, J.; Dong, B.; Wang, C. Integrative Analysis Reveals Driver Long Non-Coding RNAs in Osteosarcoma. Medicine 2019, 98, e14302. [Google Scholar] [CrossRef] [PubMed]
- Athie, A.; Marchese, F.P.; González, J.; Lozano, T.; Raimondi, I.; Juvvuna, P.K.; Abad, A.; Marin-Bejar, O.; Serizay, J.; Martínez, D.; et al. Analysis of Copy Number Alterations Reveals the lncRNA ALAL-1 as a Regulator of Lung Cancer Immune Evasion. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Zhao, M.; Zheng, C.-H.; Zhao, M.; Xia, J. Concordance between Somatic Copy Number Loss and down-Regulated Expression: A Pan-Cancer Study of Cancer Predisposition Genes. Scientific Reports 2016, 6. [Google Scholar] [CrossRef]
- Cheng, Z.; Guo, Y.; Sun, J.; Zheng, L. Four-Copy Number Alteration (CNA)-Related lncRNA Prognostic Signature for Liver Cancer. Sci. Rep. 2022, 12, 14261. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- Fisher, R.; Pusztai, L.; Swanton, C. Cancer Heterogeneity: Implications for Targeted Therapeutics. Br. J. Cancer 2013, 108, 479–485. [Google Scholar] [CrossRef]
- Chen, H.; Xu, J.; Hong, J.; Tang, R.; Zhang, X.; Fang, J.-Y. Long Noncoding RNA Profiles Identify Five Distinct Molecular Subtypes of Colorectal Cancer with Clinical Relevance. Mol. Oncol. 2014, 8, 1393–1403. [Google Scholar] [CrossRef]
- Du, Z.; Fei, T.; Verhaak, R.G.W.; Su, Z.; Zhang, Y.; Brown, M.; Chen, Y.; Liu, X.S. Integrative Genomic Analyses Reveal Clinically Relevant Long Noncoding RNAs in Human Cancer. Nat. Struct. Mol. Biol. 2013, 20, 908–913. [Google Scholar] [CrossRef]
- Zheng, M.; Hu, Y.; Gou, R.; Nie, X.; Li, X.; Liu, J.; Lin, B. Identification Three LncRNA Prognostic Signature of Ovarian Cancer Based on Genome-Wide Copy Number Variation. Biomed. Pharmacother. 2020, 124, 109810. [Google Scholar] [CrossRef] [PubMed]
- Akrami, R.; Jacobsen, A.; Hoell, J.; Schultz, N.; Sander, C.; Larsson, E. Comprehensive Analysis of Long Non-Coding RNAs in Ovarian Cancer Reveals Global Patterns and Targeted DNA Amplification. PLoS One 2013, 8, e80306. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, X.; Liu, C. The Computational Approaches of lncRNA Identification Based on Coding Potential: And Challenges. Comput. Struct. Biotechnol. J. 2020, 18, 3666–3677. [Google Scholar] [CrossRef]
- Chi, Y.; Wang, D.; Wang, J.; Yu, W.; Yang, J. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019, 8, 1015. [Google Scholar] [CrossRef]
- Zhong, L.; Zhen, M.; Sun, J.; Zhao, Q. Recent Advances on the Machine Learning Methods in Predicting ncRNA-Protein Interactions. Mol. Genet. Genomics 2021, 296, 243–258. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Luo, J.; Liang, C.; Xiao, Q.; Ding, P.; Zhang, Y. Prediction of LncRNA-Disease Associations Based on Network Consistency Projection. IEEE Access 2019, 7, 58849–58856. [Google Scholar] [CrossRef]
- Fan, X.-N.; Zhang, S.-W.; Zhang, S.-Y.; Zhu, K.; Lu, S. Prediction of lncRNA-Disease Associations by Integrating Diverse Heterogeneous Information Sources with RWR Algorithm and Positive Pointwise Mutual Information. BMC Bioinformatics 2019, 20, 87. [Google Scholar] [CrossRef]
- Zhou, J.-R.; You, Z.-H.; Cheng, L.; Ji, B.-Y. Prediction of lncRNA-Disease Associations via an Embedding Learning HOPE in Heterogeneous Information Networks. Molecular Therapy Nucleic Acids 2021, 23, 277–285. [Google Scholar] [CrossRef]
- Wang, Y.; Juan, L.; Peng, J.; Zang, T.; Wang, Y. LncDisAP: A Computation Model for LncRNA-Disease Association Prediction Based on Multiple Biological Datasets. BMC Bioinformatics 2019, 20, 582. [Google Scholar] [CrossRef]
- Yu, J.; Ping, P.; Wang, L.; Kuang, L.; Li, X.; Wu, Z. A Novel Probability Model for LncRNA–Disease Association Prediction Based on the Naïve Bayesian Classifier. Genes 2018, 9, 345. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Lei, X.; Liao, B.; Wu, F.-X. Machine Learning Approaches for Predicting Biomolecule-Disease Associations. Brief. Funct. Genomics 2021, 20, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Nath, A.; Lau, E.Y.T.; Lee, A.M.; Geeleher, P.; Cho, W.C.S.; Huang, R.S. Discovering Long Noncoding RNA Predictors of Anticancer Drug Sensitivity beyond Protein-Coding Genes. Proc. Natl. Acad. Sci. USA 2019, 116, 22020–22029. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mi, M.; Li, X.; Zheng, X.; Wu, G.; Zhang, L. A lncRNA Prognostic Signature Associated with Immune Infiltration and Tumour Mutation Burden in Breast Cancer. J. Cell. Mol. Med. 2020, 24, 12444–12456. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, L.; Feng, C.; Qin, Y.; Xiao, J.; Zhang, Z.; Ma, L. LncBook 2.0: Integrating Human Long Non-Coding RNAs with Multi-Omics Annotations. Nucleic Acids Res. 2022. [Google Scholar] [CrossRef]
- Carlevaro-Fita, J.; Lanzós, A.; Feuerbach, L.; Hong, C.; Mas-Ponte, D.; Pedersen, J.S.; PCAWG Drivers and Functional Interpretation Group; Johnson, R. PCAWG Consortium Cancer LncRNA Census Reveals Evidence for Deep Functional Conservation of Long Noncoding RNAs in Tumorigenesis. Commun Biol 2020, 3, 56. [Google Scholar] [CrossRef]
- Mularoni, L.; Sabarinathan, R.; Deu-Pons, J.; Gonzalez-Perez, A.; López-Bigas, N. OncodriveFML: A General Framework to Identify Coding and Non-Coding Regions with Cancer Driver Mutations. Genome Biol. 2016, 17, 128. [Google Scholar] [CrossRef] [PubMed]
- Lanzós, A.; Carlevaro-Fita, J.; Mularoni, L.; Reverter, F.; Palumbo, E.; Guigó, R.; Johnson, R. Discovery of Cancer Driver Long Noncoding RNAs across 1112 Tumour Genomes: New Candidates and Distinguishing Features. Sci. Rep. 2017, 7, 41544. [Google Scholar] [CrossRef]
- Chiu, H.-S.; Somvanshi, S.; Patel, E.; Chen, T.-W.; Singh, V.P.; Zorman, B.; Patil, S.L.; Pan, Y.; Chatterjee, S.S.; Sood, A.K.; et al. Pan-Cancer Analysis of lncRNA Regulation Supports Their Targeting of Cancer Genes in Each Tumor Context. Cell Rep. 2018, 23, 297–312.e12. [Google Scholar] [CrossRef]
- Mitra, R.; Adams, C.M.; Eischen, C.M. Systematic lncRNA Mapping to Genome-Wide Co-Essential Modules Uncovers Cancer Dependency on Uncharacterized lncRNAs. Elife 2022, 11. [Google Scholar] [CrossRef]
- Yang, Z.; Xu, F.; Wang, H.; Teschendorff, A.E.; Xie, F.; He, Y. Pan-Cancer Characterization of Long Non-Coding RNA and DNA Methylation Mediated Transcriptional Dysregulation. EBioMedicine 2021, 68, 103399. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Arunkumar, G.; Baek, S.; Sturgill, D.; Bui, M.; Dalal, Y. Oncogenic lncRNAs Alter Epigenetic Memory at a Fragile Chromosomal Site in Human Cancer Cells. Sci. Adv. 2022, 8, eabl5621. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Tumor Immunology and Tumor Evolution: Intertwined Histories. Immunity 2020, 52, 55–81. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, T.; Zhou, W.; Li, J.; Li, X.; Wang, Q.; Jin, X.; Yin, J.; Chen, L.; Zhang, Y.; et al. Pan-Cancer Characterization of Immune-Related lncRNAs Identifies Potential Oncogenic Biomarkers. Nat. Commun. 2020, 11, 1000. [Google Scholar] [CrossRef] [PubMed]
- Hur, K.; Kim, S.-H.; Kim, J.-M. Potential Implications of Long Noncoding RNAs in Autoimmune Diseases. Immune Netw. 2019, 19, e4. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, Y.; Yang, M.; Wang, Z.; Wang, Y.; Chaurasia, S.; Wu, Z.; Zhang, M.; Yadav, G.S.; Rathod, S.; et al. LincRNA-Immunity Landscape Analysis Identifies EPIC1 as a Regulator of Tumor Immune Evasion and Immunotherapy Resistance. Sci. Adv. 2021, 7, eabb3555. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Li, Q.; Shao, Y.; Zhang, X.; Zheng, T.; Miao, M.; Qin, L.; Wang, B.; Ye, G.; Xiao, B.; Guo, J. Plasma Long Noncoding RNA Protected by Exosomes as a Potential Stable Biomarker for Gastric Cancer. Tumour Biol. 2015, 36, 2007–2012. [Google Scholar] [CrossRef]
- Badowski, C.; He, B.; Garmire, L.X. Blood-Derived lncRNAs as Biomarkers for Cancer Diagnosis: The Good, the Bad and the Beauty. NPJ Precis. Oncol. 2022, 6, 40. [Google Scholar] [CrossRef]
- Smallegan, M.J.; Rinn, J.L. Linking Long Noncoding RNA to Drug Resistance. Proc. Natl. Acad. Sci. USA 2019, 116, 21963–21965. [Google Scholar] [CrossRef]
- Cui, H.; Kong, H.; Peng, F.; Wang, C.; Zhang, D.; Tian, J.; Zhang, L. Inferences of Individual Drug Response-Related Long Non-Coding RNAs Based on Integrating Multi-Omics Data in Breast Cancer. Mol. Ther. Nucleic Acids 2020, 20, 128–139. [Google Scholar] [CrossRef]
- Liu, H.; Wang, S.; Zhou, S.; Meng, Q.; Ma, X.; Song, X.; Wang, L.; Jiang, W. Drug Resistance-Related Competing Interactions of lncRNA and mRNA across 19 Cancer Types. Mol. Ther. Nucleic Acids 2019, 16, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA Therapeutics - Challenges and Potential Solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]
- Arun, G.; Diermeier, S.D.; Spector, D.L. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol. Med. 2018, 24, 257–277. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





