Submitted:
24 October 2023
Posted:
25 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Patient Samples
RNA and DNA Extraction and Sequencing
RNA-seq Analysis Pipeline
WES Analysis Pipeline
3. Results
3.1. RNA-seq Analysis Identifies Oxidative Phosphorylation Is Associated with LNM
| Gene Set | Size | Leading Edge Number | NES | FDR |
|---|---|---|---|---|
| HALLMARK_ANDROGEN_RESPONSE | 101 | 47 | -2.3952 | <2.2e-16 |
| HALLMARK_MYC_TARGETS_V1 | 200 | 132 | -2.2692 | <2.2e-16 |
| HALLMARK_OXIDATIVE_PHOSPHORYLATION | 199 | 117 | -2.1852 | <2.2e-16 |
| HALLMARK_MTORC1_SIGNALING | 200 | 120 | -2.0443 | <2.2e-16 |
| HALLMARK_MYC_TARGETS_V2 | 58 | 35 | -1.9282 | <2.2e-16 |
| HALLMARK_FATTY_ACID_METABOLISM | 157 | 70 | -1.8465 | 0.000112 |
| HALLMARK_GLYCOLYSIS | 199 | 93 | -1.8362 | 0.000096004 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 113 | 55 | -1.8126 | 0.00033601 |
| HALLMARK_PEROXISOME | 104 | 54 | -1.806 | 0.00037335 |
| HALLMARK_E2F_TARGETS | 200 | 103 | -1.7584 | 0.00039202 |
| HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION | 200 | 73 | 1.6463 | 0.0029856 |
| HALLMARK_MYOGENESIS | 197 | 69 | 1.7325 | 0.002488 |
| HALLMARK_KRAS_SIGNALING_UP | 198 | 79 | 1.7459 | 0.0023325 |
| HALLMARK_TNFA_SIGNALING_VIA_NFKB | 200 | 88 | 1.8804 | 0.00053315 |
| HALLMARK_INFLAMMATORY_RESPONSE | 199 | 86 | 1.9837 | <2.2e-16 |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 97 | 56 | 2.0873 | <2.2e-16 |
| HALLMARK_COMPLEMENT | 200 | 101 | 2.269 | <2.2e-16 |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 83 | 52 | 2.3259 | <2.2e-16 |
| HALLMARK_ALLOGRAFT_REJECTION | 199 | 116 | 2.4589 | <2.2e-16 |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 200 | 123 | 2.608 | <2.2e-16 |
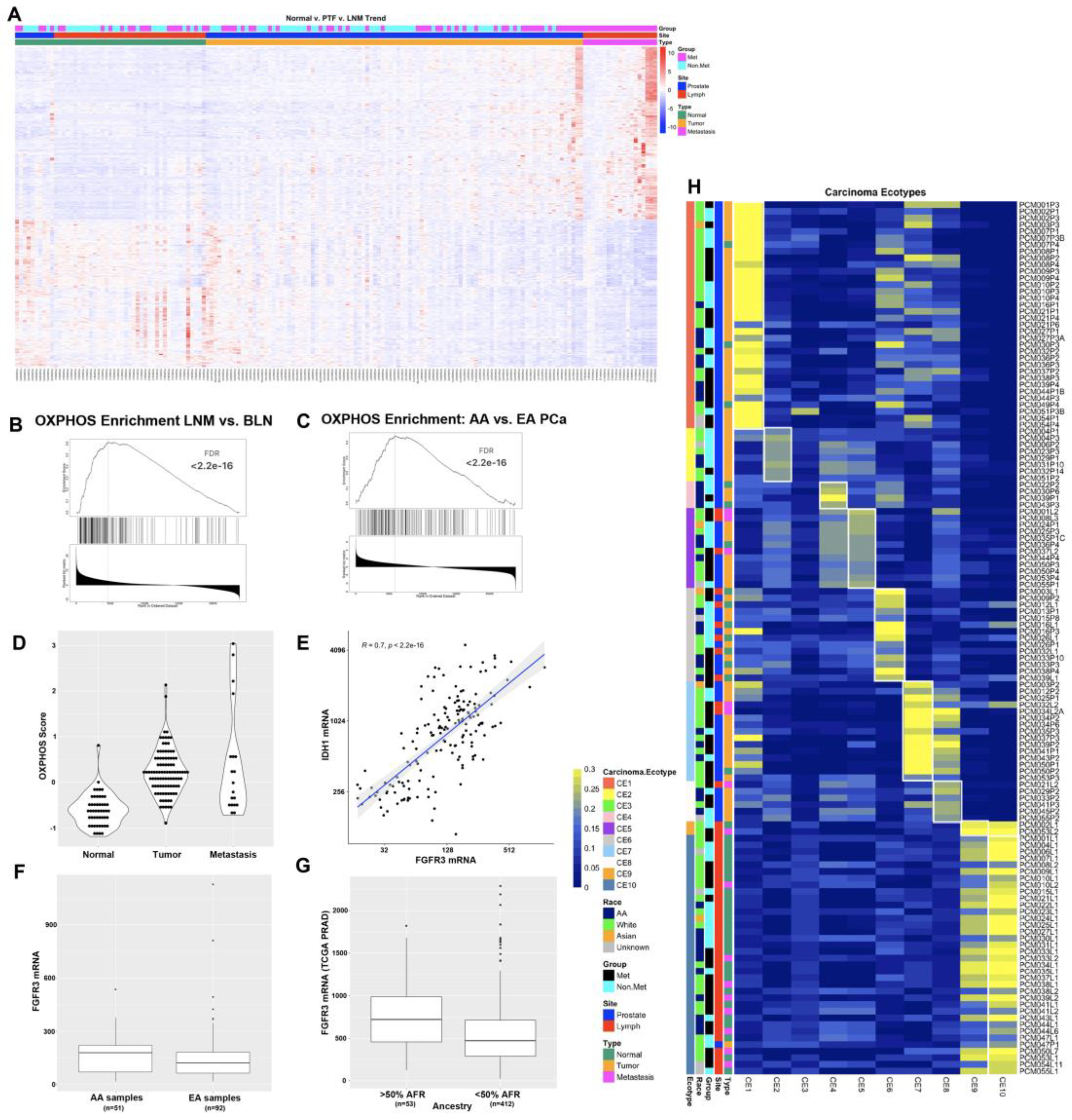
3.2. Oxidative Phosphorylation, Immune Signaling, and Hedgehog Pathways Are Differentially Expressed in AA Patients
3.3. Ecotyper Analysis Identifies Ecotypes with Activated Oxidative Phosphorylation and Androgen Signaling Are Associated with LNM
3.4. WES Analysis Determines SPOP, FLT4, and PCDH15 Mutations Are Enriched in LNM
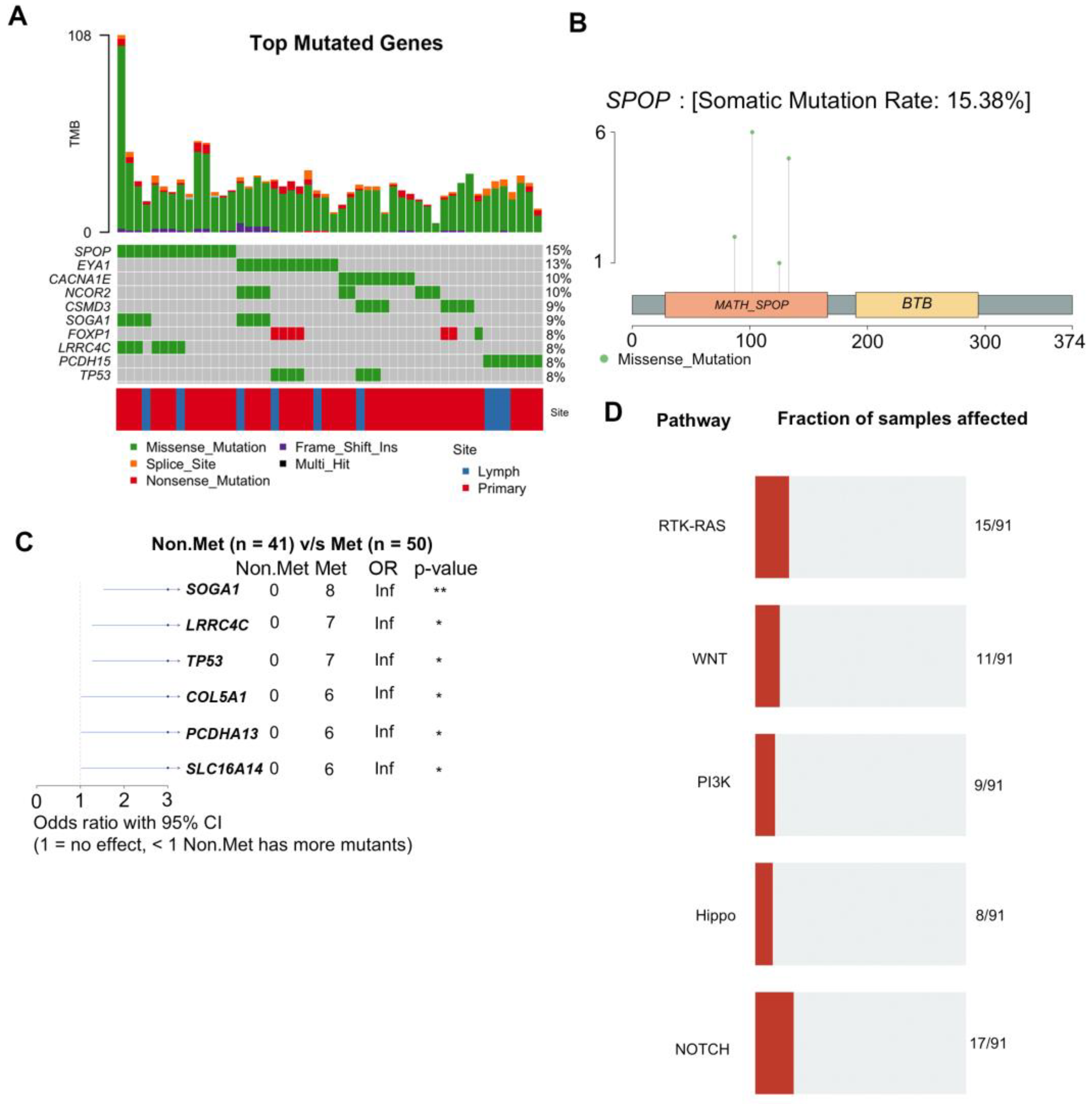
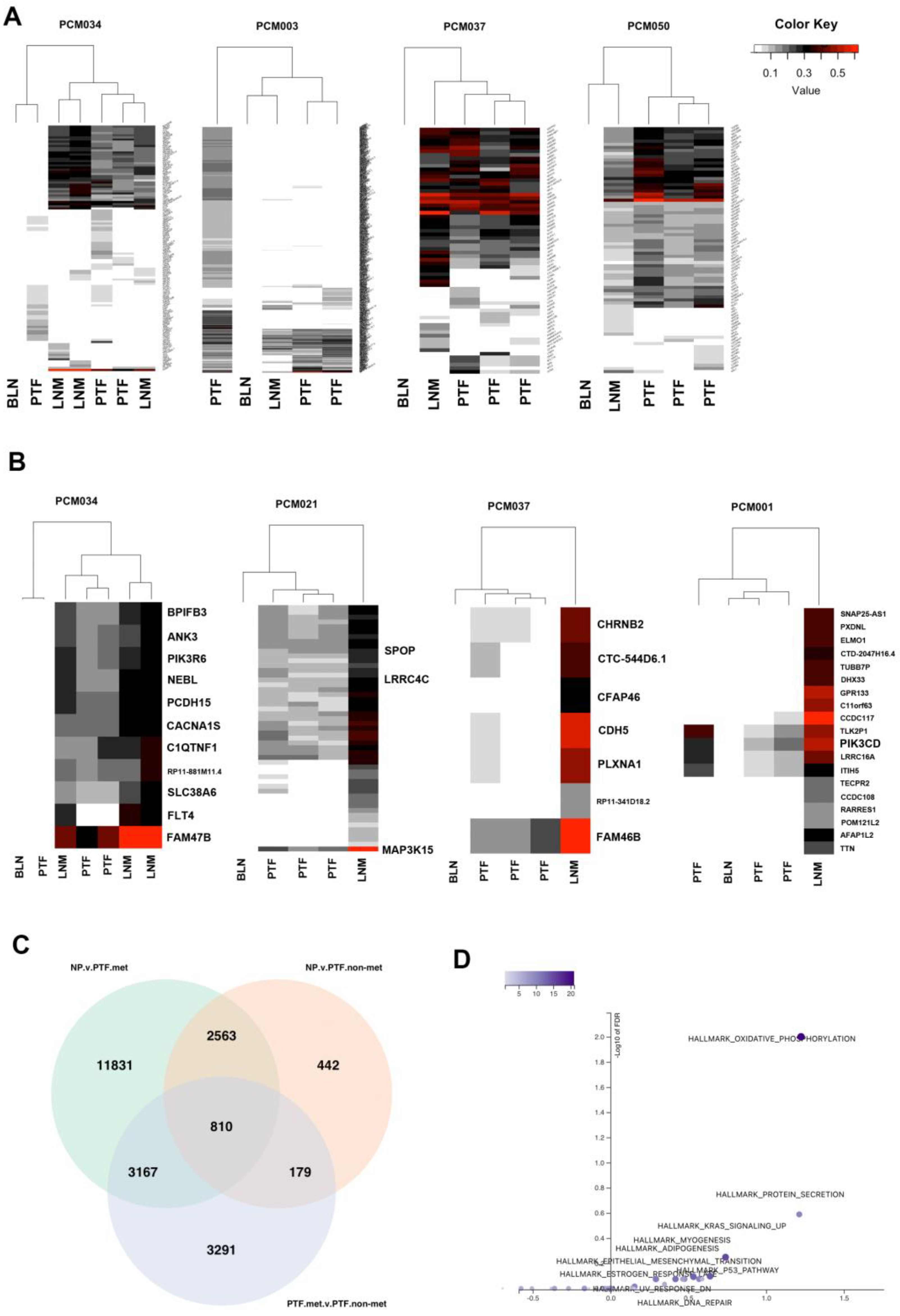
3.5. Integrative DNA/RNA Analysis Determines the Oxidative Phosphorylation Pathway, FLT4 Mutations, and PCDH15 Mutations Are Associated with Metastasis
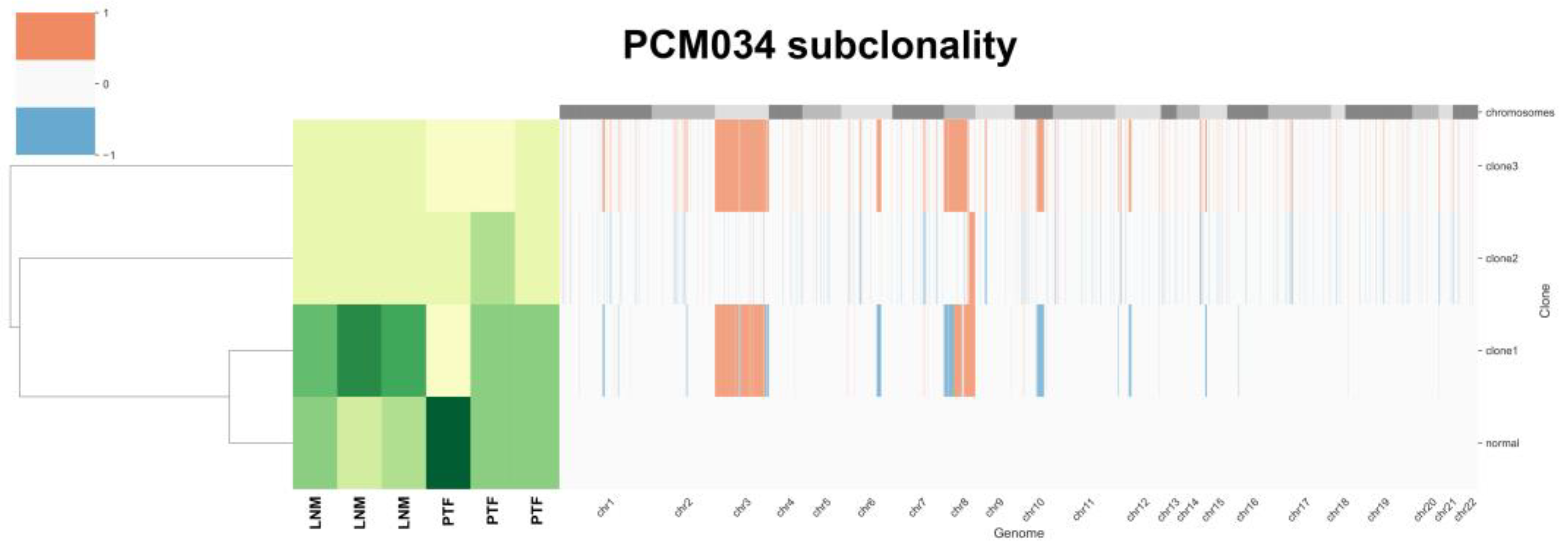
3.6. Tumor Heterogeneity Analysis Identifies Subclones Suggesting Gain of chr8 Prior to chr8p Loss
3.7. Validation in External Datasets Demonstrates EYA1 and CSMD3 Mutations Are Associated with Poor Survival
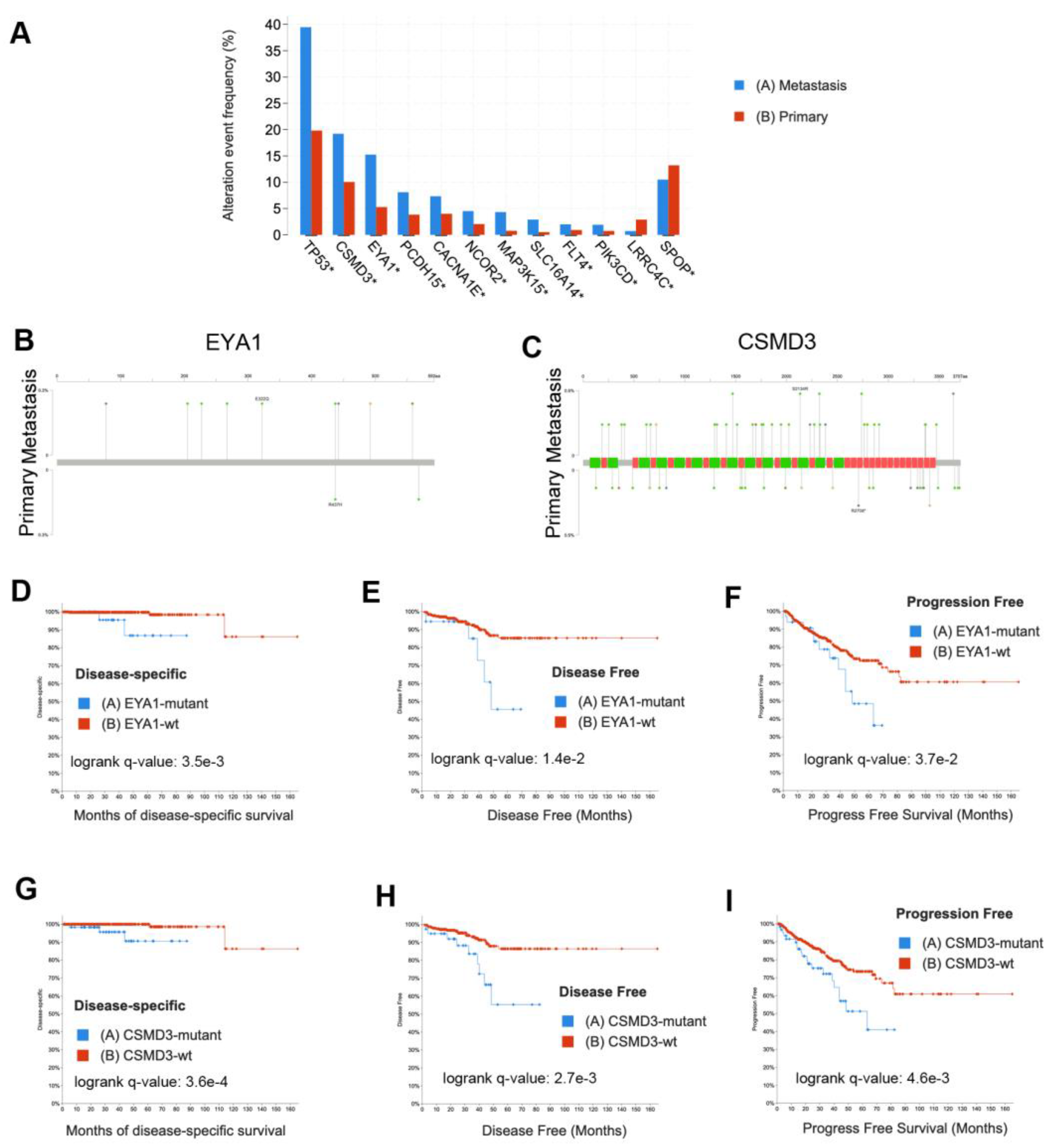
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cancer Genome Atlas Research, N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef]
- Abida, W.; Cyrta, J.; Heller, G.; Prandi, D.; Armenia, J.; Coleman, I.; Cieslik, M.; Benelli, M.; Robinson, D.; Van Allen, E.M.; et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019, 116, 11428–11436. [Google Scholar] [CrossRef] [PubMed]
- Grasso, C.S.; Wu, Y.M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef]
- He, M.X.; Cuoco, M.S.; Crowdis, J.; Bosma-Moody, A.; Zhang, Z.; Bi, K.; Kanodia, A.; Su, M.J.; Ku, S.Y.; Garcia, M.M.; et al. Transcriptional mediators of treatment resistance in lethal prostate cancer. Nat Med 2021, 27, 426–433. [Google Scholar] [CrossRef]
- Quigley, D.A.; Dang, H.X.; Zhao, S.G.; Lloyd, P.; Aggarwal, R.; Alumkal, J.J.; Foye, A.; Kothari, V.; Perry, M.D.; Bailey, A.M.; et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 2018, 174, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Seed, G.; Bertan, C.; Rescigno, P.; Dolling, D.; Figueiredo, I.; Miranda, S.; Nava Rodrigues, D.; Gurel, B.; Clarke, M.; et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. J Clin Invest 2020, 130, 1743–1751. [Google Scholar] [CrossRef]
- Russo, G.I.; Bonacci, P.; Bivona, D.; Privitera, G.F.; Broggi, G.; Caltabiano, R.; Vella, J.; Lo Giudice, A.; Asmundo, M.G.; Cimino, S.; et al. Genomic Landscape Alterations in Primary Tumor and Matched Lymph Node Metastasis in Hormone-Naive Prostate Cancer Patients. Cancers (Basel) 2022, 14. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef]
- Cheng, L.; Zincke, H.; Blute, M.L.; Bergstralh, E.J.; Scherer, B.; Bostwick, D.G. Risk of prostate carcinoma death in patients with lymph node metastasis. Cancer 2001, 91, 66–73. [Google Scholar] [CrossRef]
- Powell, I.J.; Dyson, G.; Land, S.; Ruterbusch, J.; Bock, C.H.; Lenk, S.; Herawi, M.; Everson, R.; Giroux, C.N.; Schwartz, A.G.; et al. Genes associated with prostate cancer are differentially expressed in African American and European American men. Cancer Epidemiol Biomarkers Prev 2013, 22, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.W.; Mosquera, J.M.; Garofalo, A.; Oh, C.; Baco, M.; Amin-Mansour, A.; Rabasha, B.; Bahl, S.; Mullane, S.A.; Robinson, B.D.; et al. Exome Sequencing of African-American Prostate Cancer Reveals Loss-of-Function ERF Mutations. Cancer discovery 2017, 7, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Jaratlerdsiri, W.; Chan, E.K.F.; Gong, T.; Petersen, D.C.; Kalsbeek, A.M.F.; Venter, P.A.; Stricker, P.D.; Bornman, M.R.; Hayes, V.M. Whole Genome Sequencing Reveals Elevated Tumor Mutational Burden and Initiating Driver Mutations in African Men with Treatment-Naive, High-Risk Prostate Cancer. Cancer Res 2018. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.S.; Stockert, J.A.; Hackert, V.; Yadav, K.K.; Tewari, A.K. Intratumor heterogeneity in prostate cancer. Urol Oncol 2018, 36, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Gleason, D.F. Classification of prostatic carcinomas. Cancer Chemother Rep 1966, 50, 125–128. [Google Scholar] [PubMed]
- Gleason, D.F. Histologic grading of prostate cancer: a perspective. Hum Pathol 1992, 23, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Gleason, D.F.; Mellinger, G.T. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J Urol 1974, 111, 58–64. [Google Scholar] [CrossRef]
- Aihara, M.; Wheeler, T.M.; Ohori, M.; Scardino, P.T. Heterogeneity of prostate cancer in radical prostatectomy specimens. Urology 1994, 43, 60–66. [Google Scholar] [CrossRef]
- Cheng, L.; Song, S.Y.; Pretlow, T.G.; Abdul-Karim, F.W.; Kung, H.J.; Dawson, D.V.; Park, W.S.; Moon, Y.W.; Tsai, M.L.; Linehan, W.M.; et al. Evidence of independent origin of multiple tumors from patients with prostate cancer. J Natl Cancer Inst 1998, 90, 233–237. [Google Scholar] [CrossRef]
- Mehra, R.; Han, B.; Tomlins, S.A.; Wang, L.; Menon, A.; Wasco, M.J.; Shen, R.; Montie, J.E.; Chinnaiyan, A.M.; Shah, R.B. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res 2007, 67, 7991–7995. [Google Scholar] [CrossRef]
- Boutros, P.C.; Fraser, M.; Harding, N.J.; de Borja, R.; Trudel, D.; Lalonde, E.; Meng, A.; Hennings-Yeomans, P.H.; McPherson, A.; Sabelnykova, V.Y.; et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet 2015, 47, 736–745. [Google Scholar] [CrossRef]
- Lovf, M.; Zhao, S.; Axcrona, U.; Johannessen, B.; Bakken, A.C.; Carm, K.T.; Hoff, A.M.; Myklebost, O.; Meza-Zepeda, L.A.; Lie, A.K.; et al. Multifocal Primary Prostate Cancer Exhibits High Degree of Genomic Heterogeneity. Eur Urol 2019, 75, 498–505. [Google Scholar] [CrossRef]
- Lindberg, J.; Klevebring, D.; Liu, W.; Neiman, M.; Xu, J.; Wiklund, P.; Wiklund, F.; Mills, I.G.; Egevad, L.; Grönberg, H. Exome Sequencing of Prostate Cancer Supports the Hypothesis of Independent Tumour Origins. European Urology 2013, 63, 347–353. [Google Scholar] [CrossRef]
- Cooper, C.S.; Eeles, R.; Wedge, D.C.; Van Loo, P.; Gundem, G.; Alexandrov, L.B.; Kremeyer, B.; Butler, A.; Lynch, A.G.; Camacho, N.; et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nature Genetics 2015, 47, 367–372. [Google Scholar] [CrossRef]
- Wei, L.; Wang, J.; Lampert, E.; Schlanger, S.; DePriest, A.D.; Hu, Q.; Gomez, E.C.; Murakam, M.; Glenn, S.T.; Conroy, J.; et al. Intratumoral and Intertumoral Genomic Heterogeneity of Multifocal Localized Prostate Cancer Impacts Molecular Classifications and Genomic Prognosticators. Eur Urol 2017, 71, 183–192. [Google Scholar] [CrossRef]
- Alemozaffar, M.; Akintayo, A.A.; Abiodun-Ojo, O.A.; Patil, D.; Saeed, F.; Huang, Y.; Osunkoya, A.O.; Goodman, M.M.; Sanda, M.; Schuster, D.M. [(18)F]Fluciclovine Positron Emission Tomography/Computerized Tomography for Preoperative Staging in Patients with Intermediate to High Risk Primary Prostate Cancer. J Urol 2020, 204, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Kreuger, F. TrimGalore, https://github.com, 2020.
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014, 15, 550. [Google Scholar] [CrossRef]
- Wang, J.; Vasaikar, S.; Shi, Z.; Greer, M.; Zhang, B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res 2017, 45, W130–W137. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Kuehn, H.; Gould, J.; Tamayo, P.; Mesirov, J.P. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007, 23, 3251–3253. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; O'Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra; O'Reilly Media: 2020.
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Zaccaria, S.; Raphael, B.J. Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data. Nat Commun 2020, 11, 4301. [Google Scholar] [CrossRef]
- Powell, I.J. Epidemiology and pathophysiology of prostate cancer in African-American men. J Urol 2007, 177, 444–449. [Google Scholar] [CrossRef]
- Frattini, V.; Pagnotta, S.M.; Tala; Fan, J. J.; Russo, M.V.; Lee, S.B.; Garofano, L.; Zhang, J.; Shi, P.; Lewis, G.; et al. A metabolic function of FGFR3-TACC3 gene fusions in cancer. Nature 2018, 553, 222–227. [Google Scholar] [CrossRef]
- Murari, A.; Goparaju, N.S.V.; Rhooms, S.K.; Hossain, K.F.B.; Liang, F.G.; Garcia, C.J.; Osei, C.; Liu, T.; Li, H.; Kitsis, R.N.; et al. IDH2-mediated regulation of the biogenesis of the oxidative phosphorylation system. Sci Adv 2022, 8, eabl8716. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Luca, B.A.; Steen, C.B.; Matusiak, M.; Azizi, A.; Varma, S.; Zhu, C.; Przybyl, J.; Espin-Perez, A.; Diehn, M.; Alizadeh, A.A.; et al. Atlas of clinically distinct cell states and ecosystems across human solid tumors. Cell 2021, 184, 5482–5496. [Google Scholar] [CrossRef] [PubMed]
- Abida, W.; Armenia, J.; Gopalan, A.; Brennan, R.; Walsh, M.; Barron, D.; Danila, D.; Rathkopf, D.; Morris, M.; Slovin, S.; et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis Oncol 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Armenia, J.; Wankowicz, S.A.M.; Liu, D.; Gao, J.; Kundra, R.; Reznik, E.; Chatila, W.K.; Chakravarty, D.; Han, G.C.; Coleman, I.; et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018, 50, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, G.; Nandakumar, S.; Hirani, R.; Nguyen, B.; Stopsack, K.H.; Kreitzer, C.; Rajanala, S.H.; Ghale, R.; Mazzu, Y.Z.; Pillarsetty, N.V.K.; et al. The Impact of PIK3R1 Mutations and Insulin-PI3K-Glycolytic Pathway Regulation in Prostate Cancer. Clin Cancer Res 2022, 28, 3603–3617. [Google Scholar] [CrossRef] [PubMed]
- Fraser, M.; Sabelnykova, V.Y.; Yamaguchi, T.N.; Heisler, L.E.; Livingstone, J.; Huang, V.; Shiah, Y.J.; Yousif, F.; Lin, X.; Masella, A.P.; et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 2017, 541, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Gerhauser, C.; Favero, F.; Risch, T.; Simon, R.; Feuerbach, L.; Assenov, Y.; Heckmann, D.; Sidiropoulos, N.; Waszak, S.M.; Hubschmann, D.; et al. Molecular Evolution of Early-Onset Prostate Cancer Identifies Molecular Risk Markers and Clinical Trajectories. Cancer Cell 2018, 34, 996–1011. [Google Scholar] [CrossRef] [PubMed]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, B.; Mota, J.M.; Nandakumar, S.; Stopsack, K.H.; Weg, E.; Rathkopf, D.; Morris, M.J.; Scher, H.I.; Kantoff, P.W.; Gopalan, A.; et al. Pan-cancer Analysis of CDK12 Alterations Identifies a Subset of Prostate Cancers with Distinct Genomic and Clinical Characteristics. Eur Urol 2020, 78, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Stopsack, K.H.; Nandakumar, S.; Arora, K.; Nguyen, B.; Vasselman, S.E.; Nweji, B.; McBride, S.M.; Morris, M.J.; Rathkopf, D.E.; Slovin, S.F.; et al. Differences in Prostate Cancer Genomes by Self-reported Race: Contributions of Genetic Ancestry, Modifiable Cancer Risk Factors, and Clinical Factors. Clin Cancer Res 2022, 28, 318–326. [Google Scholar] [CrossRef]
- Stopsack, K.H.; Nandakumar, S.; Wibmer, A.G.; Haywood, S.; Weg, E.S.; Barnett, E.S.; Kim, C.J.; Carbone, E.A.; Vasselman, S.E.; Nguyen, B.; et al. Oncogenic Genomic Alterations, Clinical Phenotypes, and Outcomes in Metastatic Castration-Sensitive Prostate Cancer. Clin Cancer Res 2020, 26, 3230–3238. [Google Scholar] [CrossRef]
- Liu, W.; Laitinen, S.; Khan, S.; Vihinen, M.; Kowalski, J.; Yu, G.; Chen, L.; Ewing, C.M.; Eisenberger, M.A.; Carducci, M.A.; et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nature Medicine 2009, 15, 559–565. [Google Scholar] [CrossRef]
- Kumar, A.; Coleman, I.; Morrissey, C.; Zhang, X.; True, L.D.; Gulati, R.; Etzioni, R.; Bolouri, H.; Montgomery, B.; White, T.; et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 2016, 22, 369–378. [Google Scholar] [CrossRef]
- Gundem, G.; Van Loo, P.; Kremeyer, B.; Alexandrov, L.B.; Tubio, J.M.C.; Papaemmanuil, E.; Brewer, D.S.; Kallio, H.M.L.; Högnäs, G.; Annala, M.; et al. The evolutionary history of lethal metastatic prostate cancer. Nature 2015, 520, 353–357. [Google Scholar] [CrossRef]
- An, J.; Wang, C.; Deng, Y.; Yu, L.; Huang, H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep 2014, 6, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, C.E.; Baca, S.C.; Lawrence, M.S.; Demichelis, F.; Blattner, M.; Theurillat, J.P.; White, T.A.; Stojanov, P.; Van Allen, E.; Stransky, N.; et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 2012, 44, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Niu, Q.; Ivanov, A.A.; Tsang, Y.H.; Tang, C.; Shu, C.; Li, Q.; Qian, K.; Wahafu, A.; Doyle, S.P.; et al. Systematic discovery of mutation-directed neo-protein-protein interactions in cancer. Cell 2022, 185, 1974–1985. [Google Scholar] [CrossRef]
- Das, R.; Sjostrom, M.; Shrestha, R.; Yogodzinski, C.; Egusa, E.A.; Chesner, L.N.; Chen, W.S.; Chou, J.; Dang, D.K.; Swinderman, J.T.; et al. An integrated functional and clinical genomics approach reveals genes driving aggressive metastatic prostate cancer. Nat Commun 2021, 12, 4601. [Google Scholar] [CrossRef] [PubMed]
- Long, M.D.; Jacobi, J.J.; Singh, P.K.; Llimos, G.; Wani, S.A.; Rowsam, A.M.; Rosario, S.R.; Hoogstraat, M.; Linder, S.; Kirk, J.; et al. Reduced NCOR2 expression accelerates androgen deprivation therapy failure in prostate cancer. Cell Rep 2021, 37, 110109. [Google Scholar] [CrossRef]
- Li, X.; Oghi, K.A.; Zhang, J.; Krones, A.; Bush, K.T.; Glass, C.K.; Nigam, S.K.; Aggarwal, A.K.; Maas, R.; Rose, D.W.; et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 2003, 426, 247–254. [Google Scholar] [CrossRef]
- Kong, D.; Li, A.; Liu, Y.; Cui, Q.; Wang, K.; Zhang, D.; Tang, J.; Du, Y.; Liu, Z.; Wu, G.; et al. SIX1 Activates STAT3 Signaling to Promote the Proliferation of Thyroid Carcinoma via EYA1. Front Oncol 2019, 9, 1450. [Google Scholar] [CrossRef]
- Zhao, X.; Lei, Y.; Li, G.; Cheng, Y.; Yang, H.; Xie, L.; Long, H.; Jiang, R. Integrative analysis of cancer driver genes in prostate adenocarcinoma. Mol Med Rep 2019, 19, 2707–2715. [Google Scholar] [CrossRef]
- Beuten, J.; Gelfond, J.A.; Martinez-Fierro, M.L.; Weldon, K.S.; Crandall, A.C.; Rojas-Martinez, A.; Thompson, I.M.; Leach, R.J. Association of chromosome 8q variants with prostate cancer risk in Caucasian and Hispanic men. Carcinogenesis 2009, 30, 1372–1379. [Google Scholar] [CrossRef]
- Liu, P.; Morrison, C.; Wang, L.; Xiong, D.; Vedell, P.; Cui, P.; Hua, X.; Ding, F.; Lu, Y.; James, M.; et al. Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis 2012, 33, 1270–1276. [Google Scholar] [CrossRef]
- Lu, N.; Liu, J.; Xu, M.; Liang, J.; Wang, Y.; Wu, Z.; Xing, Y.; Diao, F. CSMD3 is Associated with Tumor Mutation Burden and Immune Infiltration in Ovarian Cancer Patients. Int J Gen Med 2021, 14, 7647–7657. [Google Scholar] [CrossRef] [PubMed]
- Stearns, M.E.; Wang, M.; Hu, Y.; Kim, G.; Garcia, F.U. Expression of a flt-4 (VEGFR3) splicing variant in primary human prostate tumors. VEGF D and flt-4t(Delta773-1081) overexpression is diagnostic for sentinel lymph node metastasis. Lab Invest 2004, 84, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.Y.; Haack, H.; Crowley, D.; Barry, M.; Bronson, R.T.; Hynes, R.O. Tumor-secreted vascular endothelial growth factor-C is necessary for prostate cancer lymphangiogenesis, but lymphangiogenesis is unnecessary for lymph node metastasis. Cancer Res 2005, 65, 9789–9798. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Moss, T.J.; Laura Rubin, M.; Ning, J.; Eterovic, K.; Yu, H.; Jia, R.; Fan, X.; Tetzlaff, M.T.; Esmaeli, B. Whole-exome sequencing for ocular adnexal sebaceous carcinoma suggests PCDH15 as a novel mutation associated with metastasis. Mod Pathol 2020, 33, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Y.; Cullen, C.L.; Ricci, R.; Summers, B.S.; Rehman, S.; Ahmed, Z.M.; Foster, A.Y.; Emery, B.; Gasperini, R.; Young, K.M. Protocadherin 15 suppresses oligodendrocyte progenitor cell proliferation and promotes motility through distinct signalling pathways. Commun Biol 2022, 5, 511. [Google Scholar] [CrossRef]
- Taksler, G.B.; Keating, N.L.; Cutler, D.M. Explaining racial differences in prostate cancer mortality. Cancer 2012, 118, 4280–4289. [Google Scholar] [CrossRef]
- Mahal, B.A.; Aizer, A.A.; Ziehr, D.R.; Hyatt, A.S.; Choueiri, T.K.; Hu, J.C.; Hoffman, K.E.; Sweeney, C.J.; Beard, C.J.; D'Amico, A.V.; et al. Racial Disparities in Prostate Cancer-Specific Mortality in Men With Low-Risk Prostate Cancer. Clin Genitourin Cancer 2014. [Google Scholar] [CrossRef]
| Samples / Patients | RNAseq | WES | Both |
|---|---|---|---|
| Primary Tumor Foci | 97 | 88 | 80 |
| Metastatic Lymph Nodes (LNM) | 19 | 19 | 19 |
| Normal Prostate tissue (NP) | 10 | 0 | 0 |
| Normal Lymph Nodes (LN) | 39 | 37 | 33 |
| Total Samples | 165 | 137 | 132 |
| Metastatic Patients | 18 | 15 | 15 |
| Non-Metastatic Patients | 25 | 20 | 20 |
| White Patients | 23 | 20 | 20 |
| African-American Patients | 14 | 13 | 13 |
| Total Patients | 43 | 35 | 35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





