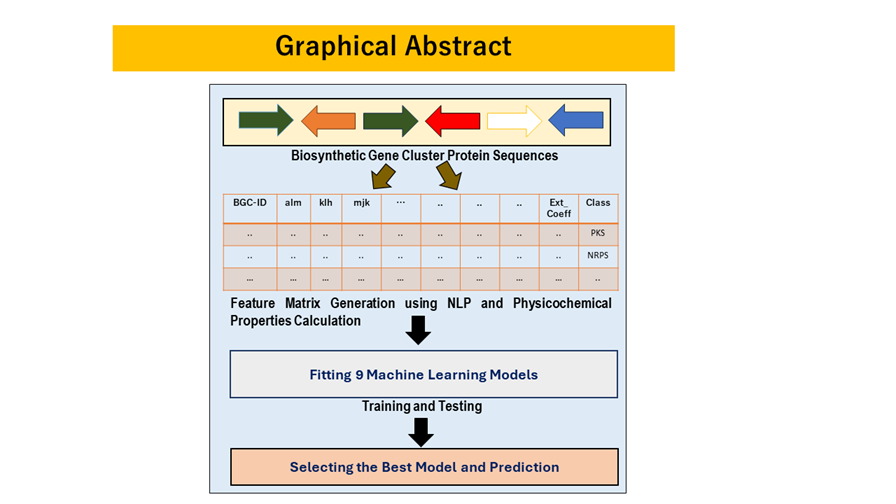
Biosynthetic gene clusters are specific genomic regions in microorganisms, like bacteria and fungi, responsible for producing bioactive compounds. Identifying these clusters is complex due to their diverse nature. This research presents a comprehensive approach to effective BGC identification. The study focuses on five classes of Natural Products: PKS , NRPS, RiPP, Terpenes, and Hybrid PKS-NRPS. Data was gathered from the MiBIG database in GBK format. Protein sequences from each file were extracted, and sequences under the same BGC ID were combined. Physicochemical properties were calculated, and sequence embeddings were generated using NLP techniques like CountVec, TFIDF, and Word2Vec specific to each NP class. An integrated feature matrix was created by merging physicochemical properties and generated embeddings. This matrix was used for training and testing of nine ML models such SVM, RF and many more. The study explored data balancing techniques with and without SMOTE and employed Grid Search for parameter optimization. This led to six datasets and 54 models. The LR model, using TFIDF with SMOTE, emerged as the most effective, achieving an accuracy of 0.96, AUC of 0.9912, and other strong metrics. This method enhances BGC identification for drug development, offering a broader understanding of their applications in medicine and biotechnology.