Submitted:
13 November 2023
Posted:
13 November 2023
You are already at the latest version
Abstract
Keywords:
Main Concept
- Rethinking Oncogene Addiction: As our grasp of cellular evolution and tumor heterogeneity deepens, the necessity to re-evaluate oncogene addiction becomes evident. The phenomena of oncogene addiction might be better understood as a cell fate outcome, occurring when cells are detached from the evolutionary process when the target oncogene is inhibited.
- Deepening Insights into Oncogenes and Tumor Suppressors: Reclassifying genes according to their heterogeneity levels has promise. Treatment strategies resulting from this approach could focus on amplifying the homogeneity of critical genes and specifying precise levels of homogeneity, which in turn is likely to enhance the predictability and efficacy of therapeutic outcomes.
- Exploring Aging's Role in Cancer Development: A better understanding of aging's role in cancer will allow us to probe the links between aging and cancer onset. A focus here should be distinguishing between normal aging decline and tumor formation. Such insights will contribute to innovative strategies to enhance broader human health.
Future PerspectivesBottom of FormFutu
- 1)
- Rethinking oncogene addiction.
- 2)
- Deepening Insights into Oncogenes and Tumor Suppressors.
- 3)
- Exploring Aging's Role in Cancer Development.
Final Remark
Contributions
Acknowledgment
Footnote
References
- Dagogo-Jack I, Shaw AT. 2018. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 15(2):81–94. [CrossRef]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. 2013. Cancer Genome Landscapes. Science. 339(6127):1546–58. [CrossRef]
- Blanke CD, Demetri GD, Von Mehren M, Heinrich MC, Eisenberg B, et al. 2008. Long-Term Results From a Randomized Phase II Trial of Standard- Versus Higher-Dose Imatinib Mesylate for Patients With Unresectable or Metastatic Gastrointestinal Stromal Tumors Expressing KIT. JCO. 26(4):620–25. [CrossRef]
- Juric D, Castel P, Griffith M, Griffith OL, Won HH, et al. 2015. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature. 518(7538):240–44. [CrossRef]
- Kwak EL, Ahronian LG, Siravegna G, Mussolin B, Borger DR, et al. 2015. Molecular Heterogeneity and Receptor Coamplification Drive Resistance to Targeted Therapy in MET -Amplified Esophagogastric Cancer. Cancer Discovery. 5(12):1271–81. [CrossRef]
- Hanahan D, Weinberg RA. 2011. Hallmarks of Cancer: The Next Generation. Cell. 144(5):646–74. [CrossRef]
- Hanahan D, Weinberg RA. 2000. The Hallmarks of Cancer. Cell. 100(1):57–70.
- Sun X, Yu Q. 2015. Intra-tumor heterogeneity of cancer cells and its implications for cancer treatment. Acta Pharmacol Sin. 36(10):1219–27. [CrossRef]
- Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, et al. 2016. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med. 22(1):105–13. [CrossRef]
- Bakhoum SF, Landau DA. 2017. Chromosomal Instability as a Driver of Tumor Heterogeneity and Evolution. Cold Spring Harb Perspect Med. 7(6):a029611. [CrossRef]
- Rao CV, Asch AS, Yamada HY. 2017. Frequently mutated genes/pathways and genomic instability as prevention targets in liver cancer. CARCIN. 38(1):2–11. [CrossRef]
- Li M, Zhang Z, Li L, Wang X. 2020. An algorithm to quantify intratumor heterogeneity based on alterations of gene expression profiles. Commun Biol. 3(1):1–19. [CrossRef]
- Geiger TR, Peeper DS. 2009. Metastasis mechanisms. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1796(2):293–308.
- Ha N-H, Faraji F, Hunter KW. 2013. Mechanisms of Metastasis. In Cancer Targeted Drug Delivery: An Elusive Dream, ed YH Bae, RJ Mrsny, K Park, pp. 435–58. New York, NY: Springer.
- Yachida S, Jones S, Bozic I, Antal T, Leary R, et al. 2010. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 467(7319):1114–17. [CrossRef]
- Yokota J. 2000. Tumor progression and metastasis. Carcinogenesis. 21(3):497–503.
- Lee W-C, Kopetz S, Wistuba II, Zhang J. 2017. Metastasis of cancer: when and how? Annals of Oncology. 28(9):2045–47.
- Patel SA, Rodrigues P, Wesolowski L, Vanharanta S. 2021. Genomic control of metastasis. Br J Cancer. 124(1):3–12. [CrossRef]
- Reiter JG, Makohon-Moore AP, Gerold JM, Heyde A, Attiyeh MA, et al. 2018. Minimal functional driver gene heterogeneity among untreated metastases. Science. 361(6406):1033–37. [CrossRef]
- Sherwood J, Dearden S, Ratcliffe M, Walker J. 2015. Mutation status concordance between primary lesions and metastatic sites of advanced non-small-cell lung cancer and the impact of mutation testing methodologies: a literature review. Journal of Experimental & Clinical Cancer Research. 34(1):92. [CrossRef]
- Xie T, Cho YB, Wang K, Huang D, Hong HK, et al. 2014. Patterns of somatic alterations between matched primary and metastatic colorectal tumors characterized by whole-genome sequencing. Genomics. 104(4):234–41. [CrossRef]
- Townson JL, Chambers AF. 2006. Dormancy of Solitary Metastatic Cells. Cell Cycle. 5(16):1744–50. [CrossRef]
- Giancotti FG. 2013. Mechanisms Governing Metastatic Dormancy and Reactivation. Cell. 155(4):750–64. [CrossRef]
- Park S-Y, Nam J-S. 2020. The force awakens: metastatic dormant cancer cells. Exp Mol Med. 52(4):569–81. [CrossRef]
- Gomis RR, Gawrzak S. 2017. Tumor cell dormancy. Molecular Oncology. 11(1):62–78.
- Summers MA, McDonald MM, Croucher PI. 2020. Cancer Cell Dormancy in Metastasis. Cold Spring Harb Perspect Med. 10(4):a037556. [CrossRef]
- Yadav AS, Pandey PR, Butti R, Radharani NNV, Roy S, et al. 2018. The Biology and Therapeutic Implications of Tumor Dormancy and Reactivation. Frontiers in Oncology. 8:. [CrossRef]
- Pradhan S, Sperduto JL, Farino CJ, Slater JH. 2018. Engineered In Vitro Models of Tumor Dormancy and Reactivation. J Biol Eng. 12(1):37. [CrossRef]
- Singh DK, Patel VG, Oh WK, Aguirre-Ghiso JA. 2021. Prostate Cancer Dormancy and Reactivation in Bone Marrow. Journal of Clinical Medicine. 10(12):2648. [CrossRef]
- Chen Q, Sun L, Chen ZJ. 2016. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat Immunol. 17(10):1142–49. [CrossRef]
- Janssen LME, Ramsay EE, Logsdon CD, Overwijk WW. 2017. The immune system in cancer metastasis: friend or foe? J Immunother Cancer. 5:79.
- Whiteside TL. 2010. Immune responses to malignancies. J Allergy Clin Immunol. 125(2 0 2):S272–83.
- Caspi RR. 2008. Immunotherapy of autoimmunity and cancer: the penalty for success. Nat Rev Immunol. 8(12):970–76. [CrossRef]
- fuji K, Hiramatsu K, Nosaka T, Naito T, Takahashi K, et al. 2021. Pembrolizumab-induced autoimmune side effects of colon and pancreas in a patient with lung cancer. Clin J Gastroenterol. 14(6):1692–99. [CrossRef]
- Amos SM, Duong CPM, Westwood JA, Ritchie DS, Junghans RP, et al. 2011. Autoimmunity associated with immunotherapy of cancer. Blood. 118(3):499–509. [CrossRef]
- Yang S, Yu K, Palmer N, Fox K, Kou SC, Kohane IS. 2020. Autoimmune Effects of Lung Cancer Immunotherapy Revealed by Data-Driven Analysis on a Nationwide Cohort. Clin Pharma and Therapeutics. 107(2):388–96. [CrossRef]
- Cheng F, Loscalzo J. 2017. Autoimmune Cardiotoxicity of Cancer Immunotherapy. Trends in Immunology. 38(2):77–78. [CrossRef]
- Weinstein IB, Joe A. 2008. Oncogene Addiction. Cancer Research. 68(9):3077–80.
- Weinstein IB, Joe AK. 2006. Mechanisms of Disease: oncogene addiction—a rationale for molecular targeting in cancer therapy. Nat Rev Clin Oncol. 3(8):448–57. [CrossRef]
- McCormick F. 2011. Cancer therapy based on oncogene addiction. Journal of Surgical Oncology. 103(6):464–67. [CrossRef]
- Lee H-J, Zhuang G, Cao Y, Du P, Kim H-J, Settleman J. 2014. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 26(2):207–21. [CrossRef]
- Pagliarini R, Shao W, Sellers WR. 2015. Oncogene addiction: pathways of therapeutic response, resistance, and road maps toward a cure. EMBO Reports. 16(3):280–96. [CrossRef]
- Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, et al. 2013. Small molecule inhibition of the KRAS–PDEδ interaction impairs oncogenic KRAS signalling. Nature. 497(7451):638–42. [CrossRef]
- Kopp F, Wagner E, Roidl A. 2013. The proto-oncogene KRAS is targeted by miR-200c. Oncotarget. 5(1):185–95. [CrossRef]
- Kim Y, Yoon JW, Xiao X, Dean NM, Monia BP, Marcusson EG. 2007. Selective down-regulation of glioma-associated oncogene 2 inhibits the proliferation of hepatocellular carcinoma cells. Cancer Res. 67(8):3583–93.
- Felsher DW, Bishop JM. 1999. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 4(2):199–207. [CrossRef]
- Schmidt AV, Monga SP, Prochownik EV, Goetzman ES. 2023. A Novel Transgenic Mouse Model Implicates Sirt2 as a Promoter of Hepatocellular Carcinoma. Int J Mol Sci. 24(16):12618. [CrossRef]
- Pollack R, Wolman S, Vogel A. 1970. Reversion of Virus-transformed Cell Lines: Hyperploidy accompanies Retention of Viral Genes. Nature. 228(5275):938–938. [CrossRef]
- Dhanyamraju PK, Schell TD, Amin S, Robertson GP. 2022. Drug-Tolerant Persister Cells in Cancer Therapy Resistance. Cancer Res. 82(14):2503–14. [CrossRef]
- De Conti G, Dias MH, Bernards R. 2021. Fighting Drug Resistance through the Targeting of Drug-Tolerant Persister Cells. Cancers. 13(5):1118.
- Álvarez-Varela A, Novellasdemunt L, Barriga FM, Hernando-Momblona X, Cañellas-Socias A, et al. 2022. Mex3a marks drug-tolerant persister colorectal cancer cells that mediate relapse after chemotherapy. Nat Cancer. 3(9):1052–70. [CrossRef]
- Kumar U, Castellanos-Uribe M, May ST, Yagüe E. 2023. Adaptive resistance is not responsible for long-term drug resistance in a cellular model of triple negative breast cancer. Gene. 850:146930. [CrossRef]
- Sánchez-Romero MA, Casadesús J. 2014. Contribution of phenotypic heterogeneity to adaptive antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 111(1):355–60. [CrossRef]
- Lim Z-F, Ma PC. 2019. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. Journal of Hematology & Oncology. 12(1):134. [CrossRef]
- Ayob AZ, Ramasamy TS. 2018. Cancer stem cells as key drivers of tumour progression. Journal of Biomedical Science. 25(1):20. [CrossRef]
- Klein CA, Blankenstein TJF, Schmidt-Kittler O, Petronio M, Polzer B, et al. 2002. Genetic heterogeneity of single disseminated tumour cells in minimal residual cancer. Lancet. 360(9334):683–89. [CrossRef]
- Berger N, Kim-Schulze S, Parekh S. 2018. Minimal Residual Disease in Multiple Myeloma: Impact on Response Assessment, Prognosis and Tumor Heterogeneity. In Biological Mechanisms of Minimal Residual Disease and Systemic Cancer, ed JA Aguirre-Ghiso, pp. 141–59. Cham: Springer International Publishing.
- Raghavendra NM, Pingili D, Kadasi S, Mettu A, Prasad SVUM. 2018. Dual or multi-targeting inhibitors: The next generation anticancer agents. European Journal of Medicinal Chemistry. 143:1277–1300. [CrossRef]
- Eder JP, Shapiro GI, Appleman LJ, Zhu AX, Miles D, et al. 2010. A phase I study of foretinib, a multi-targeted inhibitor of c-Met and vascular endothelial growth factor receptor 2. Clin Cancer Res. 16(13):3507–16. [CrossRef]
- Petrelli A, Giordano S. 2008. From single- to multi-target drugs in cancer therapy: when aspecificity becomes an advantage. Curr Med Chem. 15(5):422–32. [CrossRef]
- Foulkes WD. 2008. Inherited Susceptibility to Common Cancers. N Engl J Med. 359(20):2143–53. [CrossRef]
- Fearon ER. 1997. Human Cancer Syndromes: Clues to the Origin and Nature of Cancer. Science. 278(5340):1043–50. [CrossRef]
- Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, et al. 1990. Linkage of Early-Onset Familial Breast Cancer to Chromosome 17q21. Science. 250(4988):1684–89. [CrossRef]
- Moore N, Lyle S. 2010. Quiescent, Slow-Cycling Stem Cell Populations in Cancer: A Review of the Evidence and Discussion of Significance. Journal of Oncology. 2011:e396076. [CrossRef]
- Makena MR, Ranjan A, Thirumala V, Reddy AP. 2020. Cancer stem cells: Road to therapeutic resistance and strategies to overcome resistance. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1866(4):165339. [CrossRef]
- Shiokawa D, Sakai H, Ohata H, Miyazaki T, Kanda Y, et al. 2020. Slow-Cycling Cancer Stem Cells Regulate Progression and Chemoresistance in Colon Cancer. Cancer Res. 80(20):4451–64.
- Robinson NJ, Taylor DJ, Schiemann WP. 2019. Stem cells, immortality, and the evolution of metastatic properties in breast cancer: telomere maintenance mechanisms and metastatic evolution. J Cancer Metastasis Treat. 5:39. [CrossRef]
- Shay JW, Wright WE. 2010. Telomeres and telomerase in normal and cancer stem cells. FEBS Letters. 584(17):3819–25. [CrossRef]
- Friedmann-Morvinski D, Verma IM. 2014. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Reports. 15(3):244–53. [CrossRef]
- Visvader JE. 2011. Cells of origin in cancer. Nature. 469(7330):314–22.
- Visvader JE, Lindeman GJ. 2008. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 8(10):755–68. [CrossRef]
- Jolly MK, Celià-Terrassa T. 2019. Dynamics of Phenotypic Heterogeneity Associated with EMT and Stemness during Cancer Progression. Journal of Clinical Medicine. 8(10):1542. [CrossRef]
- Zhou F, Aroua N, Liu Y, Rohde C, Cheng J, et al. 2023. A Dynamic rRNA Ribomethylome Drives Stemness in Acute Myeloid Leukemia. Cancer Discov. 13(2):332–47.
- Jain P, Duddu AS, Jolly MK. 2022. Stochastic population dynamics of cancer stemness and adaptive response to therapies. Essays Biochem. 66(4):387–98. [CrossRef]
- Ma H, He M, Wei M. 2016. [Research progress on targeting effect and regulating mechanisms of the stemness of cancer stem cells]. Yao Xue Xue Bao. 51(2):189–96.
- Takahashi K, Okita K, Nakagawa M, Yamanaka S. 2007. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2(12):3081–89. [CrossRef]
- Chico MA, Mesas C, Doello K, Quiñonero F, Perazzoli G, et al. 2023. Cancer Stem Cells in Sarcomas: In Vitro Isolation and Role as Prognostic Markers: A Systematic Review. Cancers. 15(9):2449. [CrossRef]
- Li D, Zhang T, Gu W, Li P, Cheng X, et al. 2015. The ALDH1+ subpopulation of the human NMFH-1 cell line exhibits cancer stem-like characteristics. Oncol Rep. 33(5):2291–98. [CrossRef]
- Feng B-H, Liu A-G, Gu W-G, Deng L, Cheng X-G, et al. 2013. CD133+ subpopulation of the HT1080 human fibrosarcoma cell line exhibits cancer stem-like characteristics. Oncol Rep. 30(2):815–23. [CrossRef]
- Cell Dedifferentiation - an overview | ScienceDirect Topics. www.sciencedirect.com.
- Li M-M, Tang Y-Q, Gong Y-F, Cheng W, Li H-L, et al. 2019. Development of an oncogenic dedifferentiation SOX signature with prognostic significance in hepatocellular carcinoma. BMC Cancer. 19(1):851. [CrossRef]
- Komarova NL, Wodarz D. 2005. Drug resistance in cancer: Principles of emergence and prevention. Proc. Natl. Acad. Sci. U.S.A. 102(27):9714–19. [CrossRef]
- Colombo P-E, Fabbro M, Theillet C, Bibeau F, Rouanet P, Ray-Coquard I. 2014. Sensitivity and resistance to treatment in the primary management of epithelial ovarian cancer. Critical Reviews in Oncology/Hematology. 89(2):207–16. [CrossRef]
- Zhu P, Fan Z. 2018. Cancer stem cells and tumorigenesis. Biophys Rep. 4(4):178–88. [CrossRef]
- Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, et al. 2008. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 15(3):504–14. [CrossRef]
- Ma S, Chan K, Hu L, Lee TK, Wo JY, et al. 2007. Identification and Characterization of Tumorigenic Liver Cancer Stem/Progenitor Cells. Gastroenterology. 132(7):2542–56. [CrossRef]
- Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. 2013. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 155(7):1639–51. [CrossRef]
- Wakimoto H, Mohapatra G, Kanai R, Curry WT, Yip S, et al. 2012. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro Oncol. 14(2):132–44. [CrossRef]
- Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, et al. 2009. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 69(8):3382–89. [CrossRef]
- Sherr CJ. 2004. Principles of tumor suppression. Cell. 116(2):235–46. [CrossRef]
- Oncogene - an overview | ScienceDirect Topics. www.sciencedirect.com.
- Endo Y, Lyon S, Shen Y, Mohan N, Wu WJ. 2019. Cell proliferation and invasion are regulated differently by EGFR and MRP1 in T-DM1-resistant breast cancer cells. Sci Rep. 9(1):16383. [CrossRef]
- Gupta SC, Kim JH, Prasad S, Aggarwal BB. 2010. Regulation of survival, proliferation, invasion, angiogenesis, and metastasis of tumor cells through modulation of inflammatory pathways by nutraceuticals. Cancer Metastasis Rev. 29(3):405–34. [CrossRef]
- De Donatis A, Ranaldi F, Cirri P. 2010. Reciprocal control of cell proliferation and migration. Cell Communication and Signaling. 8(1):20. [CrossRef]
- Swami P, Thiyagarajan S, Vidger A, Indurthi VSK, Vetter SW, Leclerc E. 2020. RAGE Up-Regulation Differently Affects Cell Proliferation and Migration in Pancreatic Cancer Cells. International Journal of Molecular Sciences. 21(20):7723. [CrossRef]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, et al. 2010. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 467(7319):1114–17. [CrossRef]
- Seyfried TN, Huysentruyt LC. 2013. On the Origin of Cancer Metastasis. Crit Rev Oncog. 18(1–2):43–73. [CrossRef]
- Matassa DS, Agliarulo I, Avolio R, Landriscina M, Esposito F. 2018. TRAP1 Regulation of Cancer Metabolism: Dual Role as Oncogene or Tumor Suppressor. Genes. 9(4):195. [CrossRef]
- Robbs BK, Cruz ALS, Werneck MBF, Mognol GP, Viola JPB. 2008. Dual Roles for NFAT Transcription Factor Genes as Oncogenes and Tumor Suppressors. Molecular and Cellular Biology. 28(23):7168–81. [CrossRef]
- Uribesalgo I, Benitah SA, Di Croce L. 2012. From oncogene to tumor suppressor: The dual role of Myc in leukemia. Cell Cycle. 11(9):1757–64.
- Shen L, Shi Q, Wang W. 2018. Double agents: genes with both oncogenic and tumor-suppressor functions. Oncogenesis. 7(3):1–14. [CrossRef]
- Ayob AZ, Ramasamy TS. 2018. Cancer stem cells as key drivers of tumour progression. Journal of Biomedical Science. 25(1):20. [CrossRef]
- Damen MPF, Van Rheenen J, Scheele CLGJ. 2021. Targeting dormant tumor cells to prevent cancer recurrence. The FEBS Journal. 288(21):6286–6303. [CrossRef]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The Hallmarks of Aging. Cell. 153(6):1194–1217.
- Mc Auley MT, Guimera AM, Hodgson D, Mcdonald N, Mooney KM, et al. 2017. Modelling the molecular mechanisms of aging. Biosci Rep. 37(1):BSR20160177. [CrossRef]
- Campisi J. 2013. Aging, Cellular Senescence, and Cancer. Annu Rev Physiol. 75:685–705.
- Laconi E, Marongiu F, DeGregori J. 2020. Cancer as a disease of old age: changing mutational and microenvironmental landscapes. Br J Cancer. 122(7):943–52. [CrossRef]
- Rufini A, Tucci P, Celardo I, Melino G. 2013. Senescence and aging: the critical roles of p53. Oncogene. 32(43):5129–43. [CrossRef]
- Mori H, Funahashi Y, Yoshino Y, Kumon H, Ozaki Y, et al. Blood CDKN2A Gene Expression in Aging and Neurodegenerative Diseases. J Alzheimers Dis. 82(4):1737–44. [CrossRef]
- Zeng H, Jorapur A, Shain AH, Lang UE, Torres R, et al. 2018. Bi-allelic loss of CDKN2A initiates melanoma invasion via BRN2 activation. Cancer Cell. 34(1):56-68.e9. [CrossRef]
- Ozaki T, Nakagawara A. 2011. Role of p53 in Cell Death and Human Cancers. Cancers (Basel). 3(1):994–1013. [CrossRef]
- Endo H, Inoue M. 2019. Dormancy in cancer. Cancer Sci. 110(2):474–80.
- Dormancy - an overview | ScienceDirect Topics. www.sciencedirect.com.
- Neophytou CM, Kyriakou T-C, Papageorgis P. 2019. Mechanisms of Metastatic Tumor Dormancy and Implications for Cancer Therapy. Int J Mol Sci. 20(24):6158. [CrossRef]
- Park S-Y, Nam J-S. 2020. The force awakens: metastatic dormant cancer cells. Exp Mol Med. 52(4):569–81. [CrossRef]
- Blasco MT, Espuny I, Gomis RR. 2022. Ecology and evolution of dormant metastasis. Trends Cancer. 8(7):570–82. [CrossRef]
- Kumar R, Angelini S, Snellman E, Hemminki K. 2004. BRAF Mutations Are Common Somatic Events in Melanocytic Nevi. Journal of Investigative Dermatology. 122(2):342–48. [CrossRef]
- McNeal, A. S., Belote, R. L., Zeng, H., Urquijo, M., Barker, K., Torres, R., Curtin, M., Shain, A. H., Andtbacka, R. H., Holmen, S., Lum, D. H., McCalmont, T. H., VanBrocklin, M. W., Grossman, D., Wei, M. L., Lang, U. E., & Judson-Torres, R. L. (2021). BRAFV600E induces reversible mitotic arrest in human melanocytes via microrna-mediated suppression of AURKB. ELife, 10, e70385.
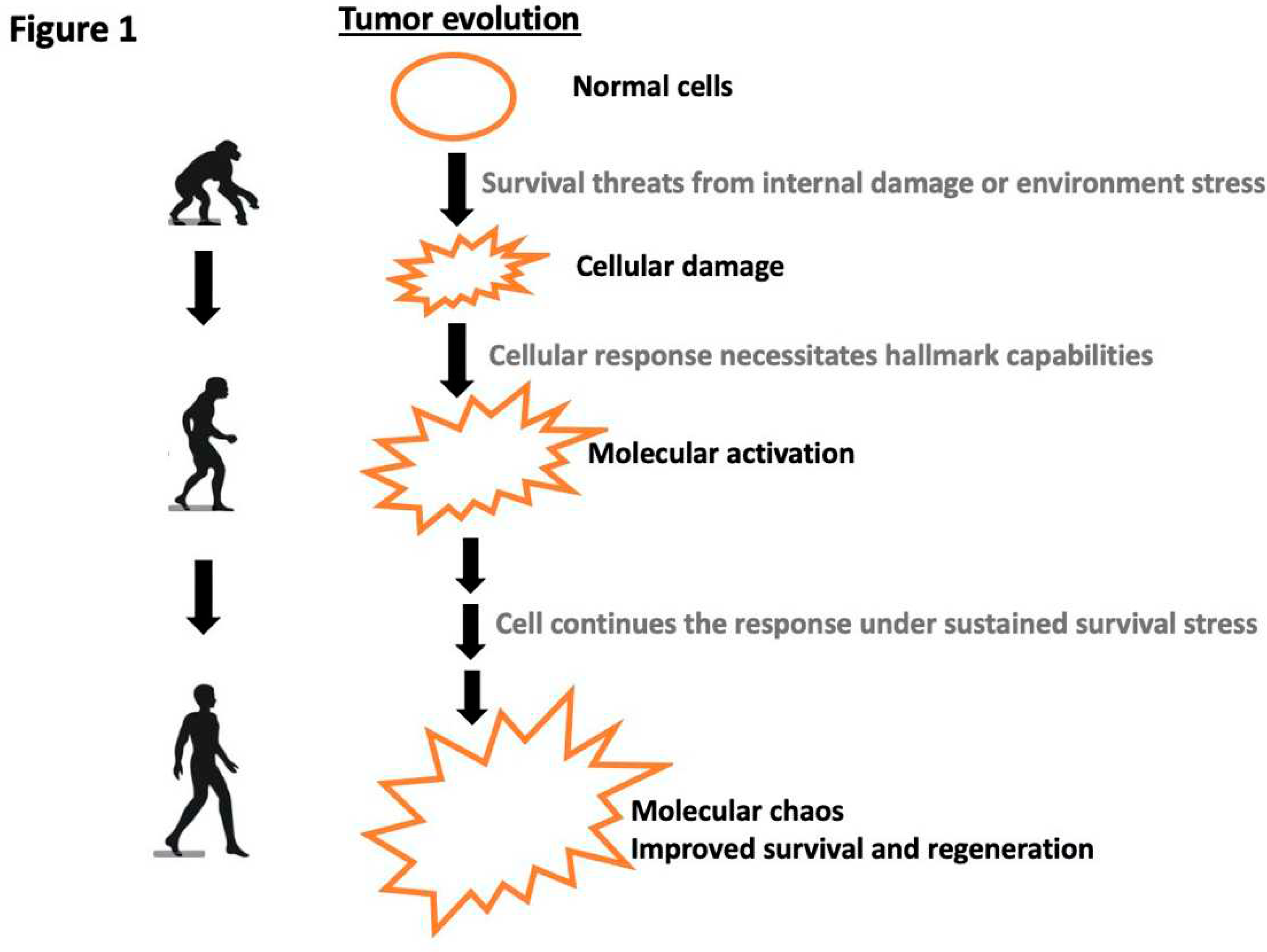
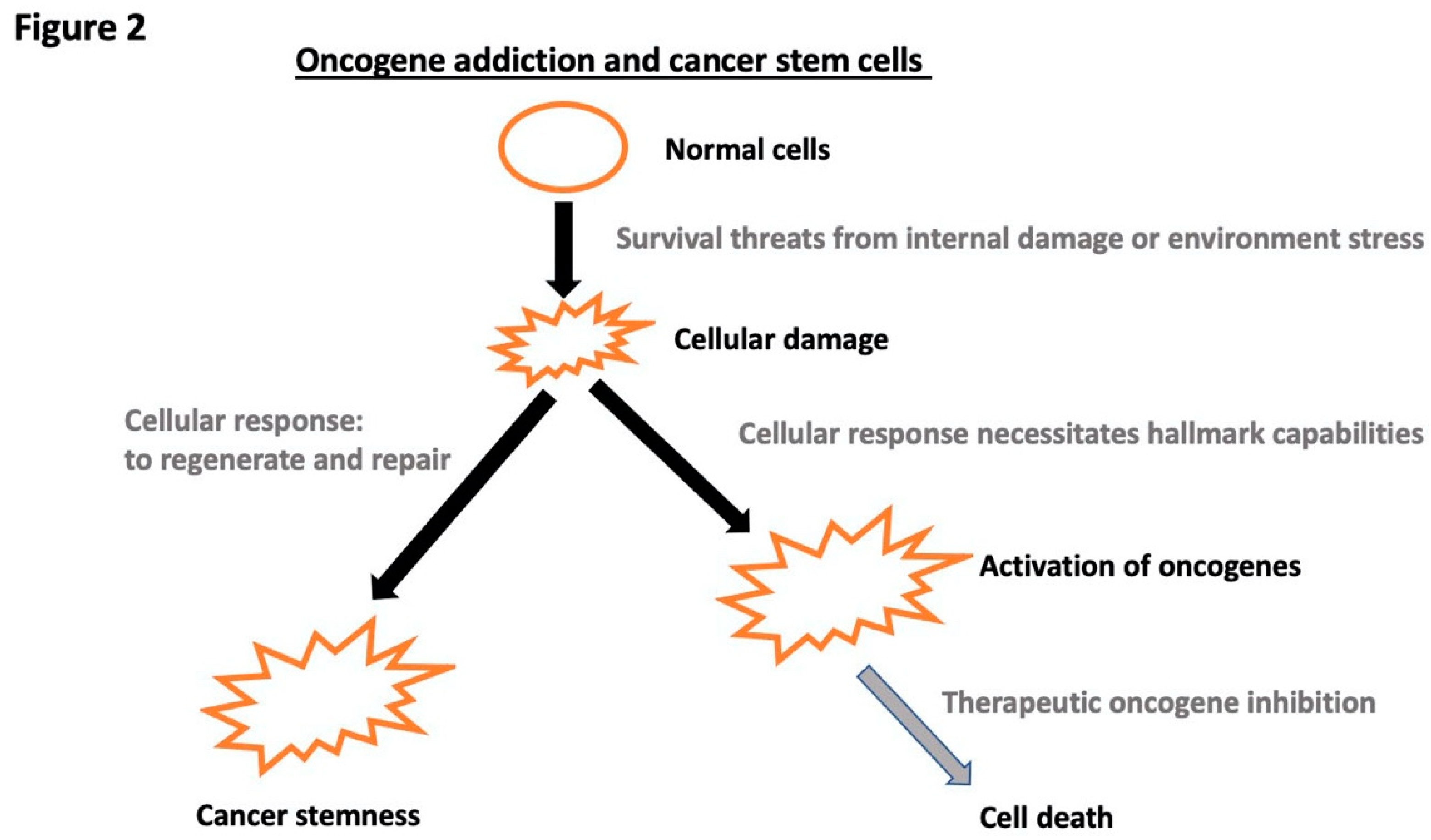
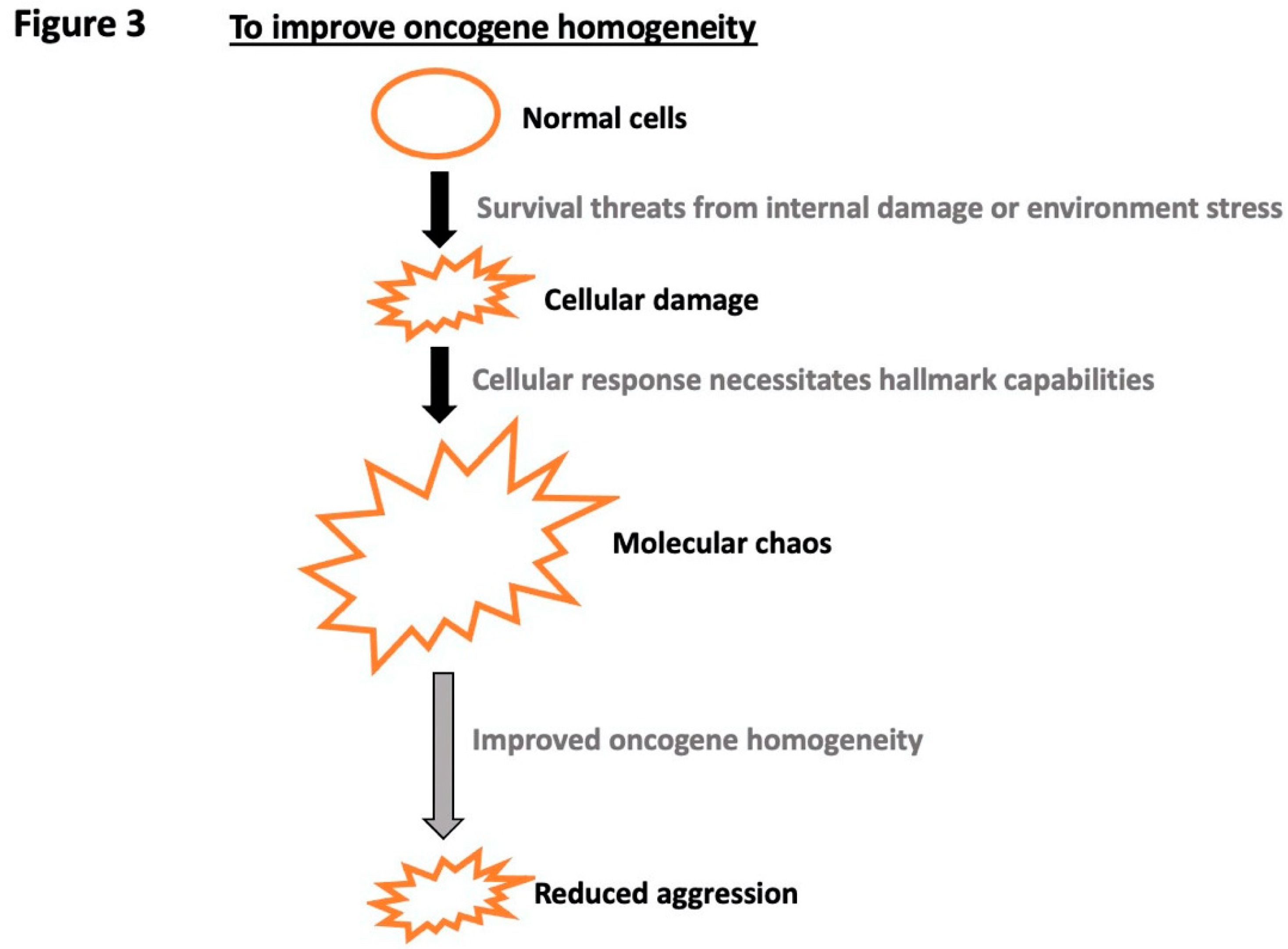
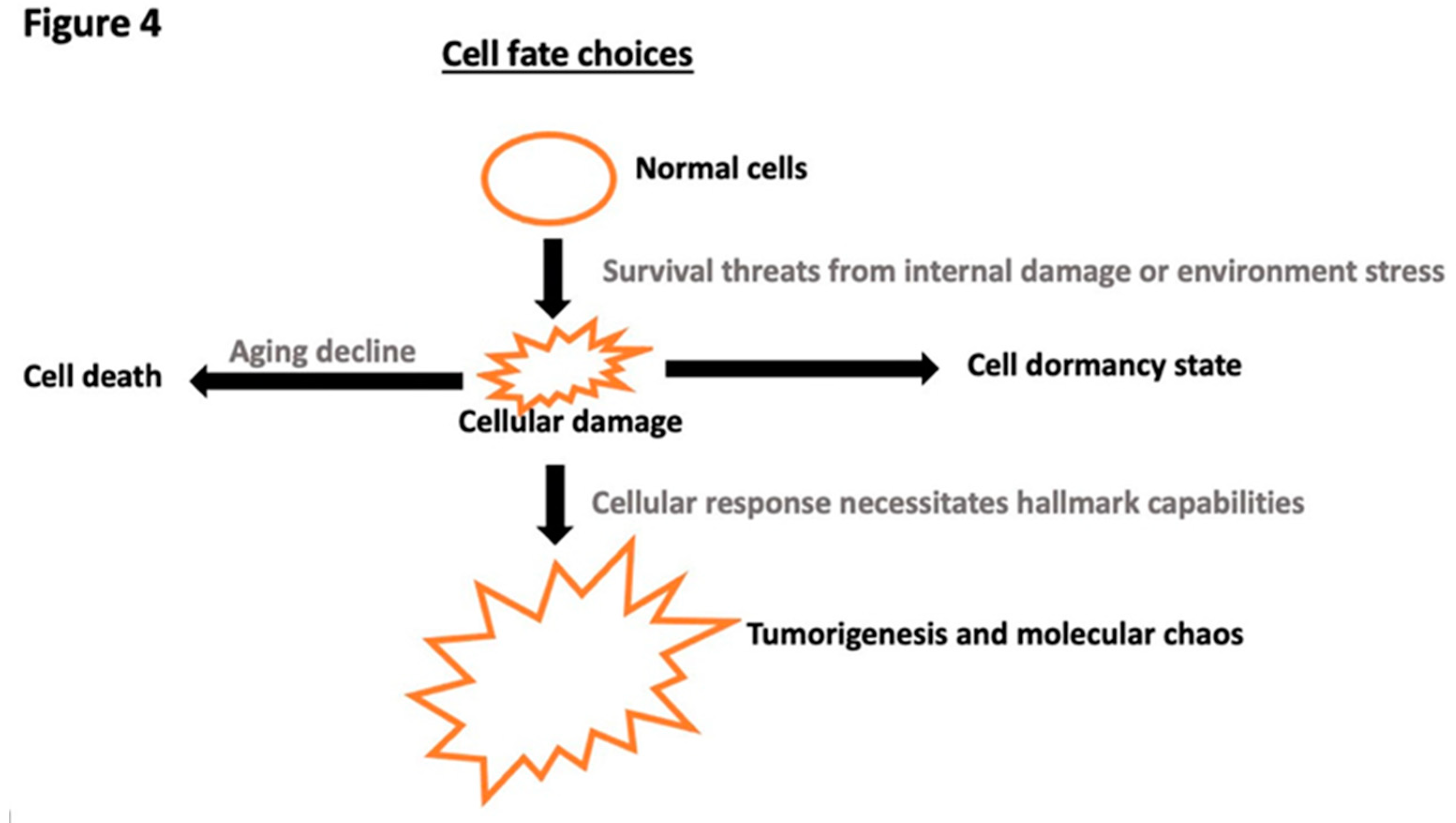
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



