Submitted:
19 October 2023
Posted:
20 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction: Limits of the Gene Biomarker Paradigm for Cancer Diagnostic and Therapy
2. Materials and methods
2.1. The best choice of tissue samples
2.2. Data filtering and normalization
2.3. Independent characteristics of gene expression
2.3.1. Normalized average expression level.
2.3.2. Relative Expression Variability
2.3.3. Expression coordination
2.3.4. Topology of the transcriptome and the Gene Master Regulator
2.4. Transcriptome alteration in cancer
2.4.1. Measures of expression regulation
2.4.2. Regulation of the control of transcript abundance
2.4.3. Regulation of expression coordination:
2.4.4. The transcriptomic distance
2.5. Functional pathways
3. Results
3.1. The global picture
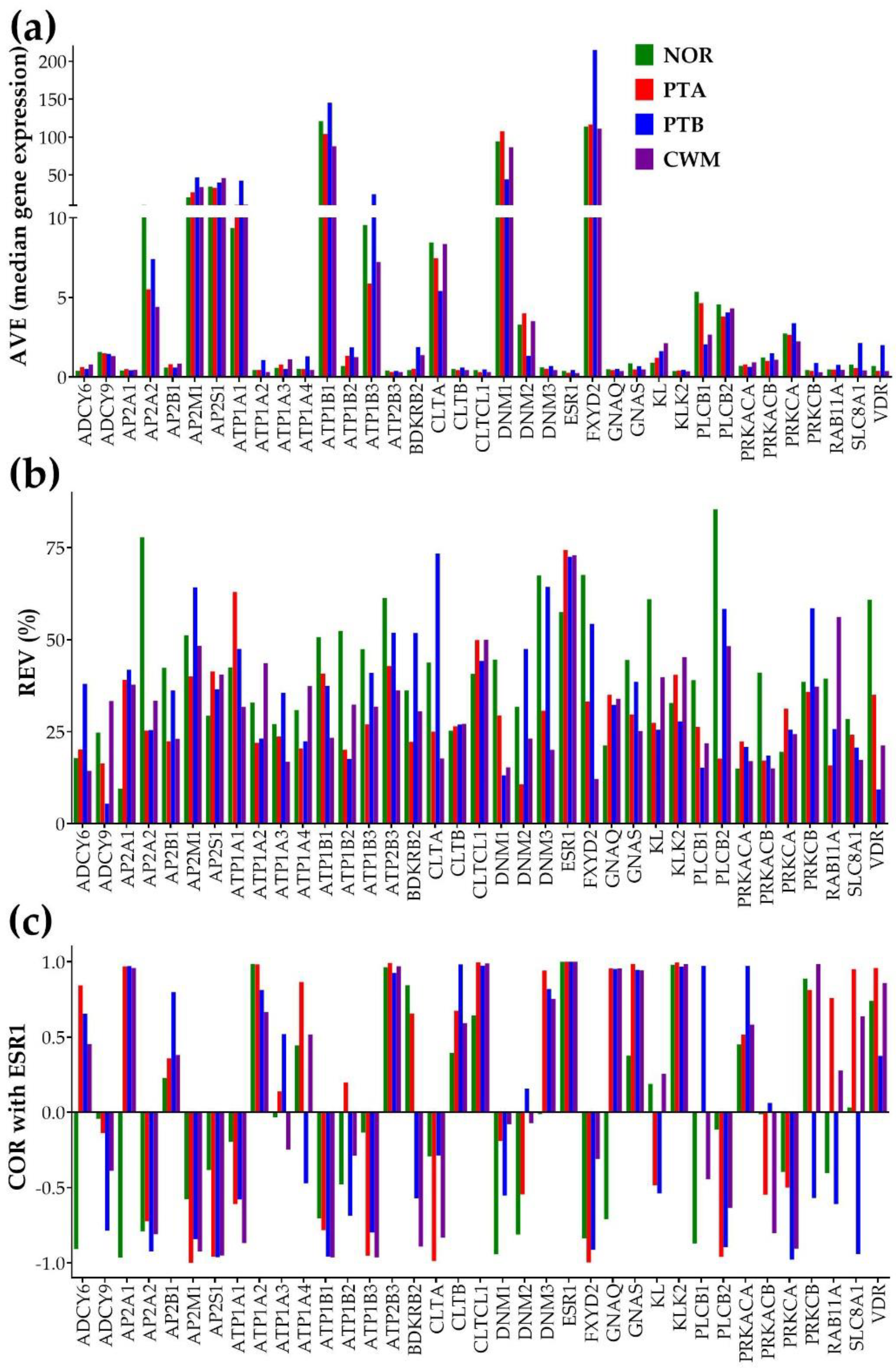
3.2. Independent characteristics of gene expression
3.4. Measures of individual gene regulation
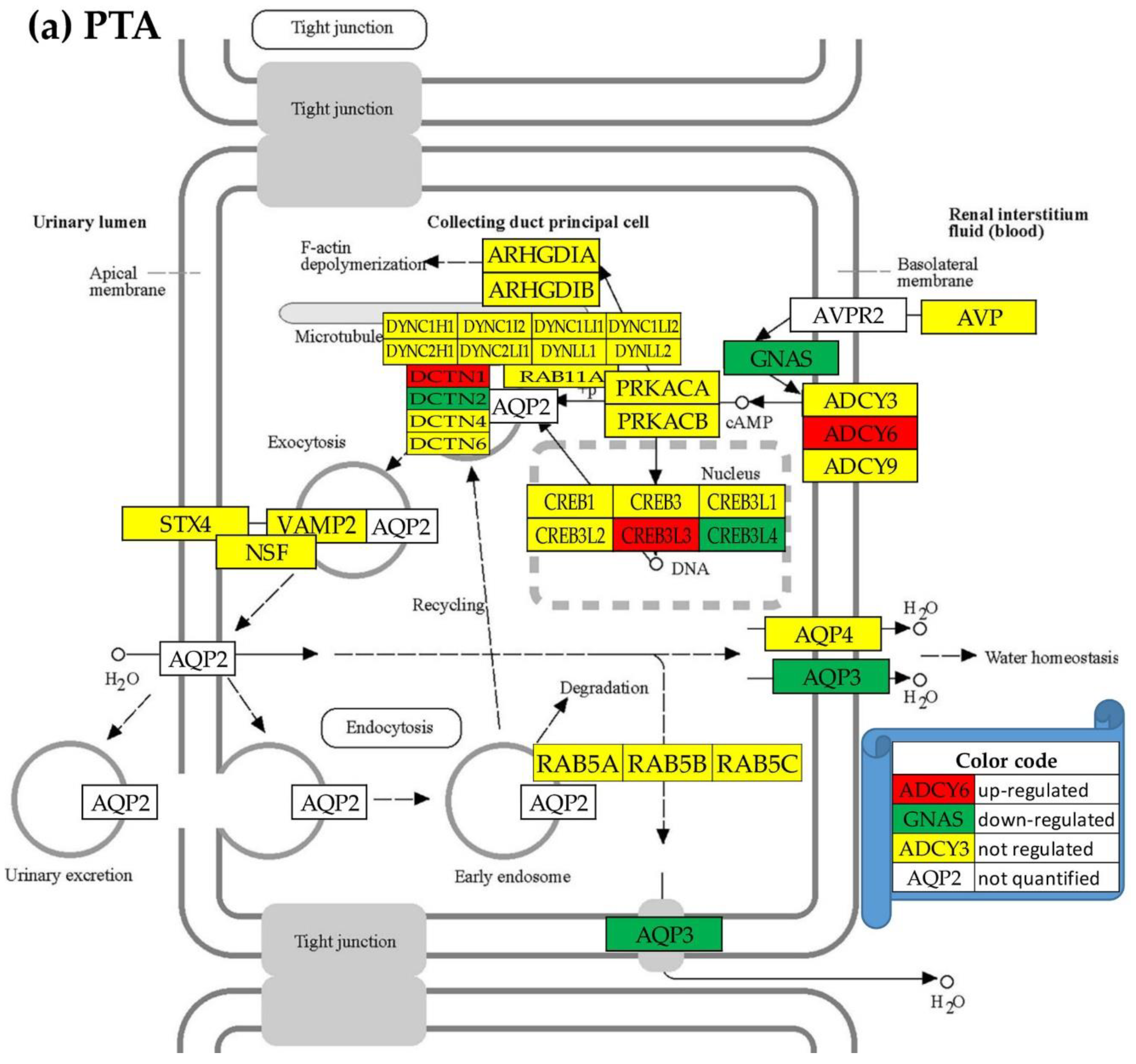
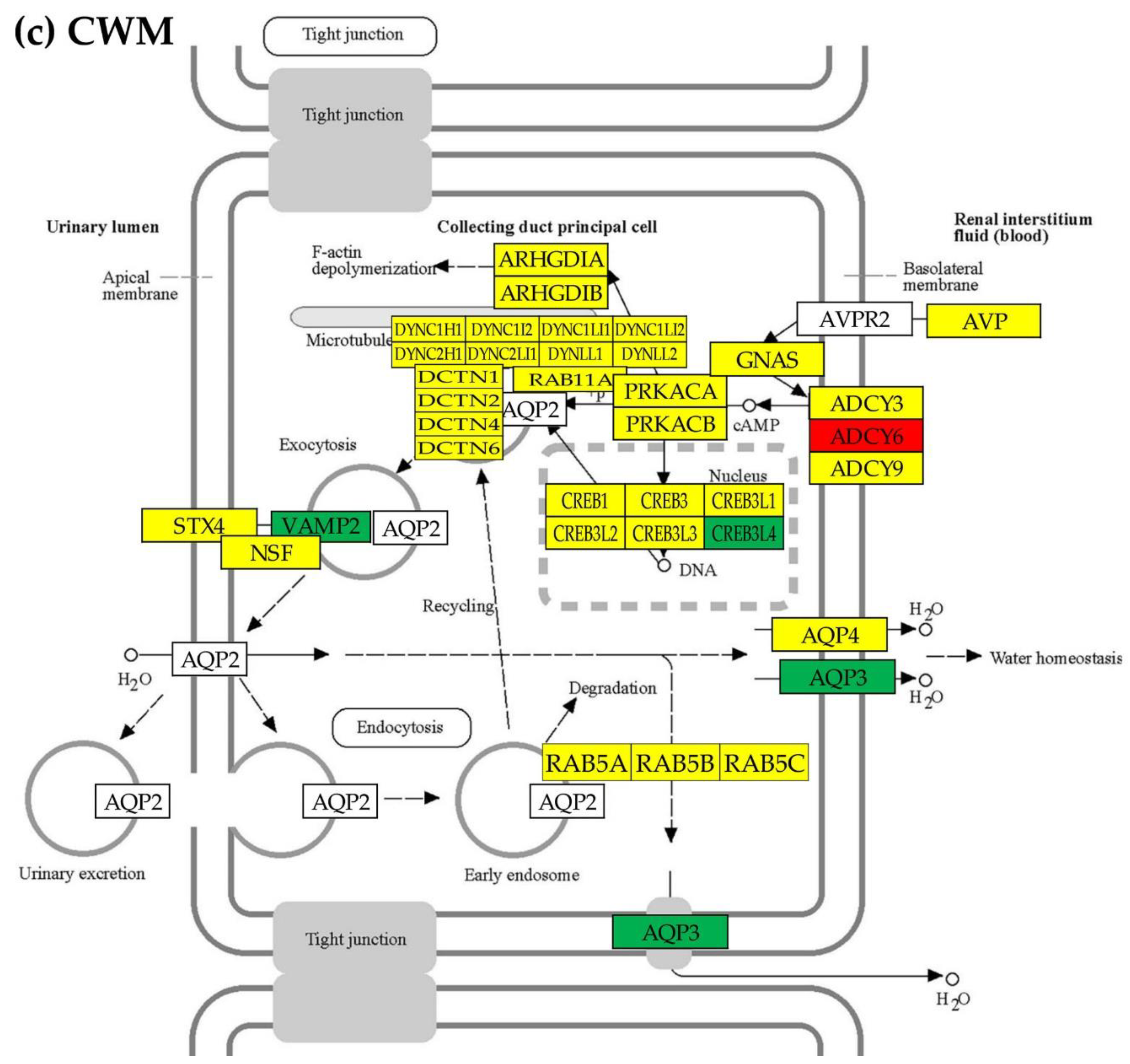
3.5. Overall regulation of the excretory pathways
3.6. Location of the regulated genes in the excretory system functional pathways
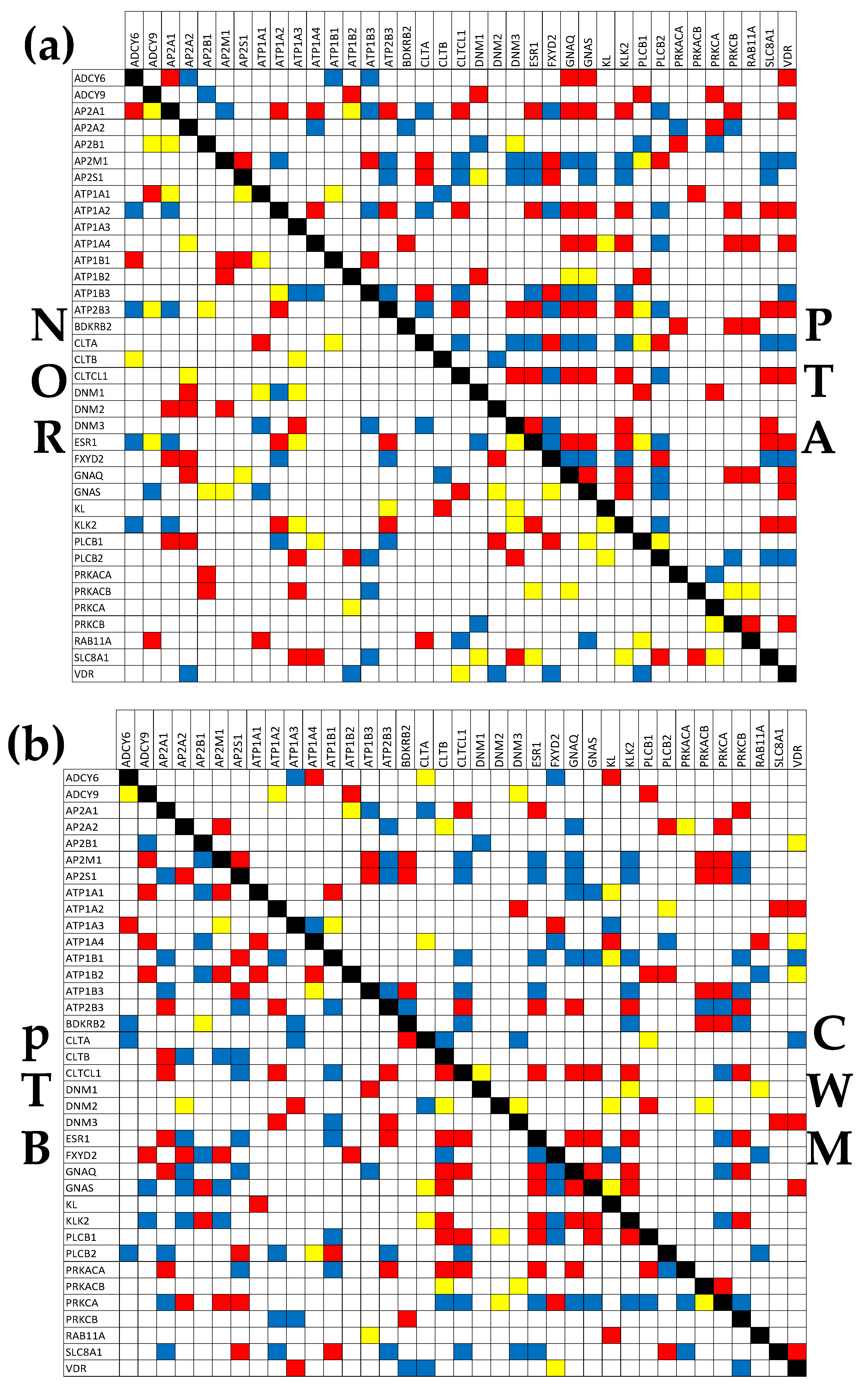
3.6. Tumor heterogeneity off the transcriptomic networks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Cancer Treatment options at Houston Methodist organization. Available on line at: https://www.houstonmethodist.org/cancer/treatment-options/. (accessed 05/03/2023).
- Pecoraro, A.; Campi, R.; Marchioni, M.; European Association of Urology Young Academic Urologists Renal Cancer Working Group. Techniques and outcomes of percutaneous tumour ablation for small renal masses. Curr Opin Urol. 2023; 33(5):360-366. [CrossRef]
- Lanza, C.; Carriero, S.; Ascenti, V.; Tintori, J.; Ricapito, F.; Lavorato, R.; Biondetti, P.; Angileri, S.A.; Piacentino, F.; Fontana F. et al. Percutaneous Application of High Power Microwave Ablation With 150 W for the Treatment of Tumors in Lung, Liver, and Kidney: A Preliminary Experience. Technol Cancer Res Treat. 2023 22:15330338231185277. [CrossRef]
- Key Statistics about Kidney Cancer. Available online: https://www.cancer.org/cancer/kidney-cancer.html ((accessed on Sept. 6th, 2023).
- Mieville, V.; Griffioen, A.W.; Benamran, D.; Nowak-Sliwinska, P. Advanced in vitro models for renal cell carcinoma therapy design. Biochim Biophys Acta Rev Cancer. 2023; 1878(5):188942. [CrossRef]
- Rotin, L.E.; Viswabandya, A.; Kumar, R.; Patriquin, C.J.; Kuo, K.H.M. A systematic review comparing allogeneic hematopoietic stem cell transplant to gene therapy in sickle cell disease. Hematology. 2023; 28(1):2163357. [CrossRef]
- Gong, Y.; Pang, H.; Yu, Z.; Wang, X.; Li, P.; Zhang, Q. Construction of inflammatory associated risk gene prognostic model of NSCLC and its correlation with chemotherapy sensitivity. Ann Med. 2023; 55(1):2200034. [CrossRef]
- Hara Y, Shiba N, Yoshida K, Yamato G, Kaburagi T, Shiraishi Y, Ohki K, Shiozawa Y, Kawamura M, Kawasaki H. et al. TP53 and RB1 alterations characterize poor prognostic subgroups in pediatric acute myeloid leukemia. Genes Chromosomes Cancer. 2023; 62(7):412-422. [CrossRef]
- Cancel-Tassin, G.; Koutros, S. Use of genomic markers to improve epidemiologic and clinical research in urology. Curr Opin Urol. 2023. [CrossRef]
- Wu, F.; Ning, H.; Sun, Y.; Wu, H.; Lyu, J. Integrative exploration of the mutual gene signatures and immune microenvironment between benign prostate hyperplasia and castration-resistant prostate cancer. Aging Male. 2023; 26(1):2183947. [CrossRef]
- Yerukala Sathipati, S.; Tsai, M.J.; Shukla, S.K.; Ho, S.Y. Artificial intelligence-driven pan-cancer analysis reveals miRNA signatures for cancer stage prediction. HGG Adv. 2023; 4(3):100190. [CrossRef]
- Yang, L.; Yang, M.; Cui, C.; Long, X.; Li, Y.; Dai, W.; Lang, T.; Zhou, Q. The myo-inositol biosynthesis rate-limiting enzyme ISYNA1 suppresses the stemness of ovarian cancer via Notch1 pathway. Cell Signal. 2023; 107:110688. [CrossRef]
- Huang, L.; Shao, J.; Xu, X.; Hong, W.; Yu, W.; Zheng, S.; Ge, X. WTAP regulates autophagy in colon cancer cells by inhibiting FLNA through N6-methyladenosine. Cell Adh Migr. 2023; 17(1):1-13. [CrossRef]
- Yang, R.Y.; Tan, J.Y.; Liu, Z.; Shen, X.L.; Hu, Y.J. Lappaol F regulates the cell cycle by activating CDKN1C/p57 in human colorectal cancer cells. Pharm Biol. 2023; 61(1):337-344. [CrossRef]
- Yavuz, M.; Takanlou, L.S.; Avcı, Ç.B.; Demircan, T. A selective androgen receptor modulator, S4, displays robust anti-cancer activity on hepatocellular cancer cells by negatively regulating PI3K/AKT/mTOR signalling pathway. Gene. 2023; 869:147390. [CrossRef]
- Ishiguro, M.; Fukushige, T.; Iwasaki, H. Establishment and Characterization of a TFE3-rearranged Renal Cell Carcinoma Cell Line (FU-UR-2) With the PRCC-TFE3 Fusion Transcript. Anticancer Res. 2023; 43(8):3463-3470. [CrossRef]
- NIH-National Cancer Institute Genomic Data Commons Data Portal. Available on line at: https://portal.gdc.cancer.gov/. Accessed 06/20/2023 .
- Sarka,r O.S.; Donninger, H.; Al Rayyan, N.; Chew, L.C.; Stamp, B.; Zhang, X.; Whitt, A.; Li, C.; Hall, M.; Mitchell, R.A. et al. Monocytic MDSCs exhibit superior immune suppression via adenosine and depletion of adenosine improves efficacy of immunotherapy. Sci Adv. 2023; 9(26):eadg3736. [CrossRef]
- Liu, C.L.; Huang, W.C.; Cheng, S.P.; Chen, M.J.; Lin, C.H.; Chang, S.C.; Chang, Y.C. Characterization of Mammary Tumors Arising from MMTV-PyVT Transgenic Mice. Curr Issues Mol Biol. 2023; 45(6):4518-4528. [CrossRef]
- Montico, F.; Lamas, C.A.; Rossetto, I.M.U.; Baseggio, A.M.; Cagnon, V.H.A.. Lobe-specific responses of TRAMP mice dorsolateral prostate following celecoxib and nintedanib therapy. J Mol Histol. 2023. [CrossRef]
- Ding, W.Y.; Kuzmuk, V.; Hunter, S.; Lay, A.; Hayes, B.; Beesley, M.; Rollason, R.; Hurcombe, JA.; Barrington, F.; Masson, C. et al. Adeno-associated virus gene therapy prevents progression of kidney disease in genetic models of nephrotic syndrome. Sci Transl Med. 2023; 15(708):eabc8226. [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Urban-Maldonado, M.; Spray, D.C. Sensitivity of the brain transcriptome to connexin ablation, Biochim Biofis Acta. 2005; 1711: 183-196. Review. [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Li, W.E.; Zoidl, G.; Dermietzel, R.; Spray, D.C. Genes controlling multiple functional pathways are transcriptionally regulated in connexin43 null mouse heart. Physiol Genomics 2005; 20: 211-223. [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Spray, D.C. Connexin-dependent transcellular transcriptomic networks in mouse brain. Prog Biophys Mol Biol. 2007; 94(1-2):168-184. Review. [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Urban-Maldonado, M.; Scemes, E.; Spray, D.C Similar transcriptomic alterations in Cx43 knock-down and knock-out astrocytes. Cell Commun. Adhes. 2008; 15:1, 195-206. [CrossRef]
- Iacobas, S.; Iacobas, D.A.; Spray, D.C.; Scemes, E. The connexin43 transcriptome during brain development: importance of genetic background. Brain Research. 2012; 1487: 131-139. [CrossRef]
- Tolkach, Y.; Kristiansen, G. The Heterogeneity of Prostate Cancer: A Practical Approach. Pathobiology 2018, 85, 108–116. [CrossRef]
- Tu, S.-M.; Zhang, M.; Wood, C.G.; Pisters, L.L. Stem Cell Theory of Cancer: Origin of Tumor Heterogeneity and Plasticity. Cancers, 2021, 13, 4006. [CrossRef]
- Berglund, E.; Maaskola, J.; Schultz, N.; Friedrich, S.; Marklund, M.; Bergenstråhle, J.; Tarish, F.; Tanoglidi, A.; Vickovic, S.;
- Larsson, L.; et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 2018, 9, 2419. [CrossRef]
- Brady, L.; Kriner, M.; Coleman, I.; Morrissey, C.; Roudier, M.; True, L.D.; Gulati, R.; Plymate, S.R.; Zhou, Z.; Birditt, B.; et al. Inter and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nat. Commun. 2021, 12, 1426. [CrossRef]
- Iacobas, S.; Iacobas, D.A. Personalized 3-Gene Panel for Prostate Cancer Target Therapy. Curr. Issues Mol. Biol. 2022, 44, 360-382. [CrossRef]
- Zhang, G.; Feng, Z.; Zeng, Q.; Huang, P. Exploring Cancer Dependency Map genes and immune subtypes in colon cancer, in which TIGD1 contributes to colon cancer progression. Aging (Albany NY). 2023; 15. [CrossRef]
- Gui, Z.; Du, J.; Wu, N.; Shen, N.; Yang, Z.; Yang, H.; Wang, X.; Zhao, N.; Zeng, Z.; Wei R. et al. Immune regulation and prognosis indicating ability of a newly constructed multi-genes containing signature in clear cell renal cell carcinoma. BMC Cancer. 2023; 23(1):649. [CrossRef]
- Qiagen Ingenuity Pathway Analysis. Available online: https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ (accessed on July 12th 2023).
- DAVID Functional Annotation Bioinformatics Microarray Analysis. Available online: https://david.ncifcrf.gov (accessed on July 12th 2023).
- Kyoto Encyclopedia of Genes and Genomes. Available online: https://www.kegg.jp/kegg/pathway.html (accessed on July 12th 2023).
- Iacobas, D.A.; Mgbemena, V.; Iacobas, S.; Menezes, K.M.; Wang, H.; Saganti, P.B. Genomic fabric remodeling in metastatic clear cell renal cell carcinoma (ccRCC): A new paradigm and proposal for a personalized gene therapy approach. Cancers 2020, 12(12), 3678; [CrossRef]
- Iacobas, D.A.; Tuli, N.; Iacobas, S.; Rasamny, J.K.; Moscatello, A.; Geliebter, J.; Tiwari, R.M. Gene master regulators of papillary and anaplastic thyroid cancer phenotypes. Oncotarget 2018, 9(2), 2410-2424. [CrossRef]
- Iacobas, S.; Iacobas, D.A. A Personalized Genomics Approach of the Prostate Cancer. Cells 2021, 10, 1644. https://www.mdpi.com/2073-4409/10/7/1644.
- Remodeling of major genomic fabrics and their interplay in metastatic clear cell renal carcinoma. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72304.
- Iacobas, D.A.; Iacobas, S.; Spray, D.C. Connexin43 and the brain transcriptome of newborn mice. Genomics. 2007, 89(1), 113-123. [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Lee, P.R.; Cohen, J.E.; Fields, R.D. Coordinated Activity of Transcriptional Networks Responding to the Pattern of Action Potential Firing in Neurons. Genes 2019, 10, 754. [CrossRef]
- Ebbing D, Gammon SD. General Chemistry – Standalone book. Student Edition. Cengage Learning: Boston, MA, U.S.A. 2015; pp. 88-100.
- Hansen J-P.; McDonald I.R. Chapter 7 – Time-dependent Correlation and Response Functions. In Theory of Simple Liquids, 4th ed.; Academic Press: London, UK, 2013. pp. 265-310.
- Eisen, M.; Spellman, P.; Brown, P.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [CrossRef]
- Butte, A.J.; Tamayo, P.; Slonim, D.; Golub, T.R.; Kohane, I.S. Discovering functional relationships between RNA expression and chemotherapeutic susceptibility using relevance networks. Proc. Natl. Acad. Sci. USA, 2000, 97, 12182–12186. [CrossRef]
- Horvath, S.; Dong, J. Geometric Interpretation of Gene Coexpression Network Analysis. PLoS Comput. Biol. 2008, 4, e1000117. [CrossRef]
- Oldham, M.C.; Langfelder, P.; Horvath, S. Network methods for describing sample relationships in genomic datasets: Application to Huntington’s disease. BMC Syst. Biol. 2012, 6, 63. [CrossRef]
- Marbach, D.; Costello, J.C.; Küner, R.; Vega, N.M.; Prill, R.J.; Camacho, D.M.; Allison, K.R.; Aderhold, A.; Bonneau, R.; Chen, Y.; et al. Wisdom of the crowds for robust gene network inference. Nat. Methods 2012, 9, 796–804. [CrossRef]
- P Value from Pearson (R) Calculator. Available on line at: https://www.socscistatistics.com/pvalues/pearsondistribution.aspx (accessed 09/01/2023).
- Mathew, R.; Iacobas, S.; Huang, J.; Iacobas, D.A. Metabolic Deregulation in Pulmonary Hypertension. Curr Issues Mol Biol. 2023; 45(6):4850-4874. [CrossRef]
- Iacobas, S.; Ede, N.; Iacobas, D.A. The Gene Master Regulators (GMR) Approach Provides Legitimate Targets for Personalized, Time-Sensitive Cancer Gene Therapy. Genes 2019, 10, 560. [CrossRef]
- Iacobas DA. Biomarkers, Master Regulators and Genomic Fabric Remodeling in a Case of Papillary Thyroid Carcinoma. Genes 2020. 11(9):E1030. [CrossRef]
- Iacobas DA. Powerful Quantifiers for Cancer Transcriptomics. World J Clinical Oncology 2020, 11(9):679-704. https://www.wjgnet.com/2218-4333/full/v11/i9/679.htm.
- Iacobas, D.A.; Xi, L. Theory and Applications of the (Cardio) Genomic Fabric Approach to Post-Ischemic and Hypoxia-Induced Heart Failure. J Pers Med. 2022; 12(8):1246. [CrossRef]
- Aldosterone-regulated sodium reabsorption. Available on line at: https://www.genome.jp/pathway/hsa04960 (accessed 09/01/2023.
- Collecting duct acid secretion. Available on line at: https://www.genome.jp/pathway/hsa04966 (accessed 09/01/2023).
- Endocrine and other factor-regulated calcium reabsorption. Available on line at: https://www.genome.jp/pathway/hsa04961 (accessed on Sept 1, 2023).
- Proximal tubule bicarbonate reclamation. Available online at: https://www.genome.jp/kegg-bin/show_pathway?hsa04964 (accessed 01/09/2023).
- Vasopressin-regulated water reabsorption. Available online at: https://www.genome.jp/kegg-bin/show_pathway?hsa04962 (accessed 09/01/2023).
- Thomas, W.; Harvey, B.J. Estrogen-induced signalling and the renal contribution to salt and water homeostasis. Steroids. 2023; 199:109299. [CrossRef]
- Li, L.; Tan, H.; Zhou, J.; Hu, F. Predicting response of immunotherapy and targeted therapy and prognosis characteristics for renal clear cell carcinoma based on m1A methylation regulators. Sci Rep. 2023; 13(1):12645. [CrossRef]
- Wang, X.X.; Myakala, K.; Libby, A.E.; Krawczyk, E.; Panov, J.; Jones, B.A.; Bhasin, K.; Shults, N.; Qi, Y.; Krausz, K.W. et al. Estrogen-Related Receptor Agonism Reverses Mitochondrial Dysfunction and Inflammation in the Aging Kidney. Am J Pathol. 2023 17:S0002-9440(23)00321-8. [CrossRef]
- Adamopoulos, P.G.; Kontos, C.K.; Diamantopoulos, M.A.; Scorilas A. Molecular cloning of novel transcripts of the adaptor-related protein complex 2 alpha 1 subunit (AP2A1) gene, using Next-Generation Sequencing. Gene. 2018, 678:55-64. [CrossRef]
- Zhang, G.; Xi, M.; Li, Y.; Wang, L.; Gao, L.; Zhang, L.; Yang, Z.; Shi, H. The ADCY9 genetic variants are associated with glioma susceptibility and patient prognosis. Genomics. 2021; 113(2):706-716. [CrossRef]
- Tang, Y.; Wang, T.; Zhang, A.; Zhu, J.; Zhou, T.; Zhou, Y.L.; Shi, J. ADCY9 functions as a novel cancer suppressor gene in lung adenocarcinoma. J Thorac Dis. 2023; 15(3):1018-1035. [CrossRef]
- Chao, X.; Jia, Y.; Feng, X.; Wang, G.; Wang, X.; Shi, H.; Zhao, F.; Jiang, C. A Case-Control Study of ADCY9 Gene Polymorphisms and the Risk of Hepatocellular Carcinoma in the Chinese Han Population. Front Oncol. 2020; 10:1450. [CrossRef]
- Li, H.; Liu, Y.; Liu, J.; Sun, Y.; Wu, J.; Xiong, Z.; Zhang, Y.; Li, B.; Jin, T. Assessment of ADCY9 polymorphisms and colorectal cancer risk in the Chinese Han population. J Gene Med. 2021; 23(2):e3298. [CrossRef]
- Chen, Q.; Cai, L.; Liang, J. Construction of prognosis model of bladder cancer based on transcriptome. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2022; 51(1):79-86. English. [CrossRef]
- Lee, Y.H.; Gyu Song, G. Genome-wide pathway analysis in pancreatic cancer. J BUON. 2015; 20(6):1565-75. PMID: 26854454.
- Guo, R.; Liu, T.; Shasaltaneh, M.D.; Wang, X.; Imani, S.; Wen, Q. Targeting Adenylate Cyclase Family: New Concept of Targeted Cancer Therapy. Front Oncol. 2022; 12:829212. [CrossRef]
- Wang, H.; Zhang, W.; Ding, Z.; Xu, T.; Zhang, X.; Xu, K. Comprehensive exploration of the expression and prognostic value of AQPs in clear cell renal cell carcinoma. Medicine (Baltimore). 2022; 101(41):e29344. [CrossRef]
- Oncomine Solutions For Next-Generation Sequencing, available online at: http://www.oncomine.org. Accessed 10/01/2024.
- The University of ALabama at Birmingham CANcer data analysis Portal, available online at: http://ualcan.path.uab.edu. Accessed 10/01/2023.
- Sinha, S.; Dwivedi, N.; Tao, S.; Jamadar, A.; Kakade, V.R.; Neil, M.O.; Weiss, R.H.; Enders, J.; Calvet, J.P.; Thomas, S.M.; et al. Targeting the vasopressin type-2 receptor for renal cell carcinoma therapy. Oncogene. 2020; 39(6):1231-1245. [CrossRef]
- Baltzer, S.; Bulatov, T.; Schmied, C.; Krämer, A.; Berger, B.T.; Oder, A.; Walker-Gray, R.; Kuschke, C.; Zühlke, K.; Eichhorst, J.; et al. Aurora Kinase A Is Involved in Controlling the Localization of Aquaporin-2 in Renal Principal Cells. Int J Mol Sci. 2022; 23(2):763. [CrossRef]
- KEGG-constructed cAMP signaling pathway. Available on line at https://www.genome.jp/pathway/hsa04024+109 (accessed 10/10/2023).
- Kim, T.H.; Park, J.M.; Kim, M.Y.; Ahn, Y.H. The role of CREB3L4 in the proliferation of prostate cancer cells. Sci Rep. 2017; 7:45300. [CrossRef]
- Giménez-Bachs, J.M.; Salinas-Sánchez, A.S.; Serrano-Oviedo, L.; Nam-Cha, S.H.; Rubio-Del, Campo, A.; Sánchez-Prieto, R. Carbonic anhydrase IX as a specific biomarker for clear cell renal cell carcinoma: comparative study of Western blot and immunohistochemistry and implications for diagnosis. Scand J Urol Nephrol. 2012; 46(5):358-64. [CrossRef]
- Tostain, J.; Li, G.; Gentil-Perret, A.; Gigante, M. Carbonic anhydrase 9 in clear cell renal cell carcinoma: a marker for diagnosis, prognosis and treatment. Eur J Cancer. 2010; 46(18):3141-8. [CrossRef]




| Primary site | # of cases | # of genes | Protein coding | # of mutations | Primary site | # of cases | # of genes | Protein coding | # of mutations | |
|---|---|---|---|---|---|---|---|---|---|---|
| Bladder | 1,725 | 20,183 | 19,692 | 114,662 | Lung | 12,262 | 21,318 | 19,790 | 443,974 | |
| Bone marrow | 11,027 | 21,474 | 19,705 | 163,756 | Ovary | 3,381 | 20,266 | 19,673 | 64,142 | |
| Brain | 1,452 | 20,343 | 19,729 | 93,128 | Pancreas | 2,776 | 19,874 | 19,502 | 36,676 | |
| Breast | 9,121 | 20,454 | 19,727 | 113,777 | Prostate | 2,387 | 19,638 | 19,402 | 27,468 | |
| Colorectal | 8,140 | 21,060 | 19,794 | 337,634 | Skin | 2,893 | 20,739 | 19,770 | 353,213 | |
| Head & neck | 2,792 | 20,535 | 19,712 | 116,274 | Stomach | 1,631 | 20,336 | 19,739 | 182,493 | |
| Kidney | 3,501 | 20,129 | 19,631 | 65,471 | Uterus | 2,803 | 21,471 | 19,781 | 769,622 |
| KEGG pathway | PTA | PTB | CWM | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Code | Genes | % up | % down | WPR | % up | % down | WPR | % up | % down | WPR |
| hsa04960 | 26 | 11.54 | 11.54 | 1.12 | 30.77 | 0.00 | 8.19 | 11.54 | 11.54 | 2.32 |
| hsa04966 | 16 | 6.25 | 12.50 | 5.20 | 12.50 | 0.00 | 16.36 | 12.50 | 0.00 | 9.69 |
| hsa04961 | 37 | 5.41 | 2.70 | 0.88 | 18.92 | 5.41 | 7.68 | 8.11 | 5.41 | 2.15 |
| hsa04964 | 18 | 16.67 | 0.00 | 0.96 | 38.89 | 0.00 | 9.04 | 11.11 | 0.00 | 2.12 |
| hsa04962 | 36 | 8.33 | 11.11 | 0.69 | 11.11 | 5.56 | 0.93 | 2.78 | 8.33 | 0.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





