1. Introduction
Sarcocystis spp. are cyst-forming intracellular protozoan parasites with an obligate two-host life cycle, with predators (definitive hosts) and their prey animals (intermediate hosts). Collectively, these species have considerable veterinary, economic, and public health importance. Presently, classification and identification of
Sarcocystis species in a given hosts mainly depend on the morphological characterization of its sarcocysts and nucleotide sequences of genetic markers [
1].
Sarcocystis cruzi in cattle (
Bos taurus) distributes worldwide, and is considered to be the most pathogenic
Sarcocystis species in the animal [
2].
Sarcocystis poephagicanis is described and named in yaks (
Bos grunniens) [
3], and this livestock is adaptive to high-altitude environments and raised mainly in Qinghai-Tibet Plateau, China, but also in adjacent areas [
4]. The two parasites presented similar morphological characteristics and life cycle (canids as definitive hosts) [
2,
3]. Owing to the relationship between them unclear,
S. poephagicanis in yaks is frequently identified as
S.
cruzi by some authors [
5,
6,
7].
Currently, molecular analysis based on nucleotide sequences has been used to infer phylogenetic relationship of
Sarcocystis species, and recommended to be a more useful and efficient tool for delineating or identifying
Sarcocystis species than the traditional morphological method, especially for those morphologically indistinguishable
Sarcocystis spp. in different hosts [
8,
9]. There are abundant nucleotide sequences of molecular markers, including 18S rDNA, 28S rDNA, and mitochondrial
cox1 genes, of
S. cruzi presently deposited in GenBank as references. However, none of nucleotide sequences of
Sarcocystis spp. in yaks have been investigated and provided in GenBank.
Therefore, the aims of present study were (1) to investigate the prevalence of S. cruzi in cattle and S. poephagicanis in yaks in China based on morphological observation, (2) to analyze the molecular characteristics of four genetic markers, namely 18S rDNA, 28S rDNA, cox1 and apicoplast large subunit ribosomal protein 6 (rpl6) of the two parasites, and (3) to infer phylogenetic relationships of the two species with other Sarcocystis spp. using 28S rDNA and cox1 sequences.
2. Materials and Methods
Muscular tissues obtained from 950 cattle and 320 yaks were separately collected from abattoirs in Kunming city, capital of Yunnan province, and Lhasa city, capital of Tibet autonomous region, both located in southwestern China, during 2021−2023. About 500 g of samples (esophagus, diaphragm, skeletal muscles, tongue, and heart) were obtained from each animal and shipped with dry ice to zoological laboratory of Yunnan university. In the laboratory, approximately 40, 3 mm muscle pieces from each collected sample were compressed between two glass slides to detect the presence of sarcocysts using a stereomicroscopy; Individual sarcocysts were extracted and isolated from muscular fibers using dissection needles and processed for light microscopy (LM), transmission electron microscopy (TEM) and DNA analysis. For observing and measuring bradyzoites filled in sarcocysts, the sarcocysts were punctured using a needle. For TEM, the sarcocysts were fixed in 2.5% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.4) at 4 °C and post-fixed in 1 % osmium tetroxide in the same buffer, then dehydrated in graded alcohols, and embedded in Epon-Alaldite mixture. Ultrathin sections were stained with uranyl acetate and lead citrate and then examined using a JEM100-CX TEM (JEOL Ltd., Tokyo, Japan) at 80 kV. For DNA isolation, individual cysts were stored in sterile water at −20 °C prior to processing.
A total of 12 individual sarcocysts, including six of
S.
cruzi and six of
S.
poephagicanis isolated from different animals, were separately subjected to genomic DNA extraction using a TIANamp Genomic DNA Kit (Tiangen Biotech Ltd., Beijing, China) according to the manufacturer’s instructions. Four genes, namely 18S rRNA, 28S rDNA,
cox1, and
rpl6, were used to characterize the two parasites. The primer pairs used were given in
Table 1. Polymerase chain reaction (PCR) amplifications were performed as previously described [
15]. The resulting PCR products were gel purified using an E.Z.N.A. ® Gel Extraction Kit (Omega Bio-Tek, Inc., Norcross, GA, USA) and ligated to the pCE2 TA/Blunt-Zero vector using a 5 min TA/Blunt-Zero Cloning Kit (Vazyme Biotech Co., Ltd. Nanjing, China) according to the manufacturer’s instructions. The ligated vectors were transformed into Trelief ® 5α Chemically Competent Cell (Tsingke Biotechnology Co., Ltd., Beijing, China). The selected positive bacterial clones were sequenced on both directions by an ABI PRISM TM 3730 XL DNA Analyzer (Applied Bio-systems, Thermo Fisher Scientific, Waltham, MA, USA).
Phylogenetic analyses were conducted separately on the nucleotide sequences of the 28S rDNA, and
cox1 sequences using MEGA X software [
16]. The maximum likelihood (ML) trees of 28S rDNA and
cox1 were generated using Hasegawa–Kishino–Yano and Kimura 2-parameter models, respectively, according to the Find Best DNA/Protein Models program integrated into MEGA X. All sites were used. The reliability of the maximum likelihood phylograms was tested via the bootstrap method using 1000 replications.
Nucleotide sequences of
Sarcocystis spp. used in the investigation were downloaded from GenBank. The 28S rDNA sequences were aligned based on the predicted secondary structure using the multiple sequence alignment algorithm of the “R-Coffee” web server [
17]. The
cox1 sequences were aligned using the program MUSCLE implemented in MEGA X. The alignments were subsequently checked visually; some sequences were slightly truncated at both ends, so that all sequences started and ended at the same nucleotide positions. The final alignment of the 28S rDNA sequences consisted of 27 nucleotide sequences and 3980 positions including gaps from 16 taxa. The final alignment of
cox1 sequences consisted of a total of 33 nucleotide sequences and 1014 positions with no gaps from 25 taxa.
Hammondia heydorni and
Toxoplasma gondii were chosen as outgroup species to root both trees.
3. Results
3.1. Prevalence of S. cruzi in cattle and S. poephagicanis in yaks
Sarcocysts of S. cruzi were found in 405 of 950 (42.6%) cattle and those of S. poephagicanis in 304 of 320 (95.0%) yaks with aid of LM. Among the examined tissues, the highest prevalence of the two parasites were recorded in heart of the two animals, i.e., 40.5% for S. cruzi in cattle and 87.8% for S. poephagicanis in yaks (
Table 2).
3.2. LM and TEM of sarcoysts of S. cruzi and S. poephagicanis
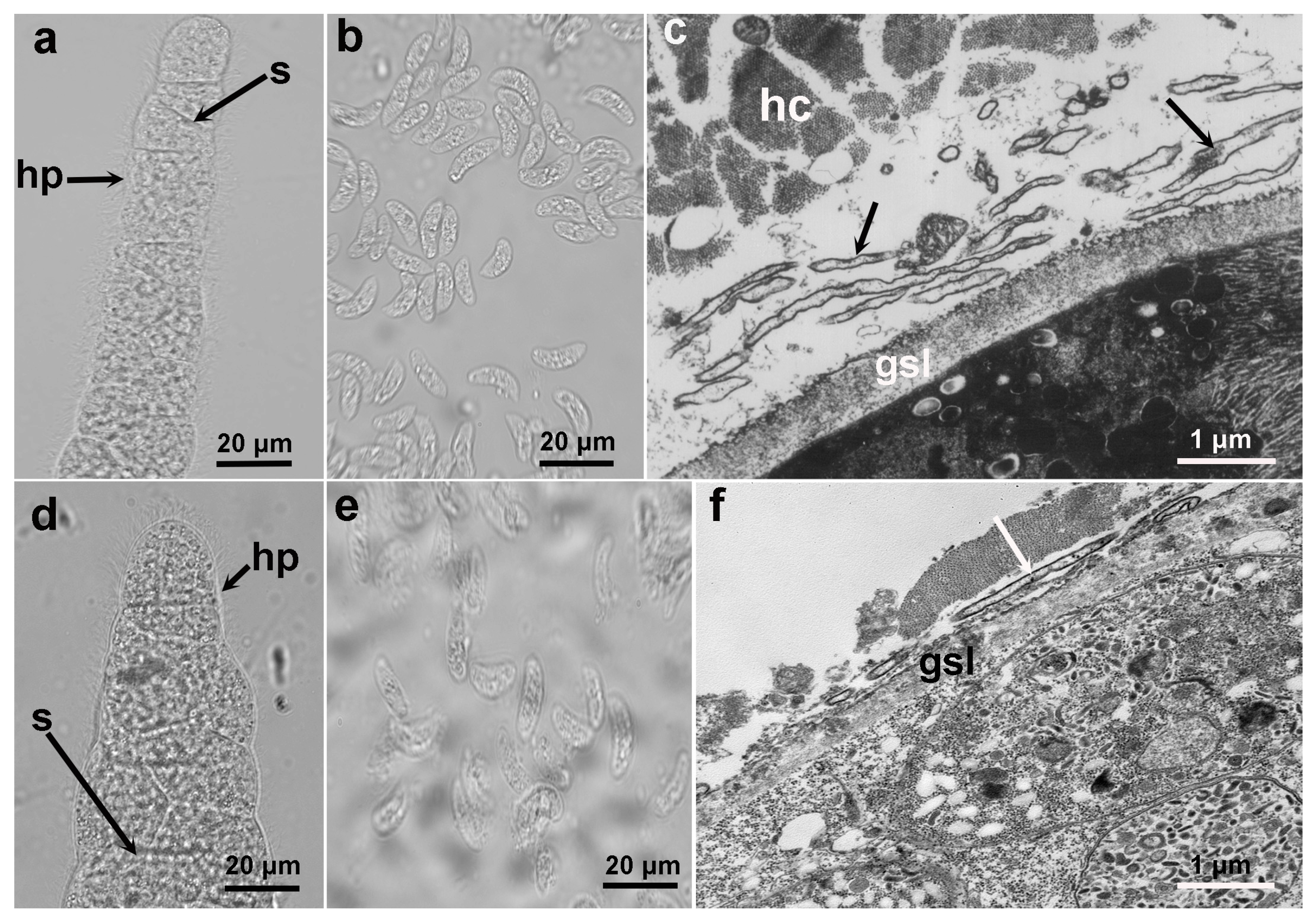
Under LM and TEM, the sarcocysts of S. cruzi and S. poephagicanis presented similar morphological characteristics (
Figure 1). The sarocysts were thin-walled and septate, and had hair-like protrusions on the surface (
Figure 1a, d). Sarcocysts of S. cruzi measured 256−1325 × 24−87 μm, and those of S. poephagicanis were 337−996 × 54−128 μm in size. The bradyzoites filled in the sarcocysts were banana-shaped, measuring 8.9−14.0 × 3.2 −4.8 μm and 8.8−16.0 × 3.2−5.6 μm in size, respectively, for S. cruzi and S. poephagicanis (
Figure 1b, e). Ultrastructuraly, the primary cyst wall contained irregularly folded, hirsute or bone-like protrusions (
Figure 1c, f). A layer of ground substances measuring 0.6−0.8 μm in thickness was located immediately beneath the primary sarcocyst wall.
3.3. Molecular characterization of 18S rDNA, 28S rDNA, cox1, and rpl6
Genomic DNA was extracted from the individual sarcocyst of the two parasites isoalted from different animals. The 18S rDNA, 28S rDNA, cox1, and rpl6 were amplified successfully using their DNAs as templates. Six nucleotide sequences of each gene for the two Sarcocystis species were analyzed in the present study. The 18S rDNA, 28S rDNA, cox1, and rpl6 sequences of S. cruzi were 1857−1869 bp, 3464−3474 bp, 1085 bp, and 864 bp in length, and shared an intraspecific identity of 99.0−100% (on average 99.4%), 98.9−99.7% (on average 99.3%), 98.8−99.9% (on average 99.3%), and.99.7−100% (on average 99.8%), respectively. The S. poephagicanis sequences of the four genes were 1871−1873 bp, 3460−3469 bp, 1085 bp, and 864 bp long, and shared an intraspecific identity of 98.8−100% (on average 99.6%), 98.0−99.6% (on average 98.8%), 98.1−100% (on average 98.8%), and 99.7−100% (on average 99.8%), respectively. Meanwhile, at the four loci, the interspecific identity was 97.9−98.6% (on average 98.3%), 97.2−98.1% (on average 97.7%), 89.5−90.4% (on average 89.9%), and 96.9−97.2% (on average 97.1%), respectively. The newly obtained sequences of the two parasites were deposited in GenBank under accession numbers, OR553288−OR553292, OR573608−OR573623, OR570876−OR570884 and OR590796−OR590800.
Figure 1.
Morphological characteristics of sarcocysts of Sarcocystis cruzi and Sarcocystis poephagicanis obtained from cattle and domestic yaks, respectively. (a) S. cruzi sarcocyst had septae (s) and surrounded by hair-like protrusions (hp) (unstained, light microscopy, LM). (b) Banana-shaped bradyzoites of S. cruzi (unstained, LM). (c) Diagonal section of a sarcocyst of S. cruzi (under transmission electron microscopy, TEM). Sarcocyst surrounded by host cell (hc), and hirsute or bone-like protrusions (arrow) presented on the surface of the ground substance layer (gsl). (d) S. poephagicanis sarcocyst. Note septae (s) and hair-like protrusions (hp) (unstained, LM). (e) Bradyzoites of S. poephagicanis. (f) TEM of a S. poephagicanis sarcocyst. Note hirsute or bone-like protrusions (arrow) and the ground substance layer (gsl).
Figure 1.
Morphological characteristics of sarcocysts of Sarcocystis cruzi and Sarcocystis poephagicanis obtained from cattle and domestic yaks, respectively. (a) S. cruzi sarcocyst had septae (s) and surrounded by hair-like protrusions (hp) (unstained, light microscopy, LM). (b) Banana-shaped bradyzoites of S. cruzi (unstained, LM). (c) Diagonal section of a sarcocyst of S. cruzi (under transmission electron microscopy, TEM). Sarcocyst surrounded by host cell (hc), and hirsute or bone-like protrusions (arrow) presented on the surface of the ground substance layer (gsl). (d) S. poephagicanis sarcocyst. Note septae (s) and hair-like protrusions (hp) (unstained, LM). (e) Bradyzoites of S. poephagicanis. (f) TEM of a S. poephagicanis sarcocyst. Note hirsute or bone-like protrusions (arrow) and the ground substance layer (gsl).
While comparing the newly obatined sequences with those previously deposited in GenBank, at the four loci (18S rDNA, 28S rDNA, cox1 and rp16), the most similar sequences with the S. cruzi sequences were those of S. cruzi (on average 99.3% identity), S. cruzi (on average 99.1% identity), S. cruzi (on average 99.1% identity) and T. gondii (72.7% identity), respectively, and the S. poephagicanis sequences had the highest similarity with those of S. cruzi (on average 98.7% identity), S. levinei (on average 97.5% identity) from water buffalo (Bubalus bubalis), S. rangi (on average 91.8% identity) from reindeer (Rangifer tarandus), and T. gondii (72.8% identity), respectively (
Table 3).
3.4. Phylogenetic Analysis
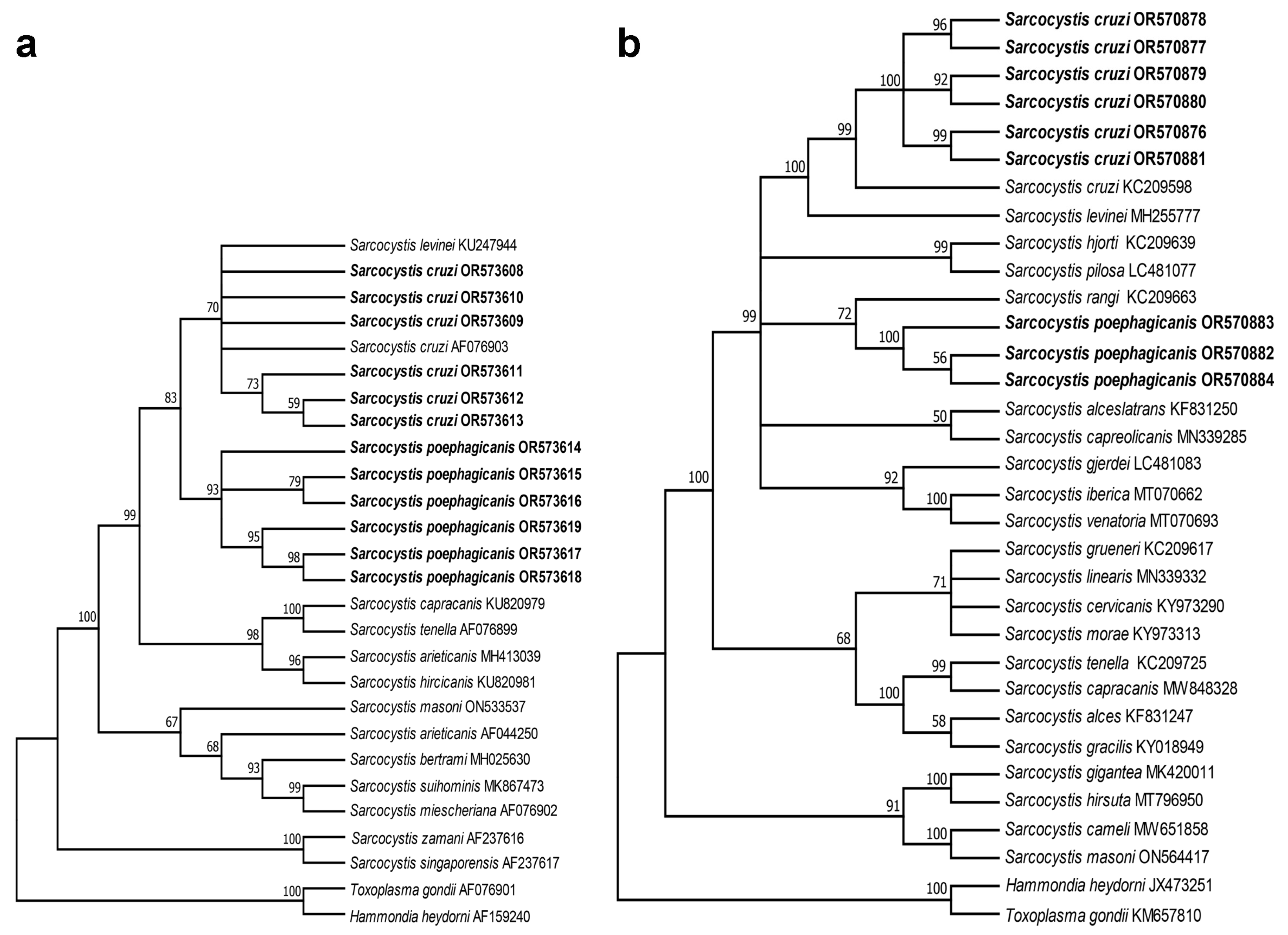
Phylogenetic analysis based on the 28S rDNA and cox1 sequences of S. cruzi and S. poephagicanis showed a similar tree topology (
Figure 2), and they were placed into a group encompassing Sarcocystis spp. in ruminants with canid as known or putative definitive hosts. In the tree inferred from 28S rDNA (
Figure 2a), S. poephagicanis newly-sequenced isolates formed an individual clade clustered with a clade formed by S. cruzi and S. levinei. In the tree inferred from cox1 (
Figure 2b), S. poephagicanis and S. rangi formed a clade, which separated the clade formed by S. cruzi and S. levinei.
4. Discussion
Presntly, at least seven Sarcocystis species are recorded in cattle, namely S. cruzi, S. heydorni, S. bovini, S. hirsuta, S. rommeli, S. hominis, and S. bovifelis [
2]. Although there are some confusions concerning the relationship of the Sarcocystis spp. in cattle, S. cruzi is undoutfully the most indisputable among these speies for its unique morphological features: thin-walled sarcocyst and the surface of its cyst wall covered with hair-like (under LM) or ribbon-like (under TEM) protrusions [
1,
2,
8,
9]. Two Sarcocystis species are discovered and named in yaks, i. e., S. poephagicanis and S. poephagi [
3]. To date, there are only two refereces [
3,
18] provided morphological characterisitics of the two parasites. In the origianl description, sarcocysts of S. poephagicanis are microscopic and thin-walled, and those of S. poephagi are macroscopic and thick-walled under LM. However, the ultrastuctural charactristics of sarcocysts of the two parasits were not accurately detailed. According to the figures provided by Wei et al.[
3,
18], we can observe the primary cyst wall of S. poephaicanis covered with short ribbon-like protrusions, and that of S. poephagi covered with closely packed long villar protrusions, similar type 7a and type 18, respectively, according to the TEM cyst wall type classified by Dubey et al. [
1]. Probabaly owing to the limitaiton of the orignal description for S. poephagicanis, and its high morphologically simialrities with S. cruzi in cattle, the thin-walled sarcocysts in yaks were frequentely regarded as S. cruzi in the epidemiological surveillance of sarcocystosis (mentioned in introduction).
Sarcocystis cruzi has been diagnosed in cattle throughout the world, and prevalence rate of its sarcocysts ranged from 29.6% to 100% [
1]. Currently, almost all accounts concerning Sarcocystis spp. in yaks are reported in China, and the prevalence of sarcocysts ranged from 14.7% to 100% [
3,
5,
6,
7,
18]. Here, with the aid of LM, the prevalence rate of S. cruzi in Chinese cattle was 42.6%, lower than 95.0% for S. poephagicanis in Chinese yaks. The difference in the prevalence rate of the two species may be due to the gradually intensive culture of cattle, but free-range farming of yak still popular in China, which cause unequal opportunities of the livestocks meeting feces of domestic dogs.
Molecular markers have been extensively used to identify Sarcocystis spp. in different animals, and different genetic genes have presented different discriminative abilities. For example, 18S rDNA has been proved unsuitable to distinguish the closely related Sarcocystis spp. in same or different ruminant animals [
19,
20], and cox1 has been recommended more suitable for distingushing the closely related speices of Sarcocystis [
15]. In the presentstudy, the four genetic markers, namely 18S rDNA, 28 rDNA, cox1 and rpl6, of the two parasites were sequenced and analyzed. At the four loci, the similarities between them were 97.9−98.6%, 97.2−98.1%, 89.5−90.4%, and 96.9−97.2% identity, respectively, which indiacted that the four genes could distingush them, and the cox1 and rpl6 were more suitable.
Phylogenetic analysis based on 28S rDNA and cox1 indicated that S. poephagicanis and S. crzui were within a group encompassing Sarcocystis spp. in ruminants with canids definitive hosts. Meanwhile, S. cruzi and S. levinei formed an individual clade separated from S. poephagicanis revealed that S. cruzi had closer relationship with S. levinei than S. poephagicanis. Sarcocysts of S. levinei are morphological undistinguishable from S. cruzi and S. poephagicanis [
1,
3]. The relationship between S. cruzi and S. levine has been resolved, and based on the divergence of their cox1 sequnences, they were supposed to repersent separated species in different hosts [
21].
5. Conclusions
In the present study, sarcocysts of S. cruzi in cattle and S. poephagicanis in yaks were detailed morphologically. Meanwhile, the four genetic markers of the two parasites were sequenced and analyzed, and the sequences of S. poephagicanis constituted the first records of Sarcocystis spp. from yaks in GenBank. Based on molecular analysis, the two morphologically indistinguishable species were supposed to represent separated species in different hosts.
Author Contributions
Conceptualization, J.H.; methodology, K.T.; software, Q.T.; validation, S.D., Y.Y. and Z.Z.; formal analysis, Z.L., M.Z. and Z.W.X.; investigation, K.T and D.L.; resources, K.T and D.L.; data curation, K.T and D.L.; writing—original draft preparation, J.H.; writing—review and editing, J.H. and J.T; supervision, J.H.; project administration, J.H.; funding acquisition, J.H. All authors have read and agreed to the published version of the manuscript.
Funding
Please add: This research was funded by the Natural Science Foundation of China, grant number 31460557, and the Research Project of Tibet Autonomous Region, grant number XZ202301ZY0006N.
Institutional Review Board Statement
The animal study protocol was approved by the Animal Ethics Committee of Yunnan University (permission number AECYU2018004).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dubey, J.P.; Calero-Bernal, R.; Rosenthal, B.M.; Speer, C.A; Fayer, R. Sarcocystosis of animals and humans, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2016; pp. 52–59, 195–214. [Google Scholar]
- Dubey, J.P.; Rosenthal, B.M. Bovine sarcocystosis: Sarcocystis species, diagnosis, prevalence, economic and public health considerations, and association of Sarcocystis species with eosinophilic myositis in cattle. Inter. J. Parasitol. 2023, 53, 625, 463–475. [Google Scholar] [CrossRef]
- Wei, T.; Chang, P.Z.; Dong, M.X.; Wang, X.Y.; Xia, A.Q. Description of two new species of Sarcocystis from the yak (Poeophagus grunniens). Sci. Agric. Sin. 1985, 4, 80–85. (In Chinese) [Google Scholar]
- Barsila, S. , Kreuzer, M.; Devkota, N.R.; Ding, L.; Marquardt, S. Adaptation to Himalayan high altitude pasture sites by yaks and different types of hybrids of yaks with cattle. Livest. Sci. 2014, 169, 125–136. [Google Scholar] [CrossRef]
- Geng, T. Prevalence of Sarcocystis spp. from domestic yaks in Xinghai county, Qinghai province, China. Chin. J. Vet. Med. 2009, 25, 49. (In Chinese) [Google Scholar]
- Zhang, G. Prevalence of Sarcocystis spp. from domestic yaks in Wulan county, Qinghai province, China. Chin. J. Vet. Med. 2010, 46, 41–42. (In Chinese) [Google Scholar]
- Chen, L. Prevalence of Sarcocystis spp. from domestic yaks in Chengduo county, Qinghai province, China. Anim. Husb. Vet. Med. 2012, 44, 111–112. (In Chinese) [Google Scholar]
- Gjerde, B. The resurrection of a species: Sarcocystis bovifelis Heydorn et al., 1975 is distinct from the current Sarcocystis hirsuta in cattle and morphologically indistinguishable from Sarcocystis sinensis in water buffaloes. Parasitol. Res. 2016, 115, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.J.; Huang, S.; Wen, T.; Esch, G.W.; Liang, Y.; Li, H.L. Morphology, molecular characteristics, and demonstration of a definitive host for Sarcocystis rommeli from cattle (Bos taurus) in China. J. Parasitol. 2017, 103, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Barta, J.R.; Martin, D.S.; Liberator, P.A.; Dashkevicz, M.; Anderson, J.W.; Feighner, S.D.; Elbrecht, A.; Perkins-Barrow, A.; Jenkins, M.C.; Danforth, H.D.; Ruff, M.D. , Profous-Juchelka, H. Phylogenetic relationships among eight Eimeria species infecting domestic fowl inferred using complete small subunit ribosomal DNA sequences. J. Parasitol. 1997, 83, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Fenger, C.K.; Granstrom, D.E.; Langemeier, J.L.; Stamper, S.; Donahue, J.M.; Patterson, J.S.; Gajadhar, A.A.; Marteniuk, J.V.; Xiaomin, Z.; Dubey, J.P. Identification of opossums (Didelphis virginiana) as the putative definitive host of Sarcocystis neurona. J. Parasitol. 1995, 81, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Mugridge, N.B.; Morrison, D.A.; Johnson, A.M.; Luton, K.; Dubey, J.P.; Votýpka, J.; Tenter, A.M. Phylogenetic relationships of the genus Frenkelia: A review of its history and new knowledge gained from comparison of large subunit ribosomal ribonucleic acid gene sequences. Int. J. Parasitol. 1999, 29, 957–972. [Google Scholar] [CrossRef] [PubMed]
- Gjerde, B. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int. J. Parasitol. 2013, 43, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Gjerde, B. Sarcocystis species in red deer revisited: With a re-description of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. Sp. based on mitochondrial cox1 sequences. Parasitology. 2014, 141, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Sun, J.; Guo, Y.; Zeng, H.; Zhang, Y; Tao, J. Infection of the Asian gray shrew Crocidura attenuata (Insectivora: Soricidae) with Sarcocystis attenuati n. sp. (Apicomplexa: Sarcocystidae) in China. Parasit. Vectors. 2022, 15, 13. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tommaso, P.D.; Moretti, S.; Xenarios, I.; Orobitg, M.; Montanyola, A.; Chang, J.M. , Taly, J.F.; Notredame, C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic. Acids. Res. 2011, 39 (Web Server issue), W13–17. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, P.C.; Dong, M.X.; Wang, X.Y. Host spectrum of two Sarcocystis species from yak (Poephagus grunniens). J. Chin. Trad. Vet. Med. 1990, 5, 8–10, 29. (In Chinese) [Google Scholar]
- Hu, J.J.; Liu, T.T. ; Liu. Q.; Esch, G.W., Chen, J.Q.; Huang, S.; Wen, T. Prevalence, morphology, and molecular characteristics of Sarcocystis spp. in domestic goats (Capra hircus) from Kunming, China. Parasitol. Res. 2016, 115, 3973–3981. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.J.; Huang, S; Wen, T; Esch, G.W.; Liang, Y.; Li. H.L. Sarcocystis spp. in domestic sheep in Kunming City, China: prevalence, morphology and molecular characteristics. Parasite 2017, 24, 30.
- Gjerde, B.; Hilali, M.; Abbas, I.E. Molecular differentiation of Sarcocystis buffalonis and Sarcocystis levinei in water buffaloes (Bubalus bubalis) from Sarcocystis hirsuta and Sarcocystis cruzi in cattle (Bos taurus). Parasitol. Res. 2016, 115, 2459–2471. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).