Submitted:
06 October 2023
Posted:
09 October 2023
You are already at the latest version
Abstract
Keywords:
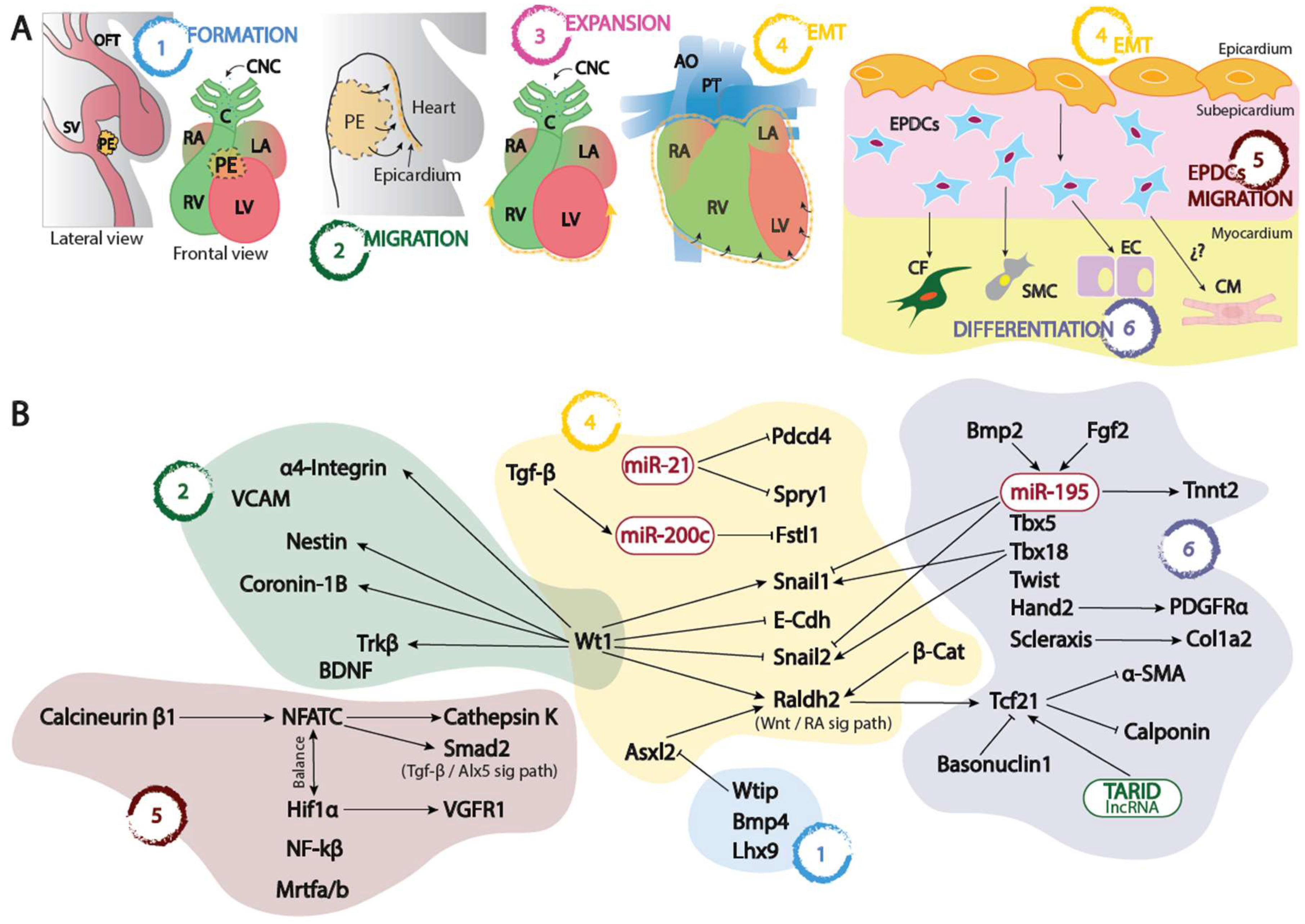
Origin of the embryonic epicardium
Derivates of the embryonic epicardium
Transcriptional regulation of the embryonic epicardium
Proepicardial development
Epicardial migration and epicardial EMT
Invading the myocardium
Differentiation of EPDCs
Other transcription factors involved in epicardial development
Post-transcriptional control of epicardial development
Conclusions and perspectives
References
- Männer, J. The development of pericardial villi in the chick embryo. Anat Embryol (Berl) 1992, 186, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Männer, J. Experimental study on the formation of the epicardium in chick embryos. Anat Embryol (Berl) 1993, 187, 281–289. [Google Scholar] [CrossRef]
- Männer, J. Perez-Pomares, J.M., Macias, D., and Munoz-Chapuli, R. The origin, formation and developmental significance of the epicardium: a review. Cells Tissues Organs 2001, 169, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Männer J, Schlueter J, Brand T. Experimental analyses of the function of the proepicardium using a new microsurgical procedure to induce loss-of-proepicardial-function in chick embryos. Dev Dyn. 2005, 233, 1454–63. [CrossRef] [PubMed]
- Lie-Venema H, van den Akker NM, Bax NA, Winter EM, Maas S, Kekarainen T, Hoeben RC, deRuiter MC, Poelmann RE, Gittenberger-de Groot AC. Origin, fate, and function of epicardium-derived cells (EPDCs) in normal and abnormal cardiac development. Scientific World Journal. 2007, 7, 1777–98. [CrossRef]
- Bax NA, Lie-Venema H, Vicente-Steijn R, Bleyl SB, Van Den Akker NM, Maas S, Poelmann RE, Gittenberger-de Groot AC. Platelet-derived growth factor is involved in the differentiation of second heart field-derived cardiac structures in chicken embryos. Dev Dyn. 2009, 238, 2658–69. [CrossRef]
- Carmona R, Guadix JA, Cano E, Ruiz-Villalba A, Portillo-Sánchez V, Pérez-Pomares JM, Muñoz-Chápuli R. The embryonic epicardium: an essential element of cardiac development. J Cell Mol Med. 2010, 14, 2066–72. [CrossRef]
- Niderla-Bielińska J, Jankowska-Steifer E, Flaht-Zabost A, Gula G, Czarnowska E, Ratajska A. Proepicardium: Current Understanding of its Structure, Induction, and Fate. Anat Rec (Hoboken). 2019, 302, 893–903. [CrossRef]
- Komiyama M, Ito K, Shimada Y. Origin and development of the epicardium in the mouse embryo. Anat Embryol (Berl) 1987, 176, 183–189. [CrossRef]
- Muñoz-Chápuli R, Macías D, González-Iriarte M, Carmona R, Atencia G, Pérez-Pomares JM. The epicardium and epicardial-derived cells: multiple functions in cardiac development. Rev Esp Cardiol. 2002, 55, 1070–82. [CrossRef]
- Balmer GM, Bollini S, Dubé KN, Martinez-Barbera JP, Williams O, Riley PR. Dynamic haematopoietic cell contribution to the developing and adult epicardium. Nat Commun. 2014, 5, 4054. [CrossRef] [PubMed]
- Mommersteeg MT, Domínguez JN, Wiese C, Norden J, de Gier-de Vries C, Burch JB, Kispert A, Brown NA, Moorman AF, Christoffels VM. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc Res. 2010, 87, 92–101. [CrossRef] [PubMed]
- Serluca, F. C. Development of the proepicardial organ in the zebrafish. Dev. Biol. 2008, 315, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Icardo, J. M., Guerrero, A, Durán, A. C., Colvee, E., Domezain, A., and Sans-Coma, V. The development of the epicardiumin the sturgeon Acipenser naccarii. Anat. Rec. (Hoboken). 2009, 292, 1593–1601. [CrossRef]
- Schulte, I., Schlueter, J, Abu-Issa, R., Brand, T., and Männer, J. Morphological and molecular left-right asymmetries in the development of the proepicardium: a comparative analysis on mouse and chick embryos. Dev. Dyn. 2007, 236, 684–695. [CrossRef]
- vanWijk B, van den Berg G, Abu-Issa R, Barnett P, van der Velden S, Schmidt M, Ruijter JM, Kirby ML, Moorman AF, van den Hoff MJ. Epicardium and myocardium separate from a common precursor pool by crosstalk between bone morphogenetic protein- and fibroblast growth factor-signaling pathways. Circ Res 2009, 105, 431–441. [CrossRef]
- Maya-Ramos L, Cleland J, Bressan M, Mikawa T. Induction of the Proepicardium. J Dev Biol. 2013, 1, 82–91. [CrossRef]
- Schlueter J, Brand T. Subpopulation of proepicardial cells is derived from the somatic mesoderm in the chick embryo. Circ Res. 2013, 113, 1128–37. [CrossRef]
- Torlopp A, Schlueter J, Brand T. 2010. Role of fibroblast growth factor signaling during proepicardium formation in the chick embryo. Dev Dyn 239: 2393–2403. [CrossRef]
- Liu J, Stainier DY. Tbx5 and Bmp signaling are essential for proepicardium specification in zebrafish. Circ Res. 2010, 106, 1818–28. [CrossRef]
- Katz TC, Singh MK, Degenhardt K, Rivera-Feliciano J, Johnson RL, Epstein JA, Tabin CJ. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Developmental Cell. 2012, 22, 639–650. [CrossRef]
- Smits AM, Dronkers E, Goumans MJ. The epicardium as a source of multipotent adult cardiac progenitor cells: Their origin, role and fate. Pharmacol Res. 2018, 127, 129–140. [CrossRef]
- Plavicki JS, Hofsteen P, Yue MS, Lanham KA, Peterson RE, Heideman W. Multiple modes of proepicardial cell migration require heartbeat. BMC Dev Biol. 2014 ;14:18. 15 May. [CrossRef]
- Li J, Miao L, Zhao C, Shaikh Qureshi WM, Shieh D, Guo H, Lu Y, Hu S, Huang A, Zhang L, et al. CDC42 is required for epicardial and pro-epicardial development by mediating FGF receptor trafficking to the plasma membrane. Development 2017, 144, 1635–1647. [CrossRef]
- Cao Y, Duca S, Cao J. Epicardium in Heart Development. Cold Spring Harb Perspect Biol. 2020, 12, a037192. [CrossRef]
- Sanchez-Fernandez C, Rodriguez-Outeiriño L, Matias-Valiente L, Ramirez de Acuña F, Hernandez-Torres F, Lozano-Velasco E, Dominguez JN, Franco D, Aranega AE. Regulation of Epicardial Cell Fate during Cardiac Development and Disease: An Overview. Int J Mol Sci. 2022, 23, 3220. [CrossRef]
- Männer, J. Does the subepicardial mesenchyme contribute myocardioblasts to the myocardium of the chick embryo heart?A quail-chick chimera study tracing the fate op the epicardial primordium. Anat. Rec. 1999, 255, 212–226. [Google Scholar] [CrossRef]
- Perez-Pomares, J.M., Phelps, A., Sedmerova, M., and Wessels, A. Epicardial-like cells on the distal arterial end of the cardiac outflow tract do not derive from the proepicardium but are derivatives of the cephalic pericardium. Dev. Dyn. 2003, 227, 56–68. [CrossRef] [PubMed]
- Vrancken Peeters, M.-P.F.M., Gittenberger-de Groot, A.C., Mentink, M.M.T., and Poelmann, R.E. Smooth muscle cells and fibroblasts of the coronary arteries derive from epithelial-mesenchymal transformation of the epicardium. Anat. Embryol. 1999, 199, 367–378. [CrossRef]
- Lie-Venema, H., Eralp, I., Maas, S., Gittenberger-de Groot, A.C., Poelmann, R.E., and DeRuiter, M.C. Myocardial heterogeneity in permissiveness for epicardium-derived cells and endothelial precursor cells along the developing heart tube at the onset of coronary vascularization. Anat. Rec. 2005; 282A, 120–129.
- Gittenberger-de Groot, A.C. , Vrancken Peeters, M. -P.F.M., Mentink, M.M.T., Gourdie, R.G., and Poelmann, R.E. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ. Res. 1998, 82, 1043–1052. [Google Scholar]
- Poelmann, R.E. , Gittenberger-de Groot, A. C., Mentink, M.M.T., Bökenkamp, R., and Hogers, B. Development of the cardiac coronary vascular endothelium, studied with antiendothelial antibodies, in chicken-quail chimeras. Circ. Res. 1993, 73, 559–568. [Google Scholar]
- Poelmann, R.E. , Lie-Venema, H. , and Gittenberger-de Groot, A.C. The role of the epicardium and neural crest as extracardiac contributors to coronary vascular development. Tex. Heart Inst. J. 2002, 29, 255–261. [Google Scholar]
- Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996, 174, 221–32. [CrossRef] [PubMed]
- Wessels A, van den Hoff MJ, Adamo RF, Phelps AL, Lockhart MM, Sauls K, Briggs LE, Norris RA, van Wijk B, Perez-Pomares JM, Dettman RW, Burch JB. Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Dev Biol. 2012, 366, 111–24. [CrossRef]
- Pérez-Pomares JM, Carmona R, González-Iriarte M, Atencia G, Wessels A, Muñoz-Chápuli R. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int J Dev Biol. 2002, 46, 1005–13.
- Carmona R, Barrena S, López Gambero AJ, Rojas A, Muñoz-Chápuli R. Epicardial cell lineages and the origin of the coronary endothelium. FASEB J. 2020, 34, 5223–5239. [CrossRef]
- Cano E, Carmona R, Ruiz-Villalba A, Rojas A, Chau YY, Wagner KD, Wagner N, Hastie ND, Muñoz-Chápuli R, Pérez-Pomares JM. Extracardiac septum transversum/proepicardial endothelial cells pattern embryonic coronary arterio- venous connections. Proc Natl Acad Sci U S A. 2016, 113, 656–61. [CrossRef]
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008, 454, 104–8. [CrossRef]
- Villa Del Campo C, Lioux G, Carmona R, Sierra R, Muñoz-Chápuli R, Clavería C, Torres M. Myc overexpression enhances of epicardial contribution to the developing heart and promotes extensive expansion of the cardiomyocyte population. Sci Rep. 2016 Oct 18;6:35366. doi: 10.1038/srep35366. Erratum in: Sci Rep. 2016 Dec 09;6:37880. PMID: 27752085; PMCID: PMC5082763.
- Christoffels VM, Grieskamp T, Norden J, Mommersteeg MT, Rudat C, Kispert A. Tbx18 and the fate of epicardial progenitors. Nature. 2009, 458, E8-E9; discussion E9-E10. [CrossRef]
- Rudat C, Kispert A. Wt1 and epicardial fate mapping. Circ Res. 2012, 111, 165–9. [CrossRef]
- Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by Developmental reprogramming of venous cells. Nature 2010, 464, 549–553. [CrossRef]
- Chen HI, Sharma B, Akerberg BN, Numi HJ, Kivelä R, Saharinen P, Aghajanian H, McKay AS, Bogard PE, Chang AH, Jacobs AH, Epstein JA, Stankunas K, Alitalo K, Red-Horse K. The sinus venosus contributes to coronary vasculature through VEGFC-stimulated angiogenesis. Development. 2014, 141, 4500–12. [CrossRef]
- Pennisi DJ, Ballard VL, Mikawa T. Epicardium is required for the full rate of myocyte proliferation and levels of expression of myocyte mitogenic factors FGF2 and its receptor, FGFR-1, but not for transmural myocardial patterning in the embryonic chick heart. Dev Dyn. 2003, 228, 161–72. [CrossRef]
- Stuckmann I, Evans S, Lassar AB. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev Biol. 2003, 255, 334–49. [CrossRef] [PubMed]
- Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Izpisua Belmonte JC, Chien KR, Ruiz-Lozano P. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci U S A. 2005, 102, 18455–60. [CrossRef]
- Lavine KJ, Ornitz DM. Fibroblast growth factors and Hedgehogs: at the heart of the epicardial signaling center. Trends Genet. 2008, 24, 33–40. [CrossRef]
- Pennisi DJ, Mikawa T. FGFR-1 is required by epicardium-derived cells for myocardial invasion and correct coronary vascular lineage differentiation. Dev Biol. 2009, 328, 148–59. [CrossRef] [PubMed]
- Cavallero S, Shen H, Yi C, Lien CL, Kumar SR, Sucov HM. CXCL12 Signaling Is Essential for Maturation of the Ventricular Coronary Endothelial Plexus and Establishment of Functional Coronary Circulation. Dev Cell. 2015, 33, 469–77. [CrossRef] [PubMed]
- Smart, N. Bollini, S, K.N. Dubé, J.M. Vieira, B. Zhou, S. Davidson, D. Yellon, J.Riegler, A.N. Price, M.F. Lythgoe, W.T. Pu, P.R. Riley, De novo cardiomyocytes from within the activated adult heart after injury, Nature 474 (2011)640–644. [CrossRef]
- Braitsch, C.M.; Yutzey, K.E. Transcriptional control of cell lineage development in epicardium-derived Cells. J Dev Biol 2013, 1, 92–111. [Google Scholar] [CrossRef]
- Ishii, Y.; Langberg, J.D.; Hurtado, R.; Lee, S.; Mikawa, T. Induction of proepicardial marker gene expression by the liver bud. Development 2007, 134, 3627–37. [Google Scholar] [CrossRef]
- Schlueter, J.; Männer, J.; Brand, T. BMP is an important regulator of proepicardial identity in the chick embryo. Dev Biol 2006, 295, 546–58. [Google Scholar] [CrossRef]
- Tandon, P.; Wilczewski, C.M.; Williams, C.E.; Conlon, F.L. The Lhx9-integrin pathway is essential for positioning of the proepicardial organ. Development 2016, 143, 831–40. [Google Scholar] [CrossRef]
- Pombal MA, Carmona R, Megías M, Ruiz A, Pérez-Pomares JM, Muñoz-Chápuli R. Epicardial development in lamprey supports an evolutionary origin of the vertebrate epicardium from an ancestral pronephric external glomerulus. Evol Dev. 2008, 10, 210–6. [CrossRef] [PubMed]
- Cano, E.; Carmona, R.; Velecela, V.; Martínez-Estrada, O.; Muñoz-Chápuli, R. The proepicardium keeps a potential for glomerular marker expression which supports its evolutionary origin from the pronephros. Evol Dev 2015, 17, 224–30. [Google Scholar] [CrossRef] [PubMed]
- Powell, R.; Bubenshchikova, E.; Fukuyo, Y.; Hsu, C.; Lakiza, O.; Nomura, H.; Renfrew, E.; Garrity, D.; Obara, T. Wtip is required for proepicardial organ specification and cardiac left/right asymmetry in zebrafish. Mol Med Rep 2016, 14, 2665–78. [Google Scholar] [CrossRef] [PubMed]
- Kreidberg, J.A.; Sariola, H.; Loring, J.M.; Maeda, M.; Pelletier, J.; Housman, D.; Jaenisch, R. WT-1 is required for early kidney development. Cell 1993, 74, 679–91. [Google Scholar] [CrossRef]
- Wagner, N.; Wagner, K.D. Every Beat You Take-The Wilms' Tumor Suppressor WT1 and the Heart. Int J Mol Sci 2021, 22, 7675. [Google Scholar] [CrossRef] [PubMed]
- Wagner, N.; Wagner, K.D.; Scholz, H.; Kirschner, K.M.; Schedl, A. Intermediate filament protein nestin is expressed in developing kidney and heart and might be regulated by the Wilms' tumor suppressor Wt1. Am J Physiol Regul Integr Comp Physiol 2006, 291, R779–87. [Google Scholar] [CrossRef]
- Hsu, W.H.; Yu, Y.R.; Hsu, S.H.; Yu, W.C.; Chu, Y.H.; Chen, Y.J.; Chen, C.M.; You, L.R. The Wilms' tumor suppressor Wt1 regulates Coronin 1B expression in the epicardium. Exp Cell Res 2013, 319, 1365–81. [Google Scholar] [CrossRef]
- Wagner N, Wagner KD, Theres H, Englert C, Schedl A, Scholz H. Coronary vessel development requires activation of the TrkB neurotrophin receptor by the Wilms' tumor transcription factor Wt1. Genes Dev. 2005, 19, 2631–42. [CrossRef]
- Martínez-Estrada, O.M.; Lettice, L.A.; Essafi, A.; Guadix, J.A.; Slight, J.; Velecela, V.; Hall, E.; Reichmann, J.; Devenney, P.S.; Hohenstein, P.; Hosen, N.; Hill, R.E.; Muñoz-Chapuli, R.; Hastie, N.D. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat Genet 2010, 42, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Casanova, J.C.; Travisano, S.; de la Pompa, J.L. Epithelial-to-mesenchymal transition in epicardium is independent of Snail1. Genesis 2013, 51, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Takeichi, M.; Nimura, K.; Mori, M.; Nakagami, H.; Kaneda, Y. The transcription factors Tbx18 and Wt1 control the epicardial epithelial-mesenchymal transition through bi-directional regulation of Slug in murine primary epicardial cells. PLoS One 2013, 8, e57829. [Google Scholar] [CrossRef] [PubMed]
- Tao, G.; Miller, L.J.; Lincoln, J. Snai1 is important for avian epicardial cell transformation and motility. Dev Dyn 2013, 242, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Guadix, J.A.; Ruiz-Villalba, A.; Lettice, L.; Velecela, V.; Muñoz-Chápuli, R.; Hastie, N.D.; Pérez-Pomares, J.M.; Martínez-Estrada, O.M. Wt1 controls retinoic acid signaling in embryonic epicardium through transcriptional activation of Raldh2. Development 2011, 138, 1093–7. [Google Scholar] [CrossRef]
- Wang, S.; Yu, J.; Jones, J.W.; Pierzchalski, K.; Kane, M.A.; Trainor, P.A.; Xavier-Neto, J.; Moise, A.R. Retinoic acid signaling promotes the cytoskeletal rearrangement of embryonic epicardial cells. FASEB J 2018, 32, 3765–3781. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, S.J.; Hutson, D.R.; Kubalak, S.W. Analysis of the proepicardium-epicardium transition during the malformation of the RXRalpha-/- epicardium. Dev Dyn 2005, 233, 1091–101. [Google Scholar] [CrossRef]
- Zamora, M.; Männer, J.; Ruiz-Lozano, P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc Natl Acad Sci U S A 2007, 104, 18109–14. [Google Scholar] [CrossRef]
- Von Gise, A.; Zhou, B.; Honor, L.B.; Ma, Q.; Petryk, A.; Pu, W.T. WT1 regulates epicardial epithelial to mesenchymal transition through β-catenin and retinoic acid signaling pathways. Dev Biol 2011, 356, 421–31. [Google Scholar] [CrossRef]
- Khan, F.F.; Li. Y.; Balyan, A.; Wang, Q.T. WTIP interacts with ASXL2 and blocks ASXL2-mediated activation of retinoic acid signaling. Biochem Biophys Res Commun 2014, 451, 101–6. [Google Scholar] [CrossRef]
- Combs, M.D.; Braitsch, C.M.; Lange, A.W.; James, J.F.; Yutzey, K.E. NFATC1 promotes epicardium-derived cell invasion into myocardium. Development 2011, 138, 1747–57. [Google Scholar] [CrossRef]
- Yang, J.; Zeini, M.; Lin, C.Y.; Lin, C.J.; Xiong, Y.; Shang, C.; Han, P.; Li, W.; Quertermous, T.; Zhou, B.; Chang, C.P. Epicardial calcineurin-NFAT signals through Smad2 to direct coronary smooth muscle cell and arterial wall development. Cardiovasc Res 2014, 101, 120–9. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Doughman, Y.; Yang, K.; Ramirez-Bergeron, D.; Watanabe, M. Epicardial HIF signaling regulates vascular precursor cell invasion into the myocardium. Dev Biol 2013, 376, 136–49. [Google Scholar] [CrossRef] [PubMed]
- DeLaughter, D.M.; Clark, C.R.; Christodoulou, D.C.; Seidman, C.E.; Baldwin, H.S.; Seidman, J.G.; Barnett, J.V. Transcriptional Profiling of Cultured, Embryonic Epicardial Cells Identifies Novel Genes and Signaling Pathways Regulated by TGFβR3 In Vitro. PLoS One 2016, 11, e0159710. [Google Scholar] [CrossRef] [PubMed]
- Trembley, M.A.; Velasquez, L.S.; de Mesy Bentley, K.L.; Small, E.M. Myocardin-related transcription factors control the motility of epicardium-derived cells and the maturation of coronary vessels. Development 2015, 142, 21–30. [Google Scholar] [CrossRef]
- Katz, T.C.; Singh, M.K.; Degenhardt, K.; Rivera-Feliciano, J.; Johnson, R.L.; Epstein, J.A.; Tabin, C.J. Distinct compartments of the proepicardial organ give rise to coronary vascular endothelial cells. Dev Cell 2012, 22, 639–50. [Google Scholar] [CrossRef] [PubMed]
- Lupu, I.E.; Redpath, A.N.; Smart, N. Spatiotemporal analysis reveals overlap of key proepicardial markers in the developing murine heart. Stem Cell Reports 2020, 14, 770–787. [Google Scholar] [CrossRef]
- Hu, H.; Lin, S.; Wang, S.; Chen, X. The role of transcription factor 21 in epicardial cell differentiation and the development of coronary heart disease. Front Cell Dev Biol 2020, 8, 457. [Google Scholar] [CrossRef]
- Braitsch, C.M.; Combs, M.D.; Quaggin, S.E.; Yutzey, K.E. Pod1/Tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Dev Biol 2012, 368, 345–57. [Google Scholar] [CrossRef]
- Tandon, P.; Miteva, Y.V.; Kuchenbrod, L.M.; Cristea, I.M.; Conlon, F.L. Tcf21 regulates the specification and maturation of proepicardial cells. Development 2013, 140, 2409–21. [Google Scholar] [CrossRef]
- Boezio, G.L.M.; Zhao, S.; Gollin, J.; Priya, R.; Mansingh, S.; Guenther, S.; Fukuda, N.; Gunawan, F.; Stainier, D.Y.R. The developing epicardium regulates cardiac chamber morphogenesis by promoting cardiomyocyte growth. Dis Model Mech 2023, 16, dmm049571. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, L.; McManus, S.A.; Moignard, V.; Sebukhan, D.; Delaune, A.; Andrews, S.; Bernard, W.G.; Morrison, M.A.; Riley, P.R.; Göttgens, B.; Gambardella Le Novère, N.; Sinha, S. BNC1 regulates cell heterogeneity in human pluripotent stem cell-derived epicardium. Development 2019, 146, dev174441. [Google Scholar] [CrossRef] [PubMed]
- Greulich, F.; Farin, H.F.; Schuster-Gossler, K.; Kispert, A. Tbx18 function in epicardial development. Cardiovasc Res 2012, 96, 476–83. [Google Scholar] [CrossRef] [PubMed]
- Greulich, F.; Rudat, C.; Farin, H.F.; Christoffels, V.M.; Kispert, A. Lack of genetic Interaction between Tbx18 and Tbx2/Tbx20 in mouse epicardial development. PLoS One 2016, 11, e0156787. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Gao, Y.; Xiao, S.; Qin, Q.; Wei, X.; Yan, Y.; Wu, L.; Deng, S.; Du, J.; Liu, Y.; She, Q. Hypoxia induced the differentiation of Tbx18-positive epicardial cells to CoSMCs. Sci Rep 2016, 6, 30468. [Google Scholar] [CrossRef] [PubMed]
- Diman, N.Y.; Brooks, G.; Kruithof, B.P.; Elemento, O.; Seidman, J.G.; Seidman, C.E.; Basson, C.T.; Hatcher, C.J. Tbx5 is required for avian and mammalian epicardial formation and coronary vasculogenesis. Circ Res 2014, 115, 834–44. [Google Scholar] [CrossRef] [PubMed]
- Shelton, E.L.; Yutzey, K.E. Twist1 function in endocardial cushion cell proliferation, migration, and differentiation during heart valve development. Dev Biol 2008, 317, 282–95. [Google Scholar] [CrossRef]
- Levay, A.K.; Peacock, J.D.; Lu, Y.; Koch, M.; Hinton, R.B. Jr; Kadler, K.E.; Lincoln, J. Scleraxis is required for cell lineage differentiation and extracellular matrix remodeling during murine heart valve formation in vivo. Circ Res 2008, 103, 948–56. [Google Scholar] [CrossRef]
- Espira, L.; Lamoureux, L.; Jones, S.C.; Gerard, R.D.; Dixon, I.M.; Czubryt, M.P. The basic helix-loop-helix transcription factor scleraxis regulates fibroblast collagen synthesis. J Mol Cell Cardiol 2009, 47, 188–95. [Google Scholar] [CrossRef]
- Barnes, R.M.; Firulli, B.A.; VanDusen, N.J.; Morikawa, Y.; Conway, S.J.; Cserjesi, P.; Vincentz, J.W.; Firulli, A.B. Hand2 loss-of-function in Hand1-expressing cells reveals distinct roles in epicardial and coronary vessel development. Circ Res 2011, 108, 940–9. [Google Scholar] [CrossRef]
- Smith, C.L.; Baek, S.T.; Sung, C.Y.; Tallquist, M.D. Epicardial-derived cell epithelial-to-mesenchymal transition and fate specification require PDGF receptor signaling. Circ Res 2011, 108, e15–26. [Google Scholar] [CrossRef] [PubMed]
- Kolander, K.D.; Holtz, M.L.; Cossette, S.M.; Duncan, S.A.; Misra, R.P. Epicardial GATA factors regulate early coronary vascular plexus formation. Dev Biol 2014, 386, 204–15. [Google Scholar] [CrossRef] [PubMed]
- Borok, M.J.; Papaioannou, V.E.; Sussel, L. Unique functions of Gata4 in mouse liver induction and heart development. Dev Biol 2016, 410, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Chu, M.; Wang, L.; Wang, H.; Shen, T.; Yang, Y.; Sun, Y.; Tang, N.; Ni, T.; Zhu, J.; Mailman, R.B.; Wang, Y. A novel role of CDX1 in embryonic epicardial development. PLoS One 2014, 9, e103271. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Hill, M.C.; Zhang, M.; Martin, T.J.; Morikawa, Y.; Wang, S.; Moise, A.R.; Wythe, J.D.; Martin, J.F. Hippo signaling plays an essential role in cell state transitions during cardiac fibroblast development. Dev Cell 2018, 45, 153–169. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Liu, B.; Deng, S.; Du, J.; She, Q. Agrin Yes-associated Protein Promotes the Proliferation of Epicardial Cells. J Cardiovasc Pharmacol 2021, 77, 94–99. [Google Scholar] [CrossRef]
- Astanina, E.; Doronzo, G.; Corà, D.; Neri, F.; Oliviero, S.; Genova, T.; Mussano, F.; Middonti, E.; Vallariello, E.; Cencioni, C.; Valdembri, D.; Serini, G.; Limana, F.; Foglio, E.; Ballabio, A.; Bussolino, F. The TFEB-TGIF1 axis regulates EMT in mouse epicardial cells. Nat Commun 2022, 13, 5191. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, 17–29. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL, Chen R, Dean C, Dinger ME, Fitzgerald KA, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. 2023, 24, 430–447. [CrossRef]
- Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021, 22, 96–118. [CrossRef]
- Liu CX, Chen LL. Circular RNAs: Characterization, cellular roles, and applications. Cell. 2022, 185, 2016–2034. [CrossRef] [PubMed]
- Lee YS, Dutta A. MicroRNAs in cancer. 2009;4:199-227. Annu Rev Pathol. 2009, 4, 199–227. [CrossRef]
- Xu J, Wu KJ, Jia QJ, Ding XF. Roles of miRNA and lncRNA in triple-negative breast cancer. J Zhejiang Univ Sci B. 2020, 21, 673–689. [CrossRef] [PubMed]
- Wu KL, Tsai YM, Lien CT, Kuo PL, Hung AJ. The Roles of MicroRNA in Lung Cancer. Int J Mol Sci. 2019, 20, 1611. [CrossRef]
- Wojciechowska A, Braniewska A, Kozar-Kamińska K. MicroRNA in cardiovascular biology and disease. Adv Clin Exp Med. 2017, 26, 865–874. [CrossRef] [PubMed]
- Huang, Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med. 2018, 22, 5768–5775. [Google Scholar] [CrossRef]
- Gomes CPC, Schroen B, Kuster GM, Robinson EL, Ford K, Squire IB, Heymans S, Martelli F, Emanueli C, Devaux Y; EU-CardioRNA COST Action (CA17129). Regulatory RNAs in Heart Failure. Circulation. 2020, 141, 313–328. [CrossRef]
- Chinchilla A, Lozano E, Daimi H, Esteban FJ, Crist C, Aranega AE, Franco D. MicroRNA profiling during mouse ventricular maturation: a role for miR-27 modulating Mef2c expression. Cardiovasc Res. 2011, 89, 98–108. [CrossRef] [PubMed]
- Bonet F, Hernandez-Torres F, Esteban FJ, Aranega A, Franco D. Comparative Analyses of MicroRNA Microarrays during Cardiogenesis: Functional Perspectives. Microarrays (Basel). 2013, 2, 81–96. [CrossRef]
- Lozano-Velasco E, Garcia-Padilla C, Del Mar Muñoz-Gallardo M, Martinez-Amaro FJ, Caño-Carrillo S, Castillo-Casas JM, Sanchez-Fernandez C, Aranega AE, Franco D. Post-Transcriptional Regulation of Molecular Determinants during Cardiogenesis. Int J Mol Sci. 2022, 23, 2839. [CrossRef]
- Caño-Carrillo S, Lozano-Velasco E, Castillo-Casas JM, Sánchez-Fernández C, Franco D. The Role of ncRNAs in Cardiac Infarction and Regeneration. J Cardiovasc Dev Dis. 2023, 10, 123. [CrossRef]
- Fang Y, Xu Y, Wang R, Hu L, Guo D, Xue F, Guo W, Zhang D, Hu J, Li Y, Zhang W, Zhang M. Recent advances on the roles of LncRNAs in cardiovascular disease. J Cell Mol Med. 2020, 24, 12246–12257. [CrossRef]
- Franco D, Aranega A, Dominguez JN. Non-coding RNAs and Atrial Fibrillation. Adv Exp Med Biol. 2020;1229:311-325. [CrossRef] [PubMed]
- Garcia-Padilla C, Lozano-Velasco E, Garcia-Lopez V, Aranega A, Franco D, Garcia-Martinez V, Lopez-Sanchez C. Comparative Analysis of Non-Coding RNA Transcriptomics in Heart Failure. Biomedicines. 2022, 10, 3076. [CrossRef]
- Zeng Y, Wu N, Zhang Z, Zhong L, Li G, Li Y. Non-coding RNA and arrhythmias: expression, function, and molecular mechanism. Europace. 2023, 25, 1296–1308. [CrossRef]
- Dueñas A, Expósito A, Aranega A, Franco D. The Role of Non-Coding RNA in Congenital Heart Diseases. J Cardiovasc Dev Dis. 2019, 6, 15. [CrossRef]
- Singh MK, Lu MM, Massera D, Epstein JA. MicroRNA-processing enzyme Dicer is required in epicardium for coronary vasculature development. J Biol Chem. 2011, 286, 41036–45. [CrossRef]
- Expósito-Villén A, E Aránega A, Franco D. Functional Role of Non-Coding RNAs during Epithelial-To-Mesenchymal Transition. Noncoding RNA. 2018, 4, 14. [CrossRef]
- Zou XZ, Liu T, Gong ZC, Hu CP, Zhang Z. MicroRNAs-mediated epithelial- mesenchymal transition in fibrotic diseases. Eur J Pharmacol. 2017 Feb 5;796:190-206. [CrossRef] [PubMed]
- Vettori S, Gay S, Distler O. Role of MicroRNAs in Fibrosis. Open Rheumatol J. 2012;6:130-9. [CrossRef]
- Gabisonia K, Prosdocimo G, Aquaro GD, Carlucci L, Zentilin L, Secco I, Ali H, Braga L, Gorgodze N, Bernini F, Burchielli S, Collesi C, Zandonà L, Sinagra G, Piacenti M, Zacchigna S, Bussani R, Recchia FA, Giacca M. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature. 2019, 569, 418–422. [CrossRef]
- Gao L, Qiu F, Cao H, Li H, Dai G, Ma T, Gong Y, Luo W, Zhu D, Qiu Z, Zhu P, Chu S, Yang H, Liu Z. Therapeutic delivery of microRNA-125a-5p oligonucleotides improves recovery from myocardial ischemia/reperfusion injury in mice and swine. Theranostics. 2023, 13, 685–703. [CrossRef] [PubMed]
- Wadley GD, Lamon S, Alexander SE, McMullen JR, Bernardo BC. Noncoding RNAs regulating cardiac muscle mass. J Appl Physiol (1985). 2019, 127, 633–644. [CrossRef] [PubMed]
- Qu S, Zeng C, Wang WE. Noncoding RNA and Cardiomyocyte Proliferation. Stem Cells Int. 2017;2017:6825427. [CrossRef]
- Abbas N, Perbellini F, Thum T. Non-coding RNAs: emerging players in cardiomyocyte proliferation and cardiac regeneration. Basic Res Cardiol. 2020, 115, 52. [CrossRef]
- Brønnum H, Andersen DC, Schneider M, Sandberg MB, Eskildsen T, Nielsen SB, Kalluri R, Sheikh SP. miR-21 promotes fibrogenic epithelial-to-mesenchymal transition of epicardial mesothelial cells involving Programmed Cell Death 4 and Sprouty-1. PLoS One. 2013, 8, e56280. [CrossRef]
- Pontemezzo E, Foglio E, Vernucci E, Magenta A, D'Agostino M, Sileno S, Astanina E, Bussolino F, Pellegrini L, Germani A, Russo MA, Limana . miR-200c-3p Regulates Epitelial-to-Mesenchymal Transition in Epicardial Mesothelial Cells by Targeting Epicardial Follistatin-Related Protein 1. Int J Mol Sci. 2021, 22, 4971. [CrossRef]
- Takahashi M, Yamagishi T, Narematsu M, Kamimura T, Kai M, Nakajima Y. Epicardium is required for sarcomeric maturation and cardiomyocyte growth in the ventricular compact layer mediated by transforming growth factor β and fibroblast growth factor before the onset of coronary circulation. Congenit Anom (Kyoto). 2014, 54, 162–71. [CrossRef] [PubMed]
- Jang J, Song G, Pettit SM, Li Q, Song X, Cai CL, Kaushal S, Li D. Epicardial HDAC3 Promotes Myocardial Growth Through a Novel MicroRNA Pathway. Circ Res. 2022, 131, 151–164. [CrossRef]
- Kruithof BP, van Wijk B, Somi S, Kruithof-de Julio M, Pérez Pomares JM, Weesie F, Wessels A, Moorman AF, van den Hoff MJ. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Dev Biol. 2006, 295, 507–22. [CrossRef] [PubMed]
- Smart N, Bollini S, Dubé KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, Pu WT, Riley PR. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011, 474, 640–4. [CrossRef]
- Dueñas A, Expósito A, Muñoz MDM, de Manuel MJ, Cámara-Morales A, Serrano-Osorio F, García-Padilla C, Hernández-Torres F, Domínguez JN, Aránega A, Franco D. MiR-195 enhances cardiomyogenic differentiation of the proepicardium/septum transversum by Smurf1 and Foxp1 modulation. Sci Rep. 2020, 10, 9334. [CrossRef]
- Garcia-Padilla C, Hernandez-Torres F, Lozano-Velasco E, Dueñas A, Muñoz-Gallardo MDM, Garcia-Valencia IS, Palencia-Vincent L, Aranega A, Franco D. The Role of Bmp- and Fgf Signaling Modulating Mouse Proepicardium Cell Fate. Front Cell Dev Biol. 2022 Jan 4;9:757781. [CrossRef]
- Boezio GLM, Zhao S, Gollin J, Priya R, Mansingh S, Guenther S, Fukuda N, Gunawan F, Stainier DYR. The developing epicardium regulates cardiac chamber morphogenesis by promoting cardiomyocyte growth. Dis Model Mech. 2023, 16, dmm049571. [CrossRef] [PubMed]
- Sun J, Peterson EA, Wang AZ, Ou J, Smith KE, Poss KD, Wang J. <i>hapln1</i> Defines an Epicardial Cell Subpopulation Required for Cardiomyocyte Expansion During Heart Morphogenesis and Regeneration. Circulation. 2022, 146, 48–63. [CrossRef]
- Huang Y, Harrison MR, Osorio A, Kim J, Baugh A, Duan C, Sucov HM, Lien CL. Igf Signaling is Required for Cardiomyocyte Proliferation during Zebrafish Heart Development and Regeneration. PLoS One. 2013, 8, e67266. [CrossRef]
- Itou J, Akiyama R, Pehoski S, Yu X, Kawakami H, Kawakami Y. Regenerative responses after mild heart injuries for cardiomyocyte proliferation in zebrafish. Dev Dyn. 2014, 243, 1477–86. [CrossRef]
- Del Campo CV, Liaw NY, Gunadasa-Rohling M, Matthaei M, Braga L, Kennedy T, Salinas G, Voigt N, Giacca M, Zimmermann WH, Riley PR. Regenerative potential of epicardium-derived extracellular vesicles mediated by conserved miRNA transfer. Cardiovasc Res. 2022, 118, 597–611. [CrossRef]
- Zhu D, Liu S, Huang K, Li J, Mei X, Li Z, Cheng K. Intrapericardial long non-coding RNA-Tcf21 antisense RNA inducing demethylation administration promotes cardiac repair. Eur Heart J. 2023, 44, 1748–1760. [CrossRef] [PubMed]
- Sanchez-Fernandez, C. Rodriguez-Outeiriño, L. Matias-Valiente, L. de Acuña, F.R., Franco, D. Aránega, A.E. Understanding Epicardial Cell Heterogeneity during Cardiogenesis and Heart Regeneration. J. Cardiovasc. Dev. Dis. 2023, 10, 376. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




