Submitted:
04 October 2023
Posted:
06 October 2023
You are already at the latest version
Abstract
Keywords:
Introduction
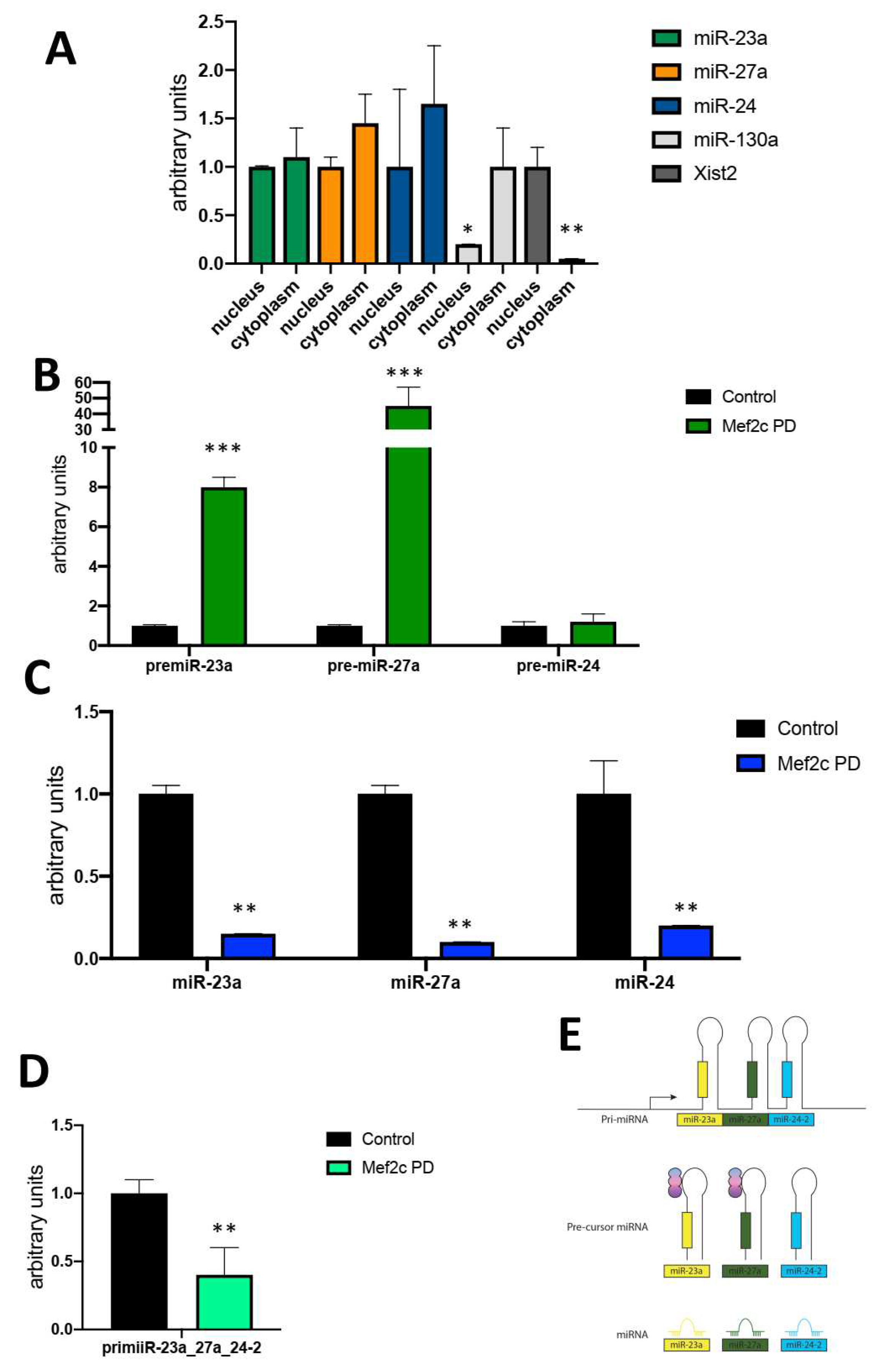
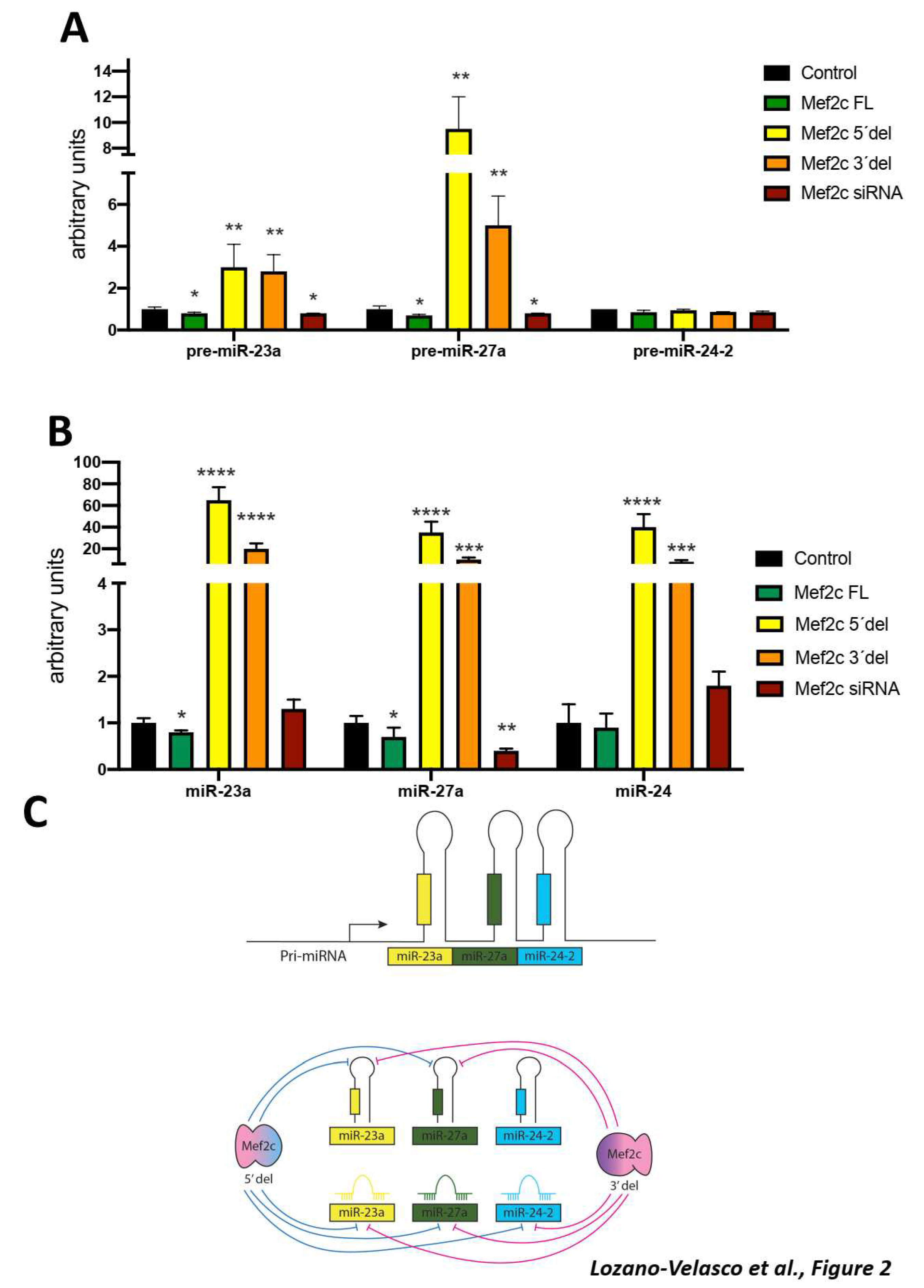
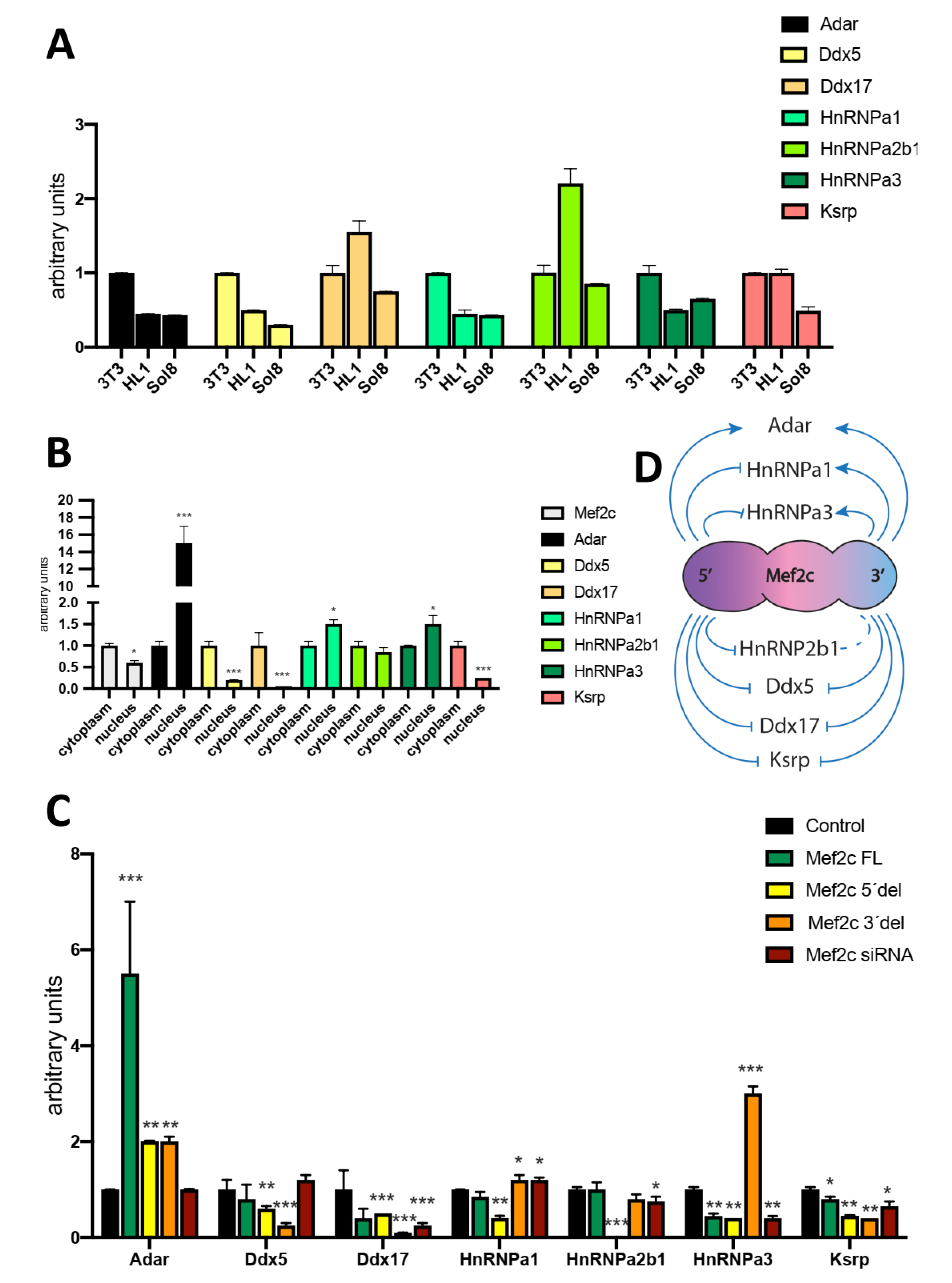
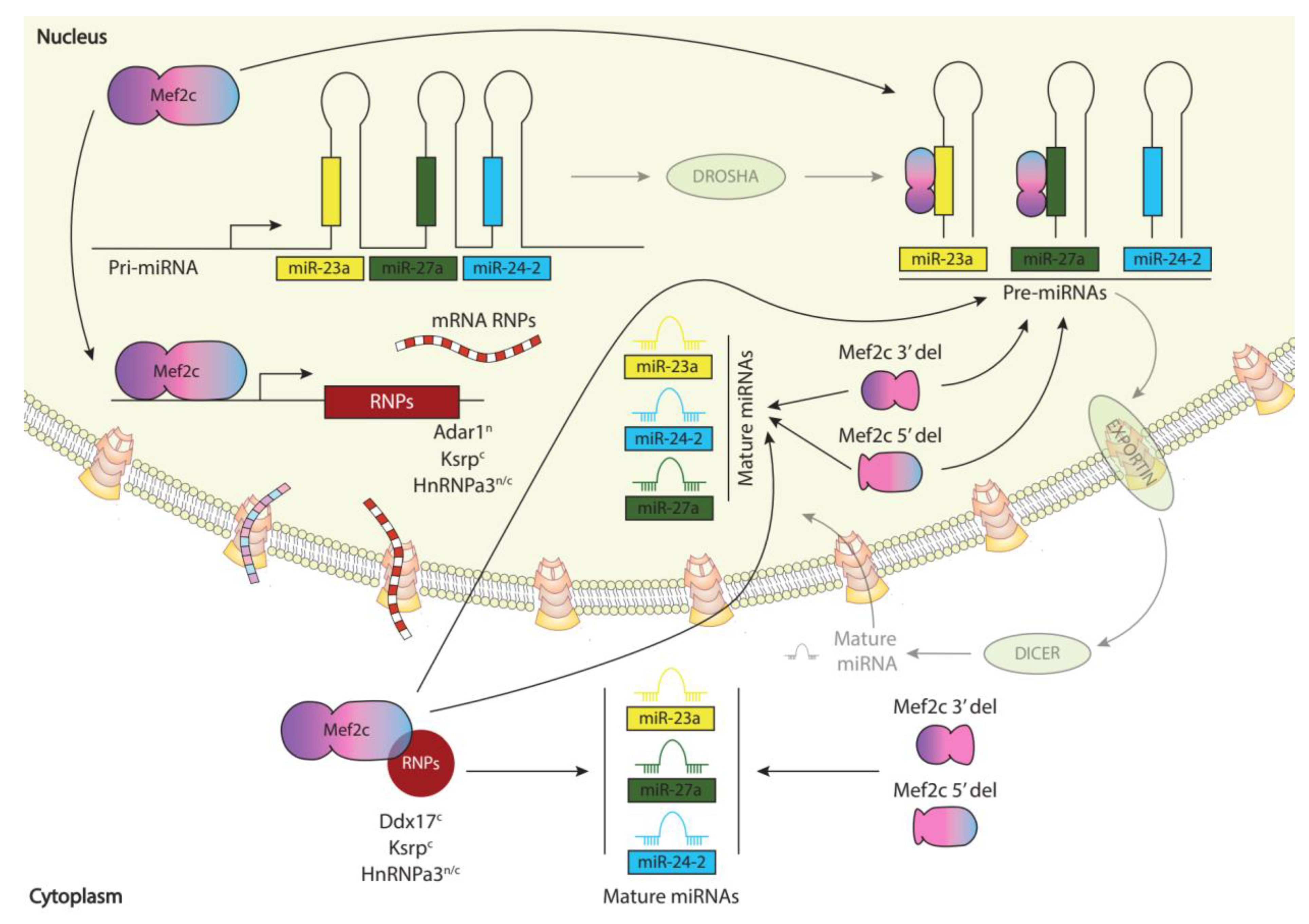
Results
Discussion
Materials & Methods
MEF2C pulldown assays
Nuclear/cytoplasmic distribution
Generation of MEF2C 3´delection and 5´delection constructs
Plasmid transfections
siRNA transfections
RNA isolation and retrotranscription
qRT-PCR analyses (mRNA)
qRT-PCR analyses (microRNA)
Statistical analyses
Supplementary Materials
Acknowledgments
References
- Wilkinson, A.C.; Göttgens, B. Transcriptional regulation of haematopoietic stem cells. Adv Exp Med Biol. 2013, 786, 187–212. [Google Scholar] [CrossRef] [PubMed]
- Bolte, C.; Whitsett, J.A.; Kalin, T.V.; Kalinichenko, V.V. Transcription Factors Regulating Embryonic Development of Pulmonary Vasculature. Adv Anat Embryol Cell Biol. 2018, 228, 1–20. [Google Scholar] [CrossRef]
- Dias, S.; Xu, W.; McGregor, S.; Kee, B. Transcriptional regulation of lymphocyte development. Curr. Opin. Genet. Dev. 2008, 18, 441–448. [Google Scholar] [CrossRef]
- Paige, S.L.; Plonowska, K.; Xu, A.; Wu, S.M. Molecular Regulation of Cardiomyocyte Differentiation. Circ. Res. 2015, 116, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Nagel, S.; Venturini, L.; Meyer, C.; Kaufmann, M.; Scherr, M.; Drexler, H.G.; Macleod, R.A.F. Transcriptional deregulation of oncogenic myocyte enhancer factor 2C in T-cell acute lymphoblastic leukemia. Leuk. Lymphoma 2011, 52, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Chan, J.K.L.; Zhu, G.; Wu, Z. Myocyte Enhancer Factor 2 Acetylation by p300 Enhances Its DNA Binding Activity, Transcriptional Activity, and Myogenic Differentiation. Mol. Cell. Biol. 2005, 25, 3575–3582. [Google Scholar] [CrossRef] [PubMed]
- Sartorelli, V.; Huang, J.; Hamamori, Y.; Kedes, L. Molecular Mechanisms of Myogenic Coactivation by p300: Direct Interaction with the Activation Domain of MyoD and with the MADS Box of MEF2C. Mol. Cell. Biol. 1997, 17, 1010–1026. [Google Scholar] [CrossRef]
- Han, J.; Jiang, Y.; Li, Z.; Kravchenko, V.V.; Ulevitch, R.J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 1997, 386, 296–299. [Google Scholar] [CrossRef]
- Liu, D.; Kang, J.S.; Derynck, R. TGF-β-activated Smad3 represses MEF2-dependent transcription in myogenic differentiation. EMBO J. 2004, 23, 1557–1566. [Google Scholar] [CrossRef]
- Aude-Garcia, C.; Collin-Faure, V.; Bausinger, H.; Hanau, D.; Rabilloud, T.; Lemercier, C. Dual roles for MEF2A and MEF2D during human macrophage terminal differentiation and c-Jun expression. Biochem. J. 2010, 430, 237–244. [Google Scholar] [CrossRef]
- Lyons, M.R.; Schwarz, C.M.; West, A.E. Members of the Myocyte Enhancer Factor 2 Transcription Factor Family Differentially RegulateBdnfTranscription in Response to Neuronal Depolarization. J. Neurosci. 2012, 32, 12780–12785. [Google Scholar] [CrossRef]
- Lin, Q.; Schwarz, J.; Bucana, C.; Olson, E.N. Control of Mouse Cardiac Morphogenesis and Myogenesis by Transcription Factor MEF2C. Science 1997, 276, 1404–1407. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.; Drake, C.J.; Schwarz, J.J. The Transcription Factor MEF2C-Null Mouse Exhibits Complex Vascular Malformations and Reduced Cardiac Expression of Angiopoietin 1 and VEGF. Dev. Biol. 1999, 211, 255–267. [Google Scholar] [CrossRef]
- Kolodziejczyk, S.M.; Wang, L.; Balazsi, K.; DeRepentigny, Y.; Kothary, R.; Megeney, L.A. MEF2 is upregulated during cardiac hypertrophy and is required for normal post-natal growth of the myocardium. Curr. Biol. 1999, 9, 1203–1206. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Nam, Y.-J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012, 485, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.-D.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593–598. [Google Scholar] [CrossRef]
- Voronova, A.; Al Madhoun, A.; Fischer, A.; Shelton, M.; Karamboulas, C.; Skerjanc, I.S. Gli2 and MEF2C activate each other's expression and function synergistically during cardiomyogenesis in vitro. Nucleic Acids Res. 2011, 40, 3329–3347. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, M.M.; Wirrig, E.E.; Phelps, A.L.; Ghatnekar, A.V.; Barth, J.L.; Norris, R.A.; Wessels, A. Mef2c Regulates Transcription of the Extracellular Matrix Protein Cartilage Link Protein 1 in the Developing Murine Heart. PLOS ONE 2013, 8, e57073. [Google Scholar] [CrossRef]
- Vanpoucke, G.; Goossens, S.; De Craene, B.; Gilbert, B.; Van Roy, F.; Berx, G. GATA-4 and MEF2C transcription factors control the tissue-specific expression of the T-catenin gene CTNNA3. Nucleic Acids Res. 2004, 32, 4155–4165. [Google Scholar] [CrossRef]
- Di Lisi, R.; Millino, C.; Calabria, E.; Altruda, F.; Schiaffino, S.; Ausoni, S. Combinatorial cis-Acting Elements Control Tissue-specific Activation of the Cardiac Troponin I Gene in Vitro and in Vivo. J. Biol. Chem. 1998, 273, 25371–25380. [Google Scholar] [CrossRef]
- Zang, M.; Li, Y.; Xue, L.; Jia, H.; Jing, H. Cooperative activation of atrial naturetic peptide promoter by dHAND and MEF2C. J. Cell. Biochem. 2004, 93, 1255–1266. [Google Scholar] [CrossRef]
- Kuisk, I.R.; Li, H.; Tran, D.; Capetanaki, Y. A Single MEF2 Site Governs Desmin Transcription in Both Heart and Skeletal Muscle during Mouse Embryogenesis. Dev. Biol. 1996, 174, 1–13. [Google Scholar] [CrossRef]
- Chen, S.L.; Wang, S.-C.M.; Hosking, B.; Muscat, G.E.O. Subcellular Localization of the Steroid Receptor Coactivators (SRCs) and MEF2 in Muscle and Rhabdomyosarcoma Cells. Mol. Endocrinol. 2001, 15, 783–796. [Google Scholar] [CrossRef] [PubMed]
- Sartorelli, V.; Huang, J.; Hamamori, Y.; Kedes, L. Molecular Mechanisms of Myogenic Coactivation by p300: Direct Interaction with the Activation Domain of MyoD and with the MADS Box of MEF2C. Mol. Cell. Biol. 1997, 17, 1010–1026. [Google Scholar] [CrossRef]
- Pagiatakis, C.; Gordon, J.W.; Ehyai, S.; McDermott, J.C. A Novel RhoA/ROCK-CPI-17-MEF2C Signaling Pathway Regulates Vascular Smooth Muscle Cell Gene Expression. J. Biol. Chem. 2012, 287, 8361–8370. [Google Scholar] [CrossRef] [PubMed]
- Grégoire, S.; Yang, X.-J. Association with Class IIa Histone Deacetylases Upregulates the Sumoylation of MEF2 Transcription Factors. Mol. Cell. Biol. 2005, 25, 2273–2287. [Google Scholar] [CrossRef] [PubMed]
- Molkentin, J.D.; Black, B.L.; Martin, J.F.; Olson, E.N. Mutational Analysis of the DNA Binding, Dimerization, and Transcriptional Activation Domains of MEF2C. Mol. Cell. Biol. 1996, 16, 2627–2636. [Google Scholar] [CrossRef] [PubMed]
- Janson, C.; Chen, Y.; Li, Y.; Leifer, D. Functional regulatory regions of human transcription factor MEF2C. Mol. Brain Res. 2001, 97, 70–82. [Google Scholar] [CrossRef]
- Dong, C.; Yang, X.-Z.; Zhang, C.-Y.; Liu, Y.-Y.; Zhou, R.-B.; Cheng, Q.-D.; Yan, E.-K.; Yin, D.-C. Myocyte enhancer factor 2C and its directly-interacting proteins: A review. Prog. Biophys. Mol. Biol. 2017, 126, 22–30. [Google Scholar] [CrossRef]
- Infantino, V.; Convertini, P.; Menga, A.; Iacobazzi, V. MEF2C exon α: Role in gene activation and differentiation. Gene 2013, 531, 355–362. [Google Scholar] [CrossRef]
- Hombach S, Kretz M. Non-coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol. 2016, 937, 3–17. [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowska, A.; Braniewska, A.; Kozar-Kamińska, K. MicroRNA in cardiovascular biology and disease. Adv. Clin. Exp. Med. 2017, 26, 868–874. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, D.-Z. microRNAs in cardiovascular development. J. Mol. Cell. Cardiol. 2012, 52, 949–957. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.-T.; Viereck, J.; Thum, T.; Clézardin, P.; Coleman, R.; Puppo, M.; Ottewell, P.; Bonnelye, E.; Paycha, F.; et al. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [PubMed]
- Barwari, T.; Joshi, A.; Mayr, M. MicroRNAs in Cardiovascular Disease. J. Am. Coll. Cardiol. 2016, 68, 2577–2584. [Google Scholar] [CrossRef]
- Wong, L.L.; Wang, J.; Liew, O.W.; Richards, A.M.; Chen, Y.-T. MicroRNA and Heart Failure. Int. J. Mol. Sci. 2016, 17, 502. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Piasecka, A.; Sekrecki, M.; Szcześniak, M.W.; Sobczak, K. MEF2C shapes the microtranscriptome during differentiation of skeletal muscles. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Chen, H.-P.; Wen, J.; Tan, S.-R.; Kang, L.-M.; Zhu, G.-C. MiR-199a-3p inhibition facilitates cardiomyocyte differentiation of embryonic stem cell through promotion of MEF2C. J. Cell. Physiol. 2019, 234, 23315–23325. [Google Scholar] [CrossRef]
- Melnik, S.; Gabler, J.; Dreher, S.I.; Hecht, N.; Hofmann, N.; Großner, T.; Richter, W. MiR-218 affects hypertrophic differentiation of human mesenchymal stromal cells during chondrogenesis via targeting RUNX2, MEF2C, and COL10A1. Stem Cell Res. Ther. 2020, 11, 1–18. [Google Scholar] [CrossRef]
- Tan, Y.; Shen, L.; Gan, M.; Fan, Y.; Cheng, X.; Zheng, T.; Niu, L.; Chen, L.; Jiang, D.; Li, X.; et al. Downregulated miR-204 Promotes Skeletal Muscle Regeneration. BioMed Res. Int. 2020, 2020, 1–9. [Google Scholar] [CrossRef]
- Chinchilla, A.; Lozano, E.; Daimi, H.; Esteban, F.J.; Crist, C.; Aranega, A.E.; Franco, D. MicroRNA profiling during mouse ventricular maturation: a role for miR-27 modulating Mef2c expression. Cardiovasc. Res. 2010, 89, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Sui, L.; Hong, X.; Yang, M.; Li, W. MiR-448 promotes vascular smooth muscle cell proliferation and migration in through directly targeting MEF2C. Environ. Sci. Pollut. Res. 2017, 24, 22294–22300. [Google Scholar] [CrossRef]
- Shen, L.; Chen, L.; Zhang, S.; Zhang, Y.; Wang, J.; Zhu, L. MicroRNA-23a reduces slow myosin heavy chain isoforms composition through myocyte enhancer factor 2C (MEF2C) and potentially influences meat quality. Meat Sci. 2016, 116, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Yelamanchili, S.V.; Chaudhuri, A.D.; Chen, L.-N.; Xiong, H.; Fox, H.S. MicroRNA-21 dysregulates the expression of MEF2C in neurons in monkey and human SIV/HIV neurological disease. Cell Death Dis. 2010, 1, e77–e77. [Google Scholar] [CrossRef]
- DeVeale, B.; Swindlehurst-Chan, J.; Blelloch, R. The roles of microRNAs in mouse development. Nat. Rev. Genet. 2021, 22, 307–323. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ransom, J.F.; Li, A.; Vedantham, V.; von Drehle, M.; Muth, A.N.; Tsuchihashi, T.; McManus, M.T.; Schwartz, R.J.; Srivastava, D. Dysregulation of Cardiogenesis, Cardiac Conduction, and Cell Cycle in Mice Lacking miRNA-1-2. Cell 2007, 129, 303–317. [Google Scholar] [CrossRef]
- Wei, Y.; Peng, S.; Wu, M.; Sachidanandam, R.; Tu, Z.; Zhang, S.; Falce, C.; A Sobie, E.; Lebeche, D.; Zhao, Y. Multifaceted roles of miR-1s in repressing the fetal gene program in the heart. Cell Res. 2014, 24, 278–292. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The Endothelial-Specific MicroRNA miR-126 Governs Vascular Integrity and Angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Fish, J.E.; Santoro, M.M.; Morton, S.U.; Yu, S.; Yeh, R.-F.; Wythe, J.D.; Ivey, K.N.; Bruneau, B.G.; Stainier, D.Y.R.; Srivastava, D. miR-126 Regulates Angiogenic Signaling and Vascular Integrity. Dev. Cell 2008, 15, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Kuo, G.; Wu, C.-Y.; Yang, H.-Y. MiR-17-92 cluster and immunity. J. Formos. Med Assoc. 2019, 118, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Y.; Qi, P.; Ma, Z. Biology of MiR-17-92 Cluster and Its Progress in Lung Cancer. Int. J. Med Sci. 2018, 15, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Liu, Z.; Zhou, L. Roles of miR-17-92 Cluster in Cardiovascular Development and Common Diseases. BioMed Res. Int. 2017, 2017, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Ikeda, G.; Tada, Y.; von Bornstädt, D.; Santoso, M.R.; Wahlquist, C.; Rhee, S.; Jeon, Y.-J.; Yu, A.C.; O’brien, C.G.; et al. miR-106a–363 cluster in extracellular vesicles promotes endogenous myocardial repair via Notch3 pathway in ischemic heart injury. Basic Res. Cardiol. 2021, 116, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Tan, W. miR-106b-25/miR-17-92clusters: Polycistrons with oncogenic roles in hepatocellular carcinoma. World J. Gastroenterol. 2014, 20, 5962–72. [Google Scholar] [CrossRef]
- Khuu, C.; Utheim, T.P.; Sehic, A. The Three Paralogous MicroRNA Clusters in Development and Disease, miR-17-92, miR-106a-363, and miR-106b-25. Scientifica 2016, 2016, 1–10. [Google Scholar] [CrossRef]
- Ru, L.; Wang, X.-M.; Niu, J.-Q. The miR-23–27–24 cluster: an emerging target in NAFLD pathogenesis. Acta Pharmacol. Sin. 2021, 43, 1167–1179. [Google Scholar] [CrossRef]
- Rogler, C.E.; Matarlo, J.S.; Kosmyna, B.; Fulop, D.; Rogler, L.E. Knockdown of miR-23, miR-27, and miR-24 Alters Fetal Liver Development and Blocks Fibrosis in Mice. Gene Expr. 2017, 17, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Wu, C.-J.; Yasuda, T.; Cruz, L.O.; Khan, A.A.; Lin, L.-L.; Nguyen, D.T.; Miller, M.; Lee, H.-M.; Kuo, M.-L.; et al. miR-23∼27∼24 clusters control effector T cell differentiation and function. J. Exp. Med. 2016, 213, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Chiang, D.Y.; Kongchan, N.; Beavers, D.L.; Alsina, K.M.; Voigt, N.; Neilson, J.R.; Jakob, H.; Martin, J.F.; Dobrev, D.; Wehrens, X.H.; et al. Loss of MicroRNA-106b-25 Cluster Promotes Atrial Fibrillation by Enhancing Ryanodine Receptor Type-2 Expression and Calcium Release. Circ. Arrhythmia Electrophysiol. 2014, 7, 1214–1222. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xue, H.; Jin, Q.-H.; Guo, J.; Chen, Y.-D. Increased expression of ryanodine receptor type-2 during atrial fibrillation by miR-106-25 cluster independent mechanism. Exp. Cell Res. 2018, 375, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Bang, C.; Fiedler, J.; Thum, T. Cardiovascular Importance of the MicroRNA-23/27/24 Family. Microcirculation 2011, 19, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Torres, F.; Aranega, A.E.; Franco, D. Identification of regulatory elements directing miR-23a–miR-27a–miR-24-2 transcriptional regulation in response to muscle hypertrophic stimuli. Biochim. et Biophys. Acta (BBA) - Gene Regul. Mech. 2014, 1839, 885–897. [Google Scholar] [CrossRef]
- Briata, P.; Chen, C.-Y.; Giovarelli, M.; Pasero, M.; Trabucchi, M.; Ramos, A.; Gherzi, R. KSRP, many functions for a single protein. Front. Biosci. 2011, 16, 1787–1796. [Google Scholar] [CrossRef]
- Gherzi, R.; Chen, C.; Trabucchi, M.; Ramos, A.; Briata, P. The role of KSRP in mRNA decay and microRNA precursor maturation. Wiley Interdiscip. Rev. RNA 2010, 1, 230–239. [Google Scholar] [CrossRef]
- Cho, C.J.; Myung, S.-J.; Chang, S. ADAR1 and MicroRNA; A Hidden Crosstalk in Cancer. Int. J. Mol. Sci. 2017, 18, 799. [Google Scholar] [CrossRef]
- Xing Z, Ma WK, Tran EJ. The DDX5/Dbp2 subfamily of DEAD-box RNA helicases. Wiley Interdiscip Rev RNA 2019, 10, e1519. [CrossRef]
- Okamoto, S.-I.; Li, Z.; Ju, C.; Schölzke, M.N.; Mathews, E.; Cui, J.; Salvesen, G.S.; Bossy-Wetzel, E.; Lipton, S.A. Dominant-interfering forms of MEF2 generated by caspase cleavage contribute to NMDA-induced neuronal apoptosis. Proc. Natl. Acad. Sci. 2002, 99, 3974–3979. [Google Scholar] [CrossRef] [PubMed]
- Bach-Elias, M.; Kokolo, M. P68 RNA Helicase (DDX5) Required for the Formation of Various Specific and Mature miRNA Active RISC Complexes. MicroRNA 2022, 11, 36–44. [Google Scholar] [CrossRef]
- Dardenne, E.; Espinoza, M.P.; Fattet, L.; Germann, S.; Lambert, M.-P.; Neil, H.; Zonta, E.; Mortada, H.; Gratadou, L.; Deygas, M.; et al. RNA Helicases DDX5 and DDX17 Dynamically Orchestrate Transcription, miRNA, and Splicing Programs in Cell Differentiation. Cell Rep. 2014, 7, 1900–1913. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.-P.; Terrone, S.; Giraud, G.; Benoit-Pilven, C.; Cluet, D.; Combaret, V.; Mortreux, F.; Auboeuf, D.; Bourgeois, C.F. The RNA helicase DDX17 controls the transcriptional activity of REST and the expression of proneural microRNAs in neuronal differentiation. Nucleic Acids Res. 2018, 46, 7686–7700. [Google Scholar] [CrossRef] [PubMed]
- Motiño, O.; Francés, D.E.; Mayoral, R.; Castro-Sánchez, L.; Fernández-Velasco, M.; Boscá, L.; García-Monzón, C.; Brea, R.; Casado, M.; Agra, N.; et al. Regulation of MicroRNA 183 by Cyclooxygenase 2 in Liver Is DEAD-Box Helicase p68 (DDX5) Dependent: Role in Insulin Signaling. Mol. Cell. Biol. 2015, 35, 2554–2567. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006, 13, 13–21. [Google Scholar] [CrossRef]
- Janknecht, R. Multi-talented DEAD-box proteins and potential tumor promoters: p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17). . 2010, 2, 223–34. [Google Scholar]
- Wong, S.K.; Lazinski, D.W. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc. Natl. Acad. Sci. 2002, 99, 15118–15123. [Google Scholar] [CrossRef]
- Bahn, J.H.; Ahn, J.; Lin, X.; Zhang, Q.; Lee, J.-H.; Civelek, M.; Xiao, X. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat. Commun. 2015, 6, 1–13. [Google Scholar] [CrossRef]
- Sakurai, M.; Shiromoto, Y.; Ota, H.; Song, C.; Kossenkov, A.V.; Wickramasinghe, J.; Showe, L.C.; Skordalakes, E.; Tang, H.-Y.; Speicher, D.W.; et al. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 2017, 24, 534–543. [Google Scholar] [CrossRef]
- Chou, C.-F.; Lin, W.-J.; Lin, C.-C.; Luber, C.A.; Godbout, R.; Mann, M.; Chen, C.-Y. DEAD Box Protein DDX1 Regulates Cytoplasmic Localization of KSRP. PLOS ONE 2013, 8, e73752. [Google Scholar] [CrossRef] [PubMed]
- Giovarelli, M.; Bucci, G.; Ramos, A.; Bordo, D.; Wilusz, C.J.; Chen, C.-Y.; Puppo, M.; Briata, P.; Gherzi, R. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc. Natl. Acad. Sci. 2014, 111, 201415098–E5028. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.P.; Huang, S.; Black, D.L.; Pollock, C.; Daily, K.; Nguyen, V.T.; Wang, C.; Lewandowska, M.A.; Bensaude, O.; Doxsey, M.E.S.J. Differentiation-induced Colocalization of the KH-type Splicing Regulatory Protein with Polypyrimidine Tract Binding Protein and the c-srcPre-mRNA. Mol. Biol. Cell 2004, 15, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [PubMed]
- Santovito, D.; Weber, C. Non-canonical features of microRNAs: paradigms emerging from cardiovascular disease. Nat. Rev. Cardiol. 2022, 19, 620–638. [Google Scholar] [CrossRef] [PubMed]
- Santovito, D.; Egea, V.; Bidzhekov, K.; Natarelli, L.; Mourão, A.; Blanchet, X.; Wichapong, K.; Aslani, M.; Brunßen, C.; Horckmans, M.; et al. Noncanonical inhibition of caspase-3 by a nuclear microRNA confers endothelial protection by autophagy in atherosclerosis. Sci. Transl. Med. 2020, 12, eaaz2294. [Google Scholar] [CrossRef]
- Penrad-Mobayed, M.; Perrin, C.; L’hôte, D.; Contremoulins, V.; Lepesant, J.-A.; Boizet-Bonhoure, B.; Poulat, F.; Baudin, X.; Veitia, R.A. A role for SOX9 in post-transcriptional processes: insights from the amphibian oocyte. Sci. Rep. 2018, 8, 7191. [Google Scholar] [CrossRef]
- Panda, A.C.; Abdelmohsen, K.; Yoon, J.-H.; Martindale, J.L.; Yang, X.; Curtis, J.; Mercken, E.M.; Chenette, D.M.; Zhang, Y.; Schneider, R.J.; et al. RNA-Binding Protein AUF1 Promotes Myogenesis by Regulating MEF2C Expression Levels. Mol. Cell. Biol. 2014, 34, 3106–3119. [Google Scholar] [CrossRef]
- Tomaselli, S.; Bonamassa, B.; Alisi, A.; Nobili, V.; Locatelli, F.; Gallo, A. ADAR Enzyme and miRNA Story: A Nucleotide that Can Make the Difference. Int. J. Mol. Sci. 2013, 14, 22796–22816. [Google Scholar] [CrossRef]
- Chawla, G.; Sokol, N.S. ADAR mediates differential expression of polycistronic microRNAs. Nucleic Acids Res. 2014, 42, 5245–5255. [Google Scholar] [CrossRef]
- Chen, T.; Xiang, J.-F.; Zhu, S.; Chen, S.; Yin, Q.-F.; Zhang, X.-O.; Zhang, J.; Feng, H.; Dong, R.; Li, X.-J.; et al. ADAR1 is required for differentiation and neural induction by regulating microRNA processing in a catalytically independent manner. Cell Res. 2015, 25, 459–476. [Google Scholar] [CrossRef]
- Widmark, A.; Sagredo, E.A.; Karlström, V.; Behm, M.; Biryukova, I.; Friedländer, M.R.; Daniel, C.; Öhman, M. ADAR1- and ADAR2-mediated regulation of maturation and targeting of miR-376b to modulate GABA neurotransmitter catabolism. J. Biol. Chem. 2022, 298, 101682. [Google Scholar] [CrossRef] [PubMed]
- Briata, P.; Chen, C.-Y.; Giovarelli, M.; Pasero, M.; Trabucchi, M.; Ramos, A.; Gherzi, R. KSRP, many functions for a single protein. Front. Biosci. 2011, 16, 1787–1796. [Google Scholar] [CrossRef] [PubMed]
- Gherzi, R.; Chen, C.-Y.; Ramos, A.; Briata, P. KSRP Controls Pleiotropic Cellular Functions. Semin. Cell Dev. Biol. 2014, 34, 2–8. [Google Scholar] [CrossRef]
- Briata, P.; Chen, C.-Y.; Ramos, A.; Gherzi, R. Functional and molecular insights into KSRP function in mRNA decay. Biochim. et Biophys. Acta (BBA) - Gene Regul. Mech. 2013, 1829, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, M.; Zhao, X.; Wang, H.; Zhu, J.; Wang, C.; Zhou, M.; Dong, H.; Zhou, R. Upregulation of KSRP by miR-27b attenuates schistosomiasis-induced hepatic fibrosis by targeting TGF-β1. FASEB J. 2020, 34, 4120–4133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Gong, A.-Y.; Eischeid, A.N.; Chen, X.-M. miR-27b Targets KSRP to Coordinate TLR4-Mediated Epithelial Defense against Cryptosporidium parvum Infection. PLOS Pathog. 2012, 8, e1002702. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Lu, Y.; Li, X.; Mao, T.; Chen, X.-M.; Zhou, R. Upregulation of KSRP by miR-27b provides IFN-γ-induced post-transcriptional regulation of CX3CL1 in liver epithelial cells. Sci. Rep. 2015, 5, 17590–17590. [Google Scholar] [CrossRef]
- Dou, R.; Liu, K.; Yang, C.; Zheng, J.; Shi, D.; Lin, X.; Wei, C.; Zhang, C.; Fang, Y.; Huang, S.; et al. EMT-cancer cells-derived exosomal miR-27b-3p promotes circulating tumour cells-mediated metastasis by modulating vascular permeability in colorectal cancer. Clin. Transl. Med. 2021, 11, e595. [Google Scholar] [CrossRef]
- Gusar, V.; Timofeeva, A.; Chagovets, V.; Kan, N.; Vysokikh, M.; Marey, M.; Karapetyan, A.; Baev, O.; Sukhikh, G. Diagnostic Potential of Exosomal HypoxamiRs in the Context of Hypoxia–Sumoylation–HypoxamiRs in Early Onset Preeclampsia at the Preclinical Stage. Life 2022, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Chien, M.-H.; Lee, W.-J.; Yang, Y.-C.; Li, Y.-L.; Chen, B.-R.; Cheng, T.-Y.; Yang, P.-W.; Wang, M.-Y.; Jan, Y.-H.; Lin, Y.-K.; et al. KSRP suppresses cell invasion and metastasis through miR-23a-mediated EGR3 mRNA degradation in non-small cell lung cancer. Biochim. et Biophys. Acta (BBA) - Gene Regul. Mech. 2017, 1860, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Song, Y.; Chan, T.H.M.; Yang, H.; Lin, C.H.; Tay, D.J.T.; Hong, H.; Tang, S.J.; Tan, K.T.; Huang, X.X.; et al. An RNA editing/dsRNA binding-independent gene regulatory mechanism of ADARs and its clinical implication in cancer. Nucleic Acids Res. 2017, 45, 10436–10451. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.F.; Schwarz, J.J.; Olson, E.N. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc. Natl. Acad. Sci. 1993, 90, 5282–5286. [Google Scholar] [CrossRef]
- Lozano-Velasco, E.; Hernández-Torres, F.; Daimi, H.; Serra, S.A.; Herraiz, A.; Hove-Madsen, L.; Aránega, A.; Franco, D. Pitx2 impairs calcium handling in a dose-dependent manner by modulating Wnt signalling. Cardiovasc. Res. 2016, 109, 55–66. [Google Scholar] [CrossRef]
- Lozano-Velasco, E.; Wangensteen, R.; Quesada, A.; Garcia-Padilla, C.; Osorio, J.A.; Ruiz-Torres, M.D.; Aranega, A.; Franco, D. Hyperthyroidism, but not hypertension, impairs PITX2 expression leading to Wnt-microRNA-ion channel remodeling. PLOS ONE 2017, 12, e0188473. [Google Scholar] [CrossRef]
- Domínguez, J.N.; Lodde, V.; Munk, R.; Abdelmohsen, K.; Gorospe, M.; Ginel, A.; Aránega, A.E.; Franco, D.; García-Padilla, C.; Jiménez-Sábado, V.; et al. Identification of atrial-enriched lncRNA Walras linked to cardiomyocyte cytoarchitecture and atrial fibrillation. FASEB J. 2021, 36, e22051–e22051. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




