Submitted:
04 October 2023
Posted:
05 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. SNP Annotation
2.2. Identification of Harmful nsSNPs
2.3. Identification of nsSNPs within MLL1 Domains
| Domains | AA change | SNP ID | Polyphen 1 and 2 | PhD-SNP, SIFT, SNAP and MAPP |
Predict SNP |
|---|---|---|---|---|---|
| C1072F | rs1085307947 | Damaging | Deleterious | Deleterious | |
| Zinc finger, CXXC-type | C1155Y | rs1057518074 | Damaging | Deleterious | Deleterious |
| C1158Y | rs1131691503 | Damaging | Deleterious | Deleterious | |
| G1180V | rs1555038115 | Damaging | Deleterious | Deleterious | |
| G1181C | rs1950071303 | Damaging | Deleterious | Deleterious | |
| C1189R | rs886041875 | Damaging | Deleterious | Deleterious | |
| C1189Y | rs1555038125 | Damaging | Deleterious | Deleterious | |
| C1194Y | rs1950106455 | Damaging | Deleterious | Deleterious | |
| PHD1-3 | C1448R | rs863224895 | Damaging | Deleterious | Deleterious |
| C1448Y C1588F |
rs1085307857 rs1555042404 |
Damaging Damaging |
Deleterious Deleterious |
Deleterious Deleterious |
|
| R1630W | rs376776245 | Damaging | Deleterious | Deleterious | |
| Bromodomain | W1635C | rs782594163 | Damaging | Deleterious | Deleterious |
| R1658W | rs373435126 | Damaging | Deleterious | Deleterious | |
| R1763P | rs781944403 | Damaging | Deleterious | Deleterious | |
| W1771R | rs1475344216 | Damaging | Deleterious | Deleterious | |
| R1892C | rs1555044474 | Damaging | Deleterious | Deleterious | |
| Y1895C | rs143373748 | Damaging | Deleterious | Deleterious | |
| G1919R | rs1555044515 | Damaging | Deleterious | Deleterious | |
| Extended PHD 4 | V1924M | rs1555044535 | Damaging | Deleterious | Deleterious |
| G1943E | rs1950444447 | Damaging | Deleterious | Deleterious | |
| L2009W | rs1555044990 | Damaging | Deleterious | Deleterious | |
| R2011W | rs781919638 | Damaging | Deleterious | Deleterious | |
| G2016D | rs1397000127 | Damaging | Deleterious | Deleterious | |
| G2027E | rs1057519403 | Damaging | Deleterious | Deleterious | |
| F/Y-rich N-terminus | R2067H | rs782768278 | Damaging | Deleterious | Deleterious |
| G2277D | rs1555046173 | Damaging | Deleterious | Deleterious | |
| R2519W | rs782129680 | Damaging | Deleterious | Deleterious | |
| R2521H | rs1555046685 | Damaging | Deleterious | Deleterious | |
| D2598V | rs368088982 | Damaging | Deleterious | Deleterious | |
| R2627C | rs1555046878 | Damaging | Deleterious | Deleterious | |
| S2652I | rs782497028 | Damaging | Deleterious | Deleterious | |
| R2659Q | rs1390104203 | Damaging | Deleterious | Deleterious | |
| Y2683H | rs1555046962 | Damaging | Deleterious | Deleterious | |
| L2700R | rs1555047001 | Damaging | Deleterious | Deleterious | |
| D2724G | rs781821970 | Damaging | Deleterious | Deleterious | |
| G2796E | rs1186580008 | Damaging | Deleterious | Deleterious | |
| G2796V | rs1186580008 | Damaging | Deleterious | Deleterious | |
| L2857Q | rs1057520696 | Damaging | Deleterious | Deleterious | |
| G3000R | rs1438502727 | Damaging | Deleterious | Deleterious | |
| S3039C | rs1279929414 | Damaging | Deleterious | Deleterious | |
| P3098L | rs1555047613 | Damaging | Deleterious | Deleterious | |
| G3129R | rs1353151956 | Damaging | Deleterious | Deleterious | |
| G3186D | rs961670105 | Damaging | Deleterious | Deleterious | |
| I3189T | rs782641177 | Damaging | Deleterious | Deleterious | |
| S3211I | rs782372675 | Damaging | Deleterious | Deleterious | |
| L3373H | rs781812638 | Damaging | Deleterious | Deleterious | |
| L3393P | rs1555048205 | Damaging | Deleterious | Deleterious | |
| C3430R | rs373345566 | Damaging | Deleterious | Deleterious | |
| N3459Y | rs782596573 | Damaging | Deleterious | Deleterious | |
| L3617P | rs146191865 | Damaging | Deleterious | Deleterious | |
| FY-rich, C-terminal | R3704Q | rs1555050212 | Damaging | Deleterious | Deleterious |
| C3743Y | rs782658611 | Damaging | Deleterious | Deleterious | |
| F3748C | rs749354451 | Damaging | Deleterious | Deleterious | |
| R3749C | rs782366377 | Damaging | Deleterious | Deleterious | |
| R3789H | rs1555052977 | Damaging | Deleterious | Deleterious | |
| R3810W | rs1555053010 | Damaging | Deleterious | Deleterious | |
| R3822C | rs1591310362 | Damaging | Deleterious | Deleterious | |
| SET domain | E3860D | rs782600332 | Damaging | Deleterious | Deleterious |
| G3863S | rs1591311290 | Damaging | Deleterious | Deleterious | |
| E3875V | rs781980190 | Damaging | Deleterious | Deleterious | |
| R3889Q | rs1555053677 | Damaging | Deleterious | Deleterious |
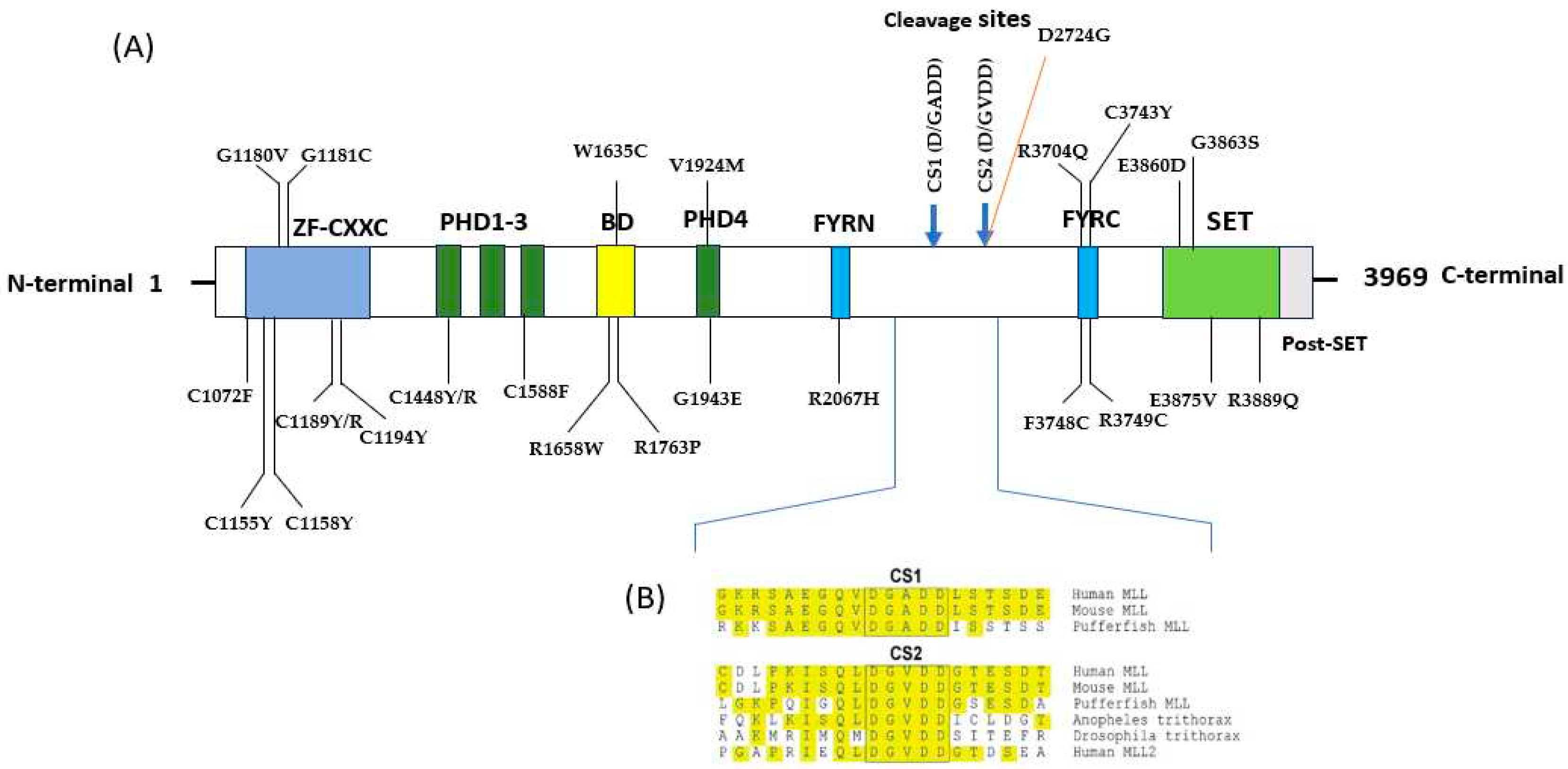
2.4. Evolutionary Conservation Analysis
| POS | SEQ | SCORE (normalized) |
COLOR | CONFIDENCE INTERVAL |
CONFIDENCE INTERVAL COLOR |
B/E | FUNCTION |
|---|---|---|---|---|---|---|---|
| 1072 | C | -0.851 | 9 | -0.937, -0.817 | 9,9 | b | s |
| 1155 | C | -0.851 | 9 | -0.937, -0.817 | 9,9 | e | f |
| 1158 | C | -0.851 | 9 | -0.937, -0.817 | 9,9 | b | s |
| 1180 | G | -0.861 | 9 | -0.937, -0.842 | 9,9 | e | f |
| 1181 | G | -0.861 | 9 | -0.937, -0.842 | 9,9 | e | f |
| 1189 | C | -0.851 | 9 | -0.937, -0.817 | 9,9 | b | s |
| 1194 | C | -0.541 | 8 | -0.760, -0.398 | 9,7 | b | / |
| 1448 | C | -0.856 | 9 | -0.937, -0.817 | 9,9 | b | s |
| 1588 | C | -0.712 | 9 | -0.864,-0.611 | 9,8 | b | s |
| 1635 | W | -0.799 | 9 | -0.937,-0.760 | 9,9 | b | s |
| 1658 | R | -0.893 | 9 | -0.943,-0.883 | 9,9 | e | f |
| 1763 | R | -0.635 | 8 | -0.790,-0.565 | 9,8 | e | f |
| 1771 | W | -0.565 | 8 | -0.817,-0.398 | 9,7 | b | / |
| 1892 | R | -0.893 | 9 | -0.943,-0.883 | 9,9 | e | f |
| 1895 | Y | -0.861 | 9 | -0.937,-0.842 | 9,9 | b | s |
| 1919 | G | -0.740 | 9 | -0.883,-0.653 | 9,8 | e | f |
| 1924 | V | -0.901 | 9 | -0.943,-0.883 | 9,9 | b | s |
| 1943 | G | -0.866 | 9 | -0.937,-0.842 | 9,9 | e | f |
| 2009 | L | -0.492 | 7 | -0.692,-0.332 | 8,7 | b | / |
| 2011 | R | -0.809 | 9 | -0.900,-0.760 | 9,9 | e | f |
| 2016 | G | -0.866 | 9 | -0.937,-0.842 | 9,9 | e | f |
| 2027 | G | -0.866 | 9 | -0.937,-0.842 | 9,9 | b | s |
| 2067 | R | -0.893 | 9 | -0.943,-0.883 | 9,9 | e | f |
| 2519 | R | -0.393 | 7 | -0.611,-0.258 | 8,6 | e | / |
| 2521 | R | -0.497 | 7 | -0.692,-0.398 | 8,7 | e | / |
| 2627 | R | -0.893 | 9 | -0.943,-0.883 | 9,9 | e | f |
| 2652 | S | -0.646 | 8 | -0.790,-0.565 | 9,8 | e | f |
| 2659 | R | -0.810 | 9 | -0.900,-0.760 | 9,9 | e | f |
| 2683 | Y | -0.728 | 9 | -0.864,-0.653 | 9,8 | b | s |
| 2700 | L | -0.497 | 7 | -0.692,-0.398 | 8,7 | b | / |
| 2724 | D | -0.893 | 9 | -0.943,-0.883 | 9,9 | e | f |
| 2857 | L | -0.864 | 9 | -0.937,-0.842 | 9,9 | b | s |
| 3039 | S | -0.842 | 9 | -0.915,-0.817 | 9,9 | e | f |
| 3098 | P | -0.752 | 9 | -0.883,-0.692 | 9,8 | e | f |
| 3129 | G | -0.313 | 7 | -0.565,-0.175 | 8,6 | b | / |
| 3189 | I | -0.827 | 9 | -0.915,-0.790 | 9,9 | b | s |
| 3373 | L | -0.610 | 8 | -0.790,-0.514 | 9,8 | b | / |
| 3393 | L | -0.729 | 9 | -0.883,-0.653 | 9,8 | e | f |
| 3430 | C | -0.269 | 6 | -0.565,-0.083 | 8,5 | b | / |
| 3704 | R | -0.886 | 9 | -0.943,-0.864 | 9,9 | e | f |
| 3743 | C | -0.846 | 9 | -0.937,-0.817 | 9,9 | b | s |
| 3748 | F | -0.856 | 9 | -0.937,-0.817 | 9,9 | b | s |
| 3749 | R | -0.698 | 8 | -0.842,-0.611 | 9,8 | e | f |
| 3789 | R | -0.887 | 9 | -0.943,-0.864 | 9,9 | e | f |
| 3810 | R | -0.887 | 9 | -0.943,-0.864 | 9,9 | e | f |
| 3822 | R | -0.794 | 9 | -0.900,-0.728 | 9,9 | b | s |
| 3860 | E | -0.882 | 9 | -0.943,-0.864 | 9,9 | e | f |
| 3863 | G | -0.857 | 9 | -0.937,-0.817 | 9,9 | b | s |
| 3875 | E | -0.882 | 9 | -0.943,-0.864 | 9,9 | e | f |
| 3889 | R | -0.793 | 9 | -0.900,-0.728 | 9,9 | b | s |
2.5. Assessment of Protein Structural Stability
| SNP ID | AA substitution | SVM2 | RI | DDG kcal/mol | SVM3 | RI | MUpro | DDG |
|---|---|---|---|---|---|---|---|---|
| rs1085307947 | C1072F | Decrease | 5 | -0.49 | Large decrease | 1 | Decrease | -0.49142 |
| rs886041875 | C1189R | Decrease | 5 | -1.05 | Large decrease | 2 | Decrease | -1.01449 |
| rs863224895 | C1448R | Decrease | 9 | -0.72 | Large decrease | 2 | Decrease | -1.05828 |
| rs373435126 | R1658W | Decrease | 1 | -0.09 | Large decrease | 1 | Decrease | -1.51963 |
| rs143373748 | Y1895C | Decrease | 6 | -0.85 | Large decrease | 0 | Decrease | -1.03143 |
| rs1555044990 | L2009W | Decrease | 4 | -1.32 | Large decrease | 1 | Decrease | -1.35469 |
| rs1397000127 | G2016D | Decrease | 8 | -0.99 | Large decrease | 3 | Decrease | -0.45411 |
| rs1057519403 | G2027E | Decrease | 5 | -1.61 | Large decrease | 4 | Decrease | -0.50805 |
| rs782768278 | R2067H | Decrease | 8 | -1.39 | Large decrease | 7 | Decrease | -1.13602 |
| rs1555046685 | R2521H | Decrease | 8 | -0.83 | Large decrease | 4 | Decrease | -1.04097 |
| rs1555046878 | R2627C | Decrease | 3 | -1.11 | Large decrease | 4 | Decrease | -0.73453 |
| rs782497028 | S2652I | Decrease | 3 | 0.13 | Large decrease | 1 | Decrease | -0.10003 |
| rs1390104203 | R2659Q | Decrease | 4 | -0.70 | Large decrease | 2 | Decrease | -0.59524 |
| rs1555047001 | L2700R | Decrease | 7 | -0.69 | Large decrease | 1 | Decrease | -1.82806 |
| rs781821970 | D2724G | Decrease | 6 | -0.89 | Large decrease | 6 | Decrease | -1.41038 |
| rs1186580008 | G2796E | Decrease | 6 | -0.75 | Large decrease | 1 | Decrease | -0.42903 |
| rs118658000 | G2796V | Decrease | 6 | -0.55 | Large decrease | 3 | Decrease | -0.47996 |
| rs1057520696 | L2857Q | Decrease | 9 | -2.31 | Large decrease | 5 | Decrease | -1.82752 |
| rs1279929414 | S3039C | Decrease | 4 | -2.02 | Large decrease | 6 | Decrease | -0.43857 |
| rs1555047613 | P3098L | Decrease | 7 | -1.11 | Large decrease | 3 | Decrease | -0.38588 |
| rs1353151956 | G3129R | Decrease | 9 | -1.35 | Large decrease | 1 | Decrease | -0.28198 |
| rs782641177 | I3189T | Decrease | 8 | -1.31 | Large decrease | 5 | Decrease | -2.11623 |
| rs781812638 | L3373H | Decrease | 7 | -1.34 | Large decrease | 5 | Decrease | -1.74765 |
| rs1555048205 | L3393P | Decrease | 3 | -1.12 | Large decrease | 2 | Decrease | -1.58189 |
| rs782658611 | C3743Y | Decrease | 1 | -0.23 | Large decrease | 3 | Decrease | -1.13086 |
| rs749354451 | F3748C | Decrease | 6 | -1.7 | Large decrease | 4 | Decrease | -1.82151 |
| rs782366377 | R3749C | Decrease | 3 | -0.85 | Large decrease | 2 | Decrease | -1.00862 |
| rs1555052977 | R3789H | Decrease | 9 | -1.44 | Large decrease | 6 | Decrease | -0.70688 |
| rs1591310362 | R3822C | Decrease | 0 | -0.85 | Large decrease | 3 | Decrease | -0.95916 |
| rs782600332 | E3860D | Decrease | 3 | -0.39 | Large decrease | 1 | Decrease | -1.01581 |
| rs1591311290 | G3863S | Decrease | 9 | -1.27 | Large decrease | 5 | Decrease | -1.34469 |
| rs1555053677 | R3889Q | Decrease | 9 | -1.25 | Large decrease | 5 | Decrease | -1.25798 |
2.6. Analysis of Protein-Protein Interactions and Functional Associations
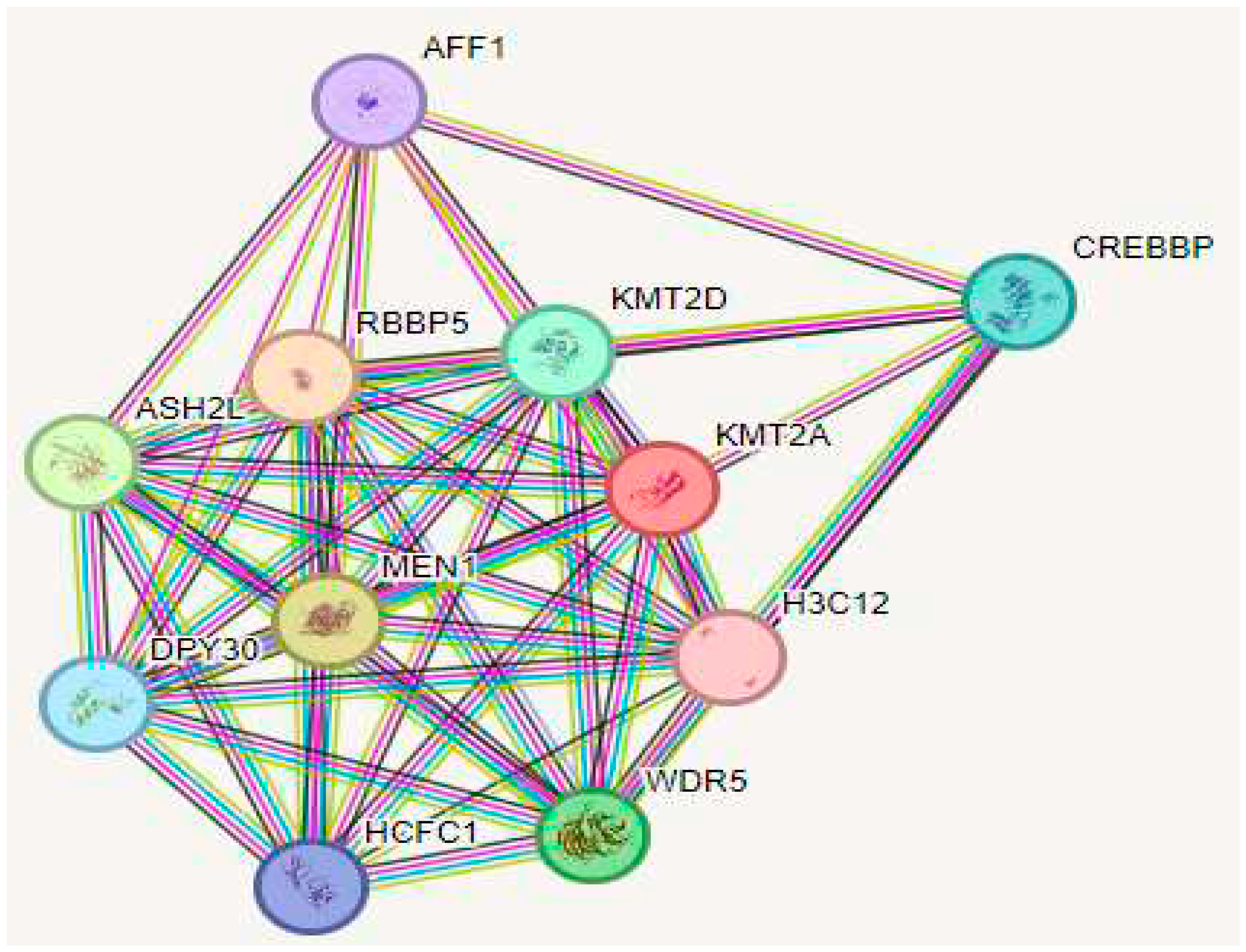
2.7. Assessment of the Functional Implications of Non-coding SNPs
| Chromosome location | dbSNP IDs | Rank | Score |
|---|---|---|---|
| chr11:118522268..118522269 | rs188913109 | 2b | 0.64591 |
| chr11:118523887..118523888 | rs539251803 | 2b | 0.6751 |
| chr11:118524166..118524167 | rs141036837 | 2b | 0.50723 |
| chr11:118524283..118524284 | rs190548021 | 2b | 1.0 |
| chr11:118524539..118524540 | rs147772025 | 3a | 0.37421 |
2.8. Prediction of Pathogenicity of nsSNPs
| AA variation | MutPred2 score | Molecular mechanism with P value less than 0.05 |
|---|---|---|
| C1072F | 0.926 | Altered Disordered interface Altered DNA binding Gain of Strand Altered Metal binding |
| C1189R | 0.825 | Altered Disordered interface Gain of Acetylation at K1185 |
| C1448R | 0.956 | Altered Metal binding Gain of Strand Altered Transmembrane protein Gain of Pyrrolidone carboxylic acid at Q1449 Loss of Sulfation at Y1447 |
| R1658W | 0.685 | Loss of Intrinsic disorder Loss of Helix Gain of Loop |
| Y1895C | 0.729 | Altered Ordered interface Loss of Proteolytic cleavage at R1892 Altered Transmembrane protein |
| L2009W | 0.662 | Altered Transmembrane protein |
| G2016D | 0.578 | Altered DNA binding Altered Transmembrane protein |
| G2027E | 0.911 | Gain of Helix |
| R2067H | 0.809 | Altered Ordered interface Altered Transmembrane protein Loss of Disulfide linkage at C2068 |
| R2521H | 0.150 | - |
| R2627C | 0.782 | Altered Disordered interface Loss of Intrinsic disorder Altered DNA binding Gain of Proteolytic cleavage at R2622 |
| S2652I | 0.321 | - |
| R2659Q | 0.290 | - |
| L2700R | 0.848 | Gain of Intrinsic disorder Gain of B-factor |
| D2724G | 0.720 | Altered Disordered interface Altered Metal binding Loss of Proteolytic cleavage at D2724 Gain of O-linked glycosylation at T2727 |
| G2796E | 0.260 | - |
| G2796V | 0.274 | - |
| L2857Q | 0.832 | Gain of Intrinsic disorder Altered Disordered interface Loss of Helix |
| S3039C | 0.249 | - |
| P3098L | 0.572 | Loss of Strand Altered Transmembrane protein Gain of Pyrrolidone carboxylic acid at Q3103 |
| G3129R | 0.546 | Loss of Strand Gain of ADP-ribosylation at G3129 Loss of B-factor |
| I3189T | 0.540 | Gain of O-linked glycosylation at S3185 |
| L3373H | 0.707 | Gain of Intrinsic disorde |
| L3393P | 0.740 | Gain of Intrinsic disorder Altered Disordered interface |
| C3743Y | 0.814 | Altered Metal binding Gain of Loop Altered Transmembrane protein |
| F3748C | 0.898 | Altered Metal binding Loss of SUMOylation at K3752 Altered Transmembrane protein |
| R3749C | 0.770 | Loss of Intrinsic disorder Loss of SUMOylation at K3752 Altered Transmembrane protein |
| R3789H | 0.512 |
Loss of Phosphorylation at Y3794 Loss of ADP-ribosylation at R3789 Gain of Sulfation at Y3794 |
| R3822C | 0.627 | Altered Disordered interface Loss of Intrinsic disorder |
|
E3860D |
0.903 |
Gain of Allosteric site at I3859 Altered Metal binding Altered Ordered interface Gain of Relative solvent accessibility Altered DNA binding Loss of Catalytic site at E3860 |
| G3863S | 0.907 | Altered Ordered interface Gain of Allosteric site at E3860 Altered Disordered interface Loss of Strand Gain of Relative solvent accessibility Altered Metal binding Altered DNA binding Gain of Catalytic site at E3860 |
|
R3889Q |
0.876 |
Altered Metal binding Altered Transmembrane protein Altered Ordered interface Loss of Allosteric site at M3887 Gain of Relative solvent accessibility Altered DNA binding Gain of Catalytic site at R3889 |
2.9. Predicting the Association of nsSNPs with Cancer Susceptibility
| CScape | CScape- somatic | ||||||
|---|---|---|---|---|---|---|---|
| Variant ID | SNP | Input | Coding score |
Message | Input | Coding score |
Message |
| rs781821970 | D2724G | 11,118504063,A,G | 0.650128 | Oncogenic | 11,118504063,A,G | 0.552102 | Driver |
| rs1555048205 | L3393P | 11,118506070,T,C | 0.5526 | Oncogenic (*HC) |
11,118506070,T,C | 0.736249 | Driver |
| rs749354451 | F3748C | 11,118519714,T,G | 0.45528 | Benign | 11,118519714,T,G | 0.612477 | Driver |
| rs1555052977 | R3789H | 11,118520001,G,A | 0.19298 | Benign | 11,118520001,G,A | 0.881861 | Driver |
| rs782600332 | E3860D | 11,118521354,G,T | 0.76665 | Oncogenic | 11,118521354,G,T | 0.249088 | Passenger |
| rs1591311290 | G3863S | 11,118521361,G,A | 0.627944 | Oncogenic | 11,118521361,G,A | 0.307779 | Passenger |
3. Discussion
4. Materials and Methods
4.1. Retrieval of SNP Data
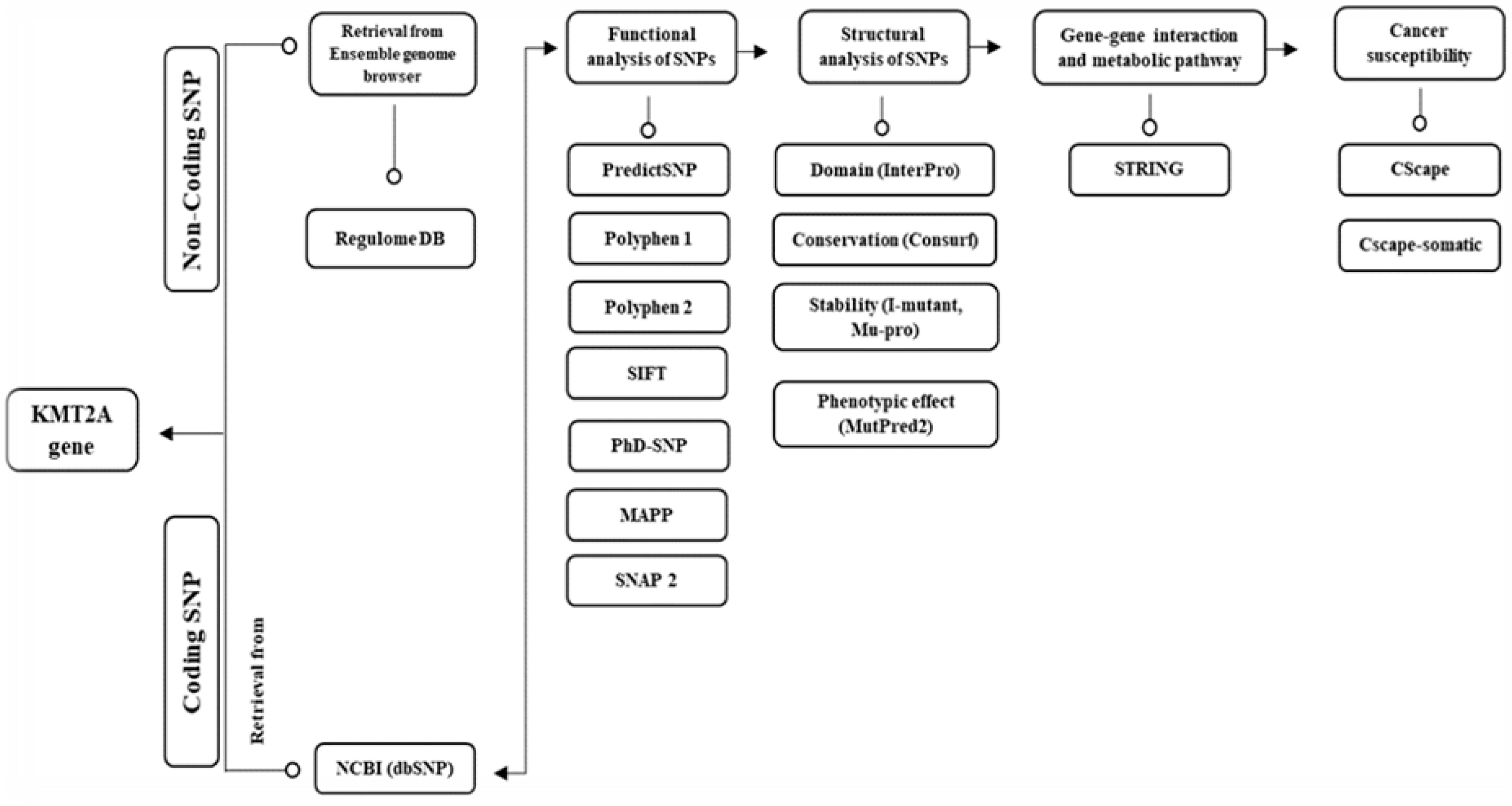
4.2. Prediction of Functional Effects of nsSNPs
4.3. Identification of nsSNPs on MLL1 Protein Domains
4.4. Analysis of Protein Evolutionary Conservation
4.5. Analysis of nsSNP Effects on Protein Stability
4.6. Protein-Protein Interaction and Functional Analysis
4.7. Analysis of the Functional Consequences of Non-Coding SNPs
4.8. Prediction of Molecular Pathogenicity of nsSNPs
4.9. Association of nsSNPs with Cancer Susceptibility
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Winters AC, Bernt KM. MLL-Rearranged Leukemias-An Update on Science and Clinical Approaches. Frontiers in pediatrics. 2017;5:4. [CrossRef]
- Castiglioni S, Di Fede E, Bernardelli C, Lettieri A, Parodi C, Grazioli P, et al. KMT2A: Umbrella Gene for Multiple Diseases. Genes. 2022 Mar;13(3). [CrossRef]
- Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992 Nov;71(4):691–700. [CrossRef]
- Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome biology. 2005;6(8):227. [CrossRef]
- Bochyńska A, Lüscher-Firzlaff J, Lüscher B. Modes of Interaction of KMT2 Histone H3 Lysine 4 Methyltransferase/COMPASS Complexes with Chromatin. Cells. 2018 Mar;7(3). [CrossRef]
- Górecki M, Kozioł I, Kopystecka A, Budzyńska J, Zawitkowska J, Lejman M. Updates in KMT2A Gene Rearrangement in Pediatric Acute Lymphoblastic Leukemia. Biomedicines. 2023 Mar;11(3). [CrossRef]
- Collins CT, Hess JL. Deregulation of the HOXA9/MEIS1 axis in acute leukemia. Current opinion in hematology. 2016 Jul;23(4):354–61.
- Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annual review of pathology. 2012;7:283–301. [CrossRef]
- Robert F, Pelletier J. Exploring the Impact of Single-Nucleotide Polymorphisms on Translation. Frontiers in genetics. 2018;9:507. [CrossRef]
- Kumar A, Rajendran V, Sethumadhavan R, Shukla P, Tiwari S, Purohit R. Computational SNP analysis: current approaches and future prospects. Cell biochemistry and biophysics. 2014 Mar;68(2):233–9. [CrossRef]
- Allemailem KS, Almatroudi A, Alrumaihi F, Makki Almansour N, Aldakheel FM, Rather RA, et al. Single nucleotide polymorphisms (SNPs) in prostate cancer: its implications in diagnostics and therapeutics. American journal of translational research. 2021;13(4):3868–89.
- Hunt SE, McLaren W, Gil L, Thormann A, Schuilenburg H, Sheppard D, et al. Ensembl variation resources. Database [Internet]. 2018 Jan 1;2018:bay119. Available from: https://doi.org/10.1093/database/bay119. [CrossRef]
- Sherry ST, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Research [Internet]. 2001;29(1):308–11. Available from: https://doi.org/10.1093/nar/29.1.308. [CrossRef]
- Bendl J, Stourac J, Salanda O, Pavelka A, Wieben ED, Zendulka J, et al. PredictSNP: Robust and Accurate Consensus Classifier for Prediction of Disease-Related Mutations. PLOS Computational Biology [Internet]. 2014;10(1):1–11. Available from: https://doi.org/10.1371/journal.pcbi.1003440. [CrossRef]
- Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic acids research. 2012 Jul;40(Web Server issue):W452-7. [CrossRef]
- Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic acids research. 2007;35(11):3823–35. [CrossRef]
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Current protocols in human genetics. 2013 Jan;Chapter 7:Unit7.20. [CrossRef]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic acids research. 2002 Sep;30(17):3894–900.
- Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics (Oxford, England). 2006 Nov;22(22):2729–34. [CrossRef]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic acids research. 2001 Jan;29(1):37–40.
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic acids research. 2016 Jul;44(W1):W344-50. [CrossRef]
- Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic acids research. 2005 Jul;33(Web Server issue):W306-10. [CrossRef]
- Cheng J, Randall A, Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins: Structure, Function, and Bioinformatics [Internet]. 2006;62(4):1125–32. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/prot.20810. [CrossRef]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids research. 2019 Jan;47(D1):D607–13.
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome research. 2012 Sep;22(9):1790–7. [CrossRef]
- Pejaver V, Urresti J, Lugo-Martinez J, Pagel K, Lin G, Nam H-J, et al. MutPred2: inferring the molecular and phenotypic impact of amino acid variants. 2017.
- Rogers MF, Gaunt TR, Campbell C. CScape-somatic: distinguishing driver and passenger point mutations in the cancer genome. Bioinformatics (Oxford, England). 2020 Jun;36(12):3637–44.
- 28. Rogers MF, Shihab HA, Gaunt TR, Campbell C. CScape: a tool for predicting oncogenic single-point mutations in the cancer genome. Scientific Reports [Internet]. 2017;7(1):11597. Available from: https://doi.org/10.1038/s41598-017-11746-4. [CrossRef]
- George Priya Doss C, Rajasekaran R, Sethumadhavan R. Computational identification and structural analysis of deleterious functional SNPs in MLL gene causing acute leukemia. Interdisciplinary sciences, computational life sciences. 2010 Sep;2(3):247–55. [CrossRef]
- Long HK, Blackledge NP, Klose RJ. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochemical Society transactions. 2013 Jun;41(3):727–40. [CrossRef]
- Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, Johnson CM, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. The EMBO journal. 2006 Oct;25(19):4503–12.
- Muntean AG, Giannola D, Udager AM, Hess JL. The PHD fingers of MLL block MLL fusion protein-mediated transformation. Blood. 2008 Dec;112(12):4690–3. [CrossRef]
- Ali M, Hom RA, Blakeslee W, Ikenouye L, Kutateladze TG. Diverse functions of PHD fingers of the MLL/KMT2 subfamily. Biochimica et biophysica acta. 2014 Feb;1843(2):366–71. [CrossRef]
- Josling GA, Selvarajah SA, Petter M, Duffy MF. The role of bromodomain proteins in regulating gene expression. Genes. 2012 May;3(2):320–43. [CrossRef]
- Filippakopoulos P, Knapp S. Chapter 10 - Bromodomains as Anticancer Targets. In: Egger G, Arimondo P, editors. Drug Discovery in Cancer Epigenetics [Internet]. Boston: Academic Press; 2016. p. 239–71. Available from: https://www.sciencedirect.com/science/article/pii/B9780128022085000102.
- Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Experimental & Molecular Medicine [Internet]. 2017;49(4):e324–e324. Available from: https://doi.org/10.1038/emm.2017.11. [CrossRef]
- Weirich S, Kudithipudi S, Jeltsch A. Somatic cancer mutations in the MLL1 histone methyltransferase modulate its enzymatic activity and dependence on the WDR5/RBBP5/ASH2L complex. Molecular oncology. 2017 Apr;11(4):373–87.
- Zhang Z-L, Yu P-F, Ling Z-Q. The role of KMT2 gene in human tumors. Histology and histopathology. 2022 Apr;37(4):323–34. [CrossRef]
- Malleshappa Gowder S, Chatterjee J, Chaudhuri T, Paul K. Prediction and analysis of surface hydrophobic residues in tertiary structure of proteins. TheScientificWorldJournal. 2014;2014:971258.
- Singh SM, Kongari N, Cabello-Villegas J, Mallela KMG. Missense mutations in dystrophin that trigger muscular dystrophy decrease protein stability and lead to cross-beta aggregates. Proceedings of the National Academy of Sciences of the United States of America. 2010 Aug;107(34):15069–74.
- Bross P, Corydon TJ, Andresen BS, Jørgensen MM, Bolund L, Gregersen N. Protein misfolding and degradation in genetic diseases. Human mutation. 1999;14(3):186–98.
- Yue P, Moult J. Identification and analysis of deleterious human SNPs. Journal of molecular biology. 2006 Mar;356(5):1263–74. [CrossRef]
- Nagi AD, Regan L. An inverse correlation between loop length and stability in a four-helix-bundle protein. Folding & design. 1997;2(1):67–75. [CrossRef]
- Tastan O, Klein-Seetharaman J, Meirovitch H. The effect of loops on the structural organization of alpha-helical membrane proteins. Biophysical journal. 2009 Mar;96(6):2299–312. [CrossRef]
- Kumar S, Warrell J, Li S, McGillivray PD, Meyerson W, Salichos L, et al. Passenger Mutations in More Than 2,500 Cancer Genomes: Overall Molecular Functional Impact and Consequences. Cell. 2020 Mar;180(5):915-927.e16.
- Stauber, R. H., Hahlbrock, A., Knauer, S. K. & Wünsch, D. Cleaving for growth: threonine aspartase 1--a protease relevant for development and disease. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 30, 1012–1022 (2016).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





