Submitted:
04 October 2023
Posted:
05 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. m6A Dysregulation in Cancer
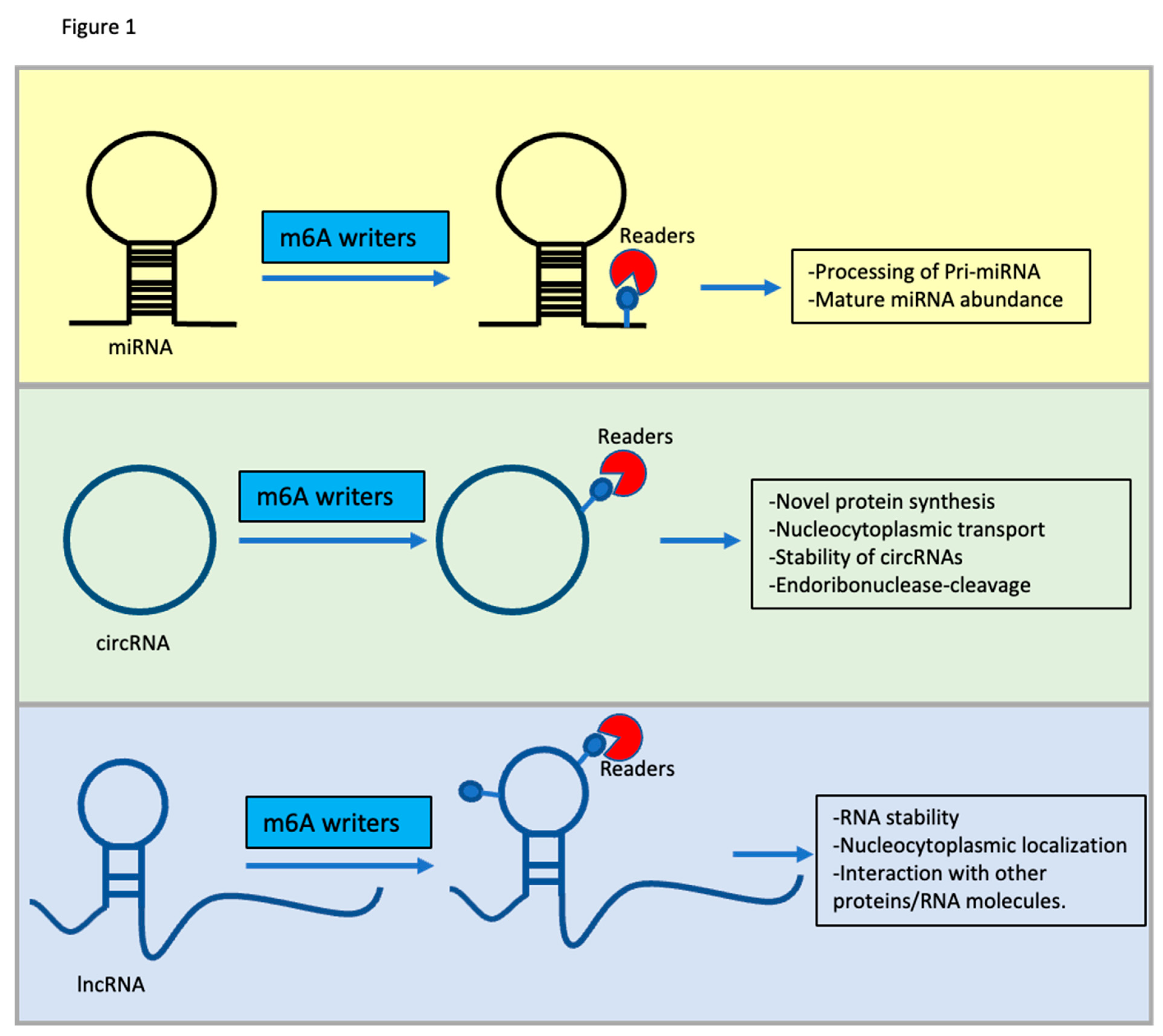
3. m6A Modification on ncRNAs in Cancer
4. MiRNAs
5. circRNAs
6. Long Non-Coding RNA (lncRNAs)
7. Conclusion and Future Directions
Acknowledgments
Conflicts of Interest
References
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of Methylated Nucleosides in Messenger RNA from Novikoff Hepatoma Cells. Proc. Natl. Acad. Sci. 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Jia, G.; Pang, X.; Wang, R.N.; Wang, X.; Li, C.J.; Smemo, S.; Dai, Q.; Bailey, K.A.; Nobrega, M.A.; et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 2013, 4, 1798. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.I.; Parisien, M.; Dai, Q.; Liu, N.; Diatchenko, L.; Sachleben, J.R.; Pan, T. N6-Methyladenosine Modification in a Long Noncoding RNA Hairpin Predisposes Its Conformation to Protein Binding. J. Mol. Biol. 2016, 428, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2013, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Leismann, J.; Spagnuolo, M.; Pradhan, M.; Wacheul, L.; Vu, M.A.; Musheev, M.; Mier, P.; A Andrade-Navarro, M.; Graille, M.; Niehrs, C.; et al. The 18S ribosomal RNA m 6 A methyltransferase Mettl5 is required for normal walking behavior in Drosophila. Embo Rep. 2020, 21, e49443. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Chen, S.; Jia, G. Detection, regulation, and functions of RNA N6-methyladenosine modification in plants. Plant Commun. 2023, 4, 100546. [Google Scholar] [CrossRef]
- Sendinc, E.; Valle-Garcia, D.; Jiao, A.; Shi, Y. Analysis of m6A RNA methylation in Caenorhabditis elegans. Cell Discov. 2020, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Baquero-Perez, B.; Geers, D.; Díez, J. From A to m6A: The Emerging Viral Epitranscriptome. Viruses 2021, 13, 1049. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Cao, S.; Huang, Q.; Xia, L.; Deng, M.; Yang, M.; Jia, G.; Liu, X.; Shi, J.; Wang, W.; et al. The RNA N6-methyladenosine modification landscape of human fetal tissues. Nature 2019, 21, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, K.; Cai, J.; Zhang, M.; Zhang, X.; Xiong, X.; Meng, H.; Xu, X.; Huang, Z.; Peng, J.; et al. Landscape and Regulation of m6A and m6Am Methylome across Human and Mouse Tissues. Mol. Cell 2019, 77, 426–440. [Google Scholar] [CrossRef]
- Qureshi, S.A.; Mumtaz, A.; Shahid, S.U.; Shabana, N. rs3751812, a common variant in fat mass and obesity-associated ( FTO ) gene, is associated with serum high- and low-density lipoprotein cholesterol in Pakistani individuals. Nutrition 2016, 39–40, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Ben-Haim, M.S.; Moshitch-Moshkovitz, S.; Rechavi, G. FTO: linking m6A demethylation to adipogenesis. Cell Res. 2014, 25, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Hsu, P.J.; Xing, X.; Fang, J.; Lu, Z.; Zou, Q.; Zhang, K.-J.; Zhang, X.; Zhou, Y.; Zhang, T.; et al. Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis. Cell Res. 2017, 27, 1216–1230. [Google Scholar] [CrossRef]
- Richard, E.M.; Polla, D.L.; Assir, M.Z.; Contreras, M.; Shahzad, M.; Khan, A.A.; Razzaq, A.; Akram, J.; Tarar, M.N.; Blanpied, T.A.; et al. Bi-allelic Variants in METTL5 Cause Autosomal-Recessive Intellectual Disability and Microcephaly. Am. J. Hum. Genet. 2019, 105, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Han, W.; Zhang, A.; Zhao, M.; Cong, W.; Jia, Y.; Wang, D.; Zhao, R. Chronic corticosterone disrupts the circadian rhythm of CRH expression and m6A RNA methylation in the chicken hypothalamus. J. Anim. Sci. Biotechnol. 2022, 13, 29. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Wang, X.; Cao, C.; Gao, Y.; Zhang, S.; Yang, Z.; Liu, Y.; Zhang, X.; Zhang, W.; Ye, L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2017, 415, 11–19. [Google Scholar] [CrossRef]
- Liu, J.; Eckert, M.A.; Harada, B.T.; Liu, S.-M.; Lu, Z.; Yu, K.; Tienda, S.M.; Chryplewicz, A.; Zhu, A.C.; Yang, Y.; et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018, 20, 1074–1083. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.; Alemu, E.A.; Mertens, C.; Gantman, E.C.; Fak, J.J.; Mele, A.; Haripal, B.; Zucker-Scharff, I.; Moore, M.J.; Park, C.Y.; et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Minerva Anestesiol. 2015, 29, 2037–2053. [Google Scholar] [CrossRef]
- Chen, K.; Lu, Z.; Wang, X.; Fu, Y.; Luo, G.; Liu, N.; Han, D.; Dominissini, D.; Dai, Q.; Pan, T.; et al. High-Resolution N6A) Map Using Photo-Crosslinking-Assisted m6A Sequencing. Angew. Chem. Int. Ed. 2014, 54, 1587–1590. [Google Scholar] [CrossRef] [PubMed]
- Molinie, B.; Wang, J.; Lim, K.S.; Hillebrand, R.; Lu, Z.-X.; Van Wittenberghe, N.; Howard, B.D.; Daneshvar, K.; Mullen, A.C.; Dedon, P.; et al. m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods 2016, 13, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Dierks, D.; Garcia-Campos, M.A.; Uzonyi, A.; Safra, M.; Edelheit, S.; Rossi, A.; Sideri, T.; Varier, R.A.; Brandis, A.; Stelzer, Y.; et al. Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nat. Methods 2021, 18, 1060–1067. [Google Scholar] [CrossRef]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef]
- Garcia-Campos, M.A.; Edelheit, S.; Toth, U.; Safra, M.; Shachar, R.; Viukov, S.; Winkler, R.; Nir, R.; Lasman, L.; Brandis, A.; et al. Deciphering the “m6A Code” via Antibody-Independent Quantitative Profiling. Cell 2019, 178, 731–747. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, L.-Q.; Zhao, Y.-L.; Yang, C.-G.; Roundtree, I.A.; Zhang, Z.; Ren, J.; Xie, W.; He, C.; Luo, G.-Z. Single-base mapping of m 6 A by an antibody-independent method. Sci. Adv. 2019, 5, eaax0250. [Google Scholar] [CrossRef]
- Hendra, C.; Pratanwanich, P.N.; Wan, Y.K.; Goh, W.S.S.; Thiery, A.; Göke, J. Detection of m6A from direct RNA sequencing using a multiple instance learning framework. Nat. Methods 2022, 19, 1590–1598. [Google Scholar] [CrossRef] [PubMed]
- Zaccara, S.; Zaccara, S.; Ries, R.J.; Ries, R.J.; Jaffrey, S.R.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Wang, S.; Liu, J.; Huang, Y.; Gong, C.; Liu, J.; Xiao, Y.; Yang, S. New sights in cancer: Component and function of N6-methyladenosine modification. Biomed. Pharmacother. 2019, 122, 109694. [Google Scholar] [CrossRef] [PubMed]
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 1997, 3, 1233–1247. [Google Scholar] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2013, 10, 93–95. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Agarwala, S.D.; Blitzblau, H.G.; Hochwagen, A.; Fink, G.R. RNA Methylation by the MIS Complex Regulates a Cell Fate Decision in Yeast. PLOS Genet. 2012, 8, e1002732. [Google Scholar] [CrossRef]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A Writers Reveals Two Distinct Classes of mRNA Methylation at Internal and 5′ Sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef]
- Patil, D.P.; Chen, C.-K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villaseñor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Minerva Anestesiol. 2018, 32, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Wang, Q.; Huang, J.; Tang, C.; et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Śledź, P.; Jinek, M. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife 2016, 5, e18434. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, Y.; Sun, B.-F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.-J.; Ping, X.-L.; Chen, Y.-S.; Wang, W.-J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef]
- Bartosovic, M.; Molares, H.C.; Gregorova, P.; Hrossova, D.; Kudla, G.; Vanacova, S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 2017, 45, 11356–11370. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Zhang, Z.; Theler, D.; Kaminska, K.H.; Hiller, M.; de la Grange, P.; Pudimat, R.; Rafalska, I.; Heinrich, B.; Bujnicki, J.M.; Allain, F.H.-T.; et al. The YTH Domain Is a Novel RNA Binding Domain. J. Biol. Chem. 2010, 285, 14701–14710. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.-S.; Hao, Y.-J.; Sun, B.-F.; Sun, H.-Y.; Li, A.; Ping, X.-L.; Lai, W.-Y.; et al. Nuclear m 6 A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Luo, G.-Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Gao, Y.; Wang, B.; Qu, Y.-X. Methylation recognition protein YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) regulates the proliferation, migration and invasion of osteosarcoma by regulating m6A level of CCR4-NOT transcription complex subunit 7 (CNOT7). Bioengineered 2022, 13, 5236–5250. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Chen, Y.-S.; Ping, X.-L.; Yang, X.; Xiao, W.; Yang, Y.; Sun, H.-Y.; Zhu, Q.; Baidya, P.; Wang, X.; et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017, 27, 444–447. [Google Scholar] [CrossRef]
- Bailey, A.S.; Batista, P.J.; Gold, R.S.; Chen, Y.G.; de Rooij, D.G.; Chang, H.Y.; Fuller, M.T. The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife 2017, 6, e26116. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef]
- Wojtas, M.N.; Pandey, R.R.; Mendel, M.; Homolka, D.; Sachidanandam, R.; Pillai, R.S. Regulation of m6A Transcripts by the 3ʹ→5ʹ RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline. Mol. Cell 2017, 68, 374–387. [Google Scholar] [CrossRef]
- Tanabe, A.; Tanikawa, K.; Tsunetomi, M.; Takai, K.; Ikeda, H.; Konno, J.; Torigoe, T.; Maeda, H.; Kutomi, G.; Okita, K.; et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016, 376, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Qiao, J.; Wang, G.; Lan, Y.; Li, G.; Guo, X.; Xi, J.; Ye, D.; Zhu, S.; Chen, W.; et al. N 6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018, 46, 3906–3920. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, J.; Feng, G.; Gao, S.; Wang, Y.; Zhang, S.; Liu, Y.; Ye, L.; Li, Y.; Zhang, X. MicroRNA-145 Modulates N6-Methyladenosine Levels by Targeting the 3′-Untranslated mRNA Region of the N6-Methyladenosine Binding YTH Domain Family 2 Protein. J. Biol. Chem. 2017, 292, 3614–3623. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sun, Y.; Davis, A.E.; Shah, S.H.; Hamed, L.K.; Wu, M.-R.; Lin, C.-H.; Ding, J.B.; Wang, S. Mettl14-mediated m6A modification ensures the cell-cycle progression of late-born retinal progenitor cells. Cell Rep. 2023, 42, 112596–112596. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Liu, W.; Zhang, Y.; Yang, Y.; Jiang, X.; Wu, S.; Shao, L. METTL3-mediated m6A modification regulates cell cycle progression of dental pulp stem cells. Stem Cell Res. Ther. 2021, 12, 159. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhong, Y.; Cao, G.; Shi, H.; Liu, Y.; Li, L.; Yin, P.; Chen, J.; Xiao, Z.; Du, B. METTL3 promotes cell cycle progression via m6A/YTHDF1-dependent regulation of CDC25B translation. Int. J. Biol. Sci. 2022, 18, 3223–3236. [Google Scholar] [CrossRef]
- Jia, R.; Chai, P.; Wang, S.; Sun, B.; Xu, Y.; Yang, Y.; Ge, S.; Jia, R.; Yang, Y.-G.; Fan, X. m6A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol. Cancer 2019, 18, 161. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, B.; Shi, J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene 2019, 722, 144076. [Google Scholar] [CrossRef]
- Visvanathan, A.; Patil, V.; Arora, A.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Somasundaram, K. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 2017, 37, 522–533. [Google Scholar] [CrossRef]
- Li, T.; Hu, P.-S.; Zuo, Z.; Lin, J.-F.; Li, X.; Wu, Q.-N.; Chen, Z.-H.; Zeng, Z.-L.; Wang, F.; Zheng, J.; et al. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer 2019, 18, 112. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Wang, X.; Cao, C.; Gao, Y.; Zhang, S.; Yang, Z.; Liu, Y.; Zhang, X.; Zhang, W.; Ye, L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2017, 415, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m 6 A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.; Lin, S.; Zhang, W.; Liu, Q.; Wang, L.; Ramirez-Moya, J.; Du, P.; Kim, W.; Tang, S.; Sliz, P.; et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 2018, 561, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kang, M.; Zhang, B.; Meng, F.; Song, J.; Kaneko, H.; Shimamoto, F.; Tang, B. RETRACTED ARTICLE: m6A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol. Cancer 2019, 18, 185. [Google Scholar] [CrossRef] [PubMed]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; Mackay, M.; et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Weng, H.; Huang, H.; Wu, H.; Qin, X.; Zhao, B.S.; Dong, L.; Shi, H.; Skibbe, J.; Shen, C.; Hu, C.; et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell 2017, 22, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chai, P.; Jia, R.; Jia, R. Novel insights on m6A RNA methylation in tumorigenesis: a double-edged sword. Mol. Cancer 2018, 17, 101. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Ye, M.; Liu, B.; Wei, M.; Ma, D.; Dong, K. m6A Modification: A Double-Edged Sword in Tumor Development. Front. Oncol. 2021, 11, 679367. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; He, Q.; Feng, Y.; Chen, M.; Zhang, D. A m6Avalue predictive of prostate cancer stemness, tumor immune landscape and immunotherapy response. NAR Cancer 2022, 4, zcac010. [Google Scholar] [CrossRef]
- Xie, J.; Ba, J.; Zhang, M.; Wan, Y.; Jin, Z.; Yao, Y. The m6A methyltransferase METTL3 promotes the stemness and malignant progression of breast cancer by mediating m6A modification on SOX2. J. BUON 2021, 26, 444–449. [Google Scholar] [PubMed]
- Fang, Z.; Mei, W.; Qu, C.; Lu, J.; Shang, L.; Cao, F.; Li, F. Role of m6A writers, erasers and readers in cancer. Exp. Hematol. Oncol. 2022, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Yang, F.; Zhan, H.; Situ, J.; Li, W.; Mao, Y.; Luo, Y. RNA m6A Methyltransferase METTL3 Promotes The Growth Of Prostate Cancer By Regulating Hedgehog Pathway. OncoTargets Ther. 2019, 12, 9143–9152. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Du, Y.; Wang, L.; Liu, X. The M6A methyltransferase METTL3 promotes the development and progression of prostate carcinoma via mediating MYC methylation. J. Cancer 2020, 11, 3588–3595. [Google Scholar] [CrossRef]
- Chen, Y.; Pan, C.; Wang, X.; Xu, D.; Ma, Y.; Hu, J.; Chen, P.; Xiang, Z.; Rao, Q.; Han, X. Silencing of METTL3 effectively hinders invasion and metastasis of prostate cancer cells. Theranostics 2021, 11, 7640–7657. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, L.; Wang, C.; Lu, S.; Yang, X.; He, Y.; Zhang, C.Z.; Yun, J. Loss of RDM1 enhances hepatocellular carcinoma progression via p53 and Ras/Raf/ERK pathways. Mol. Oncol. 2019, 14, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wei, L.; Law, C.; Tsang, F.H.; Shen, J.; Cheng, C.L.; Tsang, L.; Ho, D.W.; Chiu, D.K.; Lee, J.M.; et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2017, 67, 2254–2270. [Google Scholar] [CrossRef]
- Gao, Q.; Zheng, J.; Ni, Z.; Sun, P.; Yang, C.; Cheng, M.; Wu, M.; Zhang, X.; Yuan, L.; Zhang, Y.; et al. The m6A Methylation-Regulated AFF4 Promotes Self-Renewal of Bladder Cancer Stem Cells. Stem Cells Int. 2020, 2020, 8849218. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Li, X.; Sun, H.; Gao, Z.; Zhu, Z.; Yuan, K. Role of WTAP in Cancer: From Mechanisms to the Therapeutic Potential. Biomolecules 2022, 12, 1224. [Google Scholar] [CrossRef]
- Kuai, Y.; Gong, X.; Ding, L.; Li, F.; Lei, L.; Gong, Y.; Liu, Q.; Tan, H.; Zhang, X.; Liu, D.; et al. Wilms’ tumor 1-associating protein plays an aggressive role in diffuse large B-cell lymphoma and forms a complex with BCL6 via Hsp90. Cell Commun. Signal. 2018, 16, 50. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhao, K.; Zeng, H.; Li, Z.; Chen, K.; Zhang, Z.; Li, E.; Wu, Z. N6-methyladenosine (m6A) methyltransferase WTAP accelerates the Warburg effect of gastric cancer through regulating HK2 stability. Biomed. Pharmacother. 2020, 133, 111075. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.-Y.; Gao, J.; Sun, X.; Cao, M.-D.; Shi, L.; Xia, T.-S.; Zhou, W.-B.; Wang, S.; Ding, Q.; Wei, J.-F. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene 2019, 38, 6123–6141. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Li, H.; Zhang, D.; Xu, L.; Liu, H.; Hao, X.; Yan, X.; Liao, H.; Chen, X.; Xie, K.; et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 2019, 18, 186. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.-J.; Shao, Y.-C.; Yang, Y.; Song, W.-J.; He, X.; Zeng, Y.-F.; Huang, S.-R.; Wei, L.; Zhang, J.-W. Analysis of N6-Methyladenosine Methyltransferase Reveals METTL14 and ZC3H13 as Tumor Suppressor Genes in Breast Cancer. Front. Oncol. 2020, 10, 578963. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, S.; He, C.; Xue, P.; Zhang, L.; He, Z.; Zang, L.; Feng, B.; Sun, J.; Zheng, M. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer 2020, 19, 46. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; He, L.; Gao, X.; Tang, N.; Lin, L.; Yu, X.; Wang, D. The m6A Methyltransferase METTL14-Mediated N6-Methyladenosine Modification of PTEN mRNA Inhibits Tumor Growth and Metastasis in Stomach Adenocarcinoma. Front. Oncol. 2021, 11, 699749. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhuang, Y.; Zhang, J.; Chen, M.; Wu, S. METTL14 Inhibits Hepatocellular Carcinoma Metastasis Through Regulating EGFR/PI3K/AKT Signaling Pathway in an m6A-Dependent Manner. Cancer Manag. Res. 2020, 12, 13173–13184. [Google Scholar] [CrossRef]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N 6 -Methyladenosine RNA Demethylase. Cancer Cell 2016, 31, 127–141. [Google Scholar] [CrossRef]
- Bian, X.; Shi, D.; Xing, K.; Zhou, H.; Lu, L.; Yu, D.; Wu, W. AMD1 upregulates hepatocellular carcinoma cells stemness by FTO mediated mRNA demethylation. Clin. Transl. Med. 2021, 11, e352. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Lin, Z.; Wan, A.; Chen, H.; Liang, H.; Sun, L.; Wang, Y.; Li, X.; Xiong, X.-F.; Wei, B.; et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Cancer 2019, 18, 46. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ren, D.; Du, Z.; Wang, H.; Zhang, H.; Jin, Y. m 6 A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem. Biophys. Res. Commun. 2018, 502, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Gao, Q.; Wang, S. Kinase GSK3β functions as a suppressor in colorectal carcinoma through the FTO-mediated MZF1/c-Myc axis. J. Cell. Mol. Med. 2021, 25, 2655–2665. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.; Dong, L.; Li, C.; Yin, Z.; Rao, S.; Zhou, Q. The m6A eraser FTO facilitates proliferation and migration of human cervical cancer cells. Cancer Cell Int. Erratum in: Cancer Cell Int. 2020, 20, 423. 2019, 19, 321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, L.; Lou, W.; Su, J.; Huang, J.; Liu, A.; Xu, Y.; He, H.; Gao, Y.; Xu, D.; et al. Aberrant activation of m6A demethylase FTO renders HIF2α low/− clear cell renal cell carcinoma sensitive to BRD9 inhibitors. Sci. Transl. Med. 2021, 13, eabf6045. [Google Scholar] [CrossRef] [PubMed]
- Jiao, M.; Tian, R.; Liu, G.; Liu, X.; Wei, Q.; Yan, J.; Wang, K.; Yang, P. Circular RNA and Messenger RNA Expression Profile and Competing Endogenous RNA Network in Subchondral Bone in Osteonecrosis of the Femoral Head. DNA Cell Biol. 2021, 40, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Rong, Z.-X.; Li, Z.; He, J.-J.; Liu, L.-Y.; Ren, X.-X.; Gao, J.; Mu, Y.; Guan, Y.-D.; Duan, Y.-M.; Zhang, X.-P.; et al. Downregulation of Fat Mass and Obesity Associated (FTO) Promotes the Progression of Intrahepatic Cholangiocarcinoma. Front. Oncol. 2019, 9, 369. [Google Scholar] [CrossRef] [PubMed]
- Nagaki, Y.; Motoyama, S.; Yamaguchi, T.; Hoshizaki, M.; Sato, Y.; Sato, T.; Koizumi, Y.; Wakita, A.; Kawakita, Y.; Imai, K.; et al. m6A demethylase ALKBH5 promotes proliferation of esophageal squamous cell carcinoma associated with poor prognosis. Genes Cells 2020, 25, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Zhang, L.; Liu, J.; Wan, Y.; Jiang, Y.; Yang, J.; Sun, R.; Ma, X.; Sun, G.; Meng, H.; et al. ALKBH5-HOXA10 loop-mediated JAK2 m6A demethylation and cisplatin resistance in epithelial ovarian cancer. J. Exp. Clin. Cancer Res. 2021, 40, 284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, F.; Wang, Z.; Yang, X.; Yu, H.; Si, S.; Lu, J.; Zhou, Z.; Lu, Q.; Wang, Z.; et al. ALKBH5 promotes the proliferation of renal cell carcinoma by regulating AURKB expression in an m6A-dependent manner. Ann. Transl. Med. 2020, 8, 646. [Google Scholar] [CrossRef]
- Qiu, X.; Yang, S.; Wang, S.; Wu, J.; Zheng, B.; Wang, K.; Shen, S.; Jeong, S.; Li, Z.; Zhu, Y.; et al. M6A Demethylase ALKBH5 Regulates PD-L1 Expression and Tumor Immunoenvironment in Intrahepatic Cholangiocarcinoma. Cancer Res 2021, 81, 4778–4793. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, Y.; Chen, J.; Peng, C.; Zhang, Y.; Tong, R.; Cheng, Q.; Yang, B.; Feng, X.; Lu, Y.; et al. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m6A-guided epigenetic inhibition of LYPD1. Mol. Cancer 2020, 19, 123. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Yang, Y.; Kang, M.; Wang, Y.; Wang, Y.; Bi, Y.; He, S.; Shimamoto, F. m6A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol. Cancer 2020, 19, 3. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Fan, S.; Wu, M.; Zuo, Z.; Li, X.; Jiang, L.; Shen, Q.; Xu, P.; Zeng, L.; Zhou, Y.; et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat. Commun. 2019, 10, 4892. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.; Shi, L.; Ye, Y.; Shi, H.; Zeng, L.; Tiwary, S.; Huse, J.T.; Huo, L.; Ma, L.; Ma, Y.; et al. YTHDF3 Induces the Translation of m6A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 2020, 38, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Einstein, J.M.; Perelis, M.; Chaim, I.A.; Meena, J.K.; Nussbacher, J.K.; Tankka, A.T.; Yee, B.A.; Li, H.; Madrigal, A.A.; Neill, N.J.; et al. Inhibition of YTHDF2 triggers proteotoxic cell death in MYC-driven breast cancer. Mol. Cell 2021, 81, 3048–3064. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Liao, D.; Zhang, M.; Zeng, C.; Li, X.; Zhang, R.; Ma, H.; Kang, T. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2018, 442, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Waly, A.A.; El-Ekiaby, N.; Assal, R.A.; Abdelrahman, M.M.; Hosny, K.A.; El Tayebi, H.M.; Esmat, G.; Breuhahn, K.; Abdelaziz, A.I. Methylation in MIRLET7A3 Gene Induces the Expression of IGF-II and Its mRNA Binding Proteins IGF2BP-2 and 3 in Hepatocellular Carcinoma. Front. Physiol. 2019, 9, 1918. [Google Scholar] [CrossRef]
- Zhang, L.; Wan, Y.; Zhang, Z.; Jiang, Y.; Gu, Z.; Ma, X.; Nie, S.; Yang, J.; Lang, J.; Cheng, W.; et al. IGF2BP1 overexpression stabilizes PEG10 mRNA in an m6A-dependent manner and promotes endometrial cancer progression. Theranostics 2021, 11, 1100–1114. [Google Scholar] [CrossRef]
- Xu, Y.; Guo, Z.; Peng, H.; Guo, L.; Wang, P. IGF2BP3 promotes cell metastasis and is associated with poor patient survival in nasopharyngeal carcinoma. J. Cell. Mol. Med. 2021, 26, 410–421. [Google Scholar] [CrossRef]
- Tan, B.; Zhou, K.; Liu, W.; Prince, E.; Qing, Y.; Li, Y.; Han, L.; Qin, X.; Su, R.; Pokharel, S.P.; et al. RNA N6-methyladenosine reader YTHDC1 is essential for TGF-beta-mediated metastasis of triple negative breast cancer. Theranostics 2022, 12, 5727–5743. [Google Scholar] [CrossRef]
- Su, Y.; Wang, B.; Huang, J.; Huang, M.; Lin, T. YTHDC1 positively regulates PTEN expression and plays a critical role in cisplatin resistance of bladder cancer. Cell Prolif. 2023, 56, e13404. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; You, G.; Yin, X.; Zhu, G.; Wang, W.; Yu, Y.; Zhu, J. Overexpression of YTHDC2 contributes to the progression of prostate cancer and predicts poor outcomes in patients with prostate cancer. J. Biochem. Mol. Toxicol. 2023, 37, e23308. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Xu, H.; Bai, G.; Hu, H.; Wang, D.; Li, H.; Wang, Z. ELAVL1 promotes prostate cancer progression by interacting with other m6A regulators. Front. Oncol. 2022, 12, 939784. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, D.; Sun, L.; Qin, H.; Fan, A.; Meng, L.; Graves-Deal, R.; Glass, S.E.; Franklin, J.L.; Liu, Q.; et al. Interaction of lncRNA MIR100HG with hnRNPA2B1 facilitates m6A-dependent stabilization of TCF7L2 mRNA and colorectal cancer progression. Mol. Cancer 2022, 21, 74. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Kou, R.; Meng, J.; Jiang, D.; Zhong, R.; Dong, M. m6A genotypes and prognostic signature for assessing the prognosis of patients with acute myeloid leukemia. BMC Med Genom. 2023, 16, 191. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Yu, B.; Tao, D.; Xu, X.; Xu, Y.; Wang, J.; Jiao, Y.; Wang, L. The role of m6A methylation in therapy resistance in cancer. Mol. Cancer 2023, 22, 91. [Google Scholar] [CrossRef] [PubMed]
- Smolarz, B.; Durczyński, A.; Romanowicz, H.; Szyłło, K.; Hogendorf, P. miRNAs in Cancer (Review of Literature). Int. J. Mol. Sci. 2022, 23, 2805. [Google Scholar] [CrossRef]
- Yoshida, T.; Asano, Y.; Ui-Tei, K. Modulation of MicroRNA Processing by Dicer via Its Associated dsRNA Binding Proteins. Non-Coding RNA 2021, 7, 57. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Zhang, J.; Bai, R.; Li, M.; Ye, H.; Wu, C.; Wang, C.; Li, S.; Tan, L.; Mai, D.; Li, G.; et al. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat. Commun. 2019, 10, 1858. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.M.; Hwang, S.J.; Masuda, K.; Choi, K.-M.; Jeong, M.-R.; Nam, D.-H.; Gorospe, M.; Kim, H.H. Heterogeneous Nuclear Ribonucleoprotein C1/C2 Controls the Metastatic Potential of Glioblastoma by Regulating PDCD4. Mol. Cell. Biol. 2012, 32, 4237–4244. [Google Scholar] [CrossRef] [PubMed]
- Klinge, C.M.; Piell, K.M.; Tooley, C.S.; Rouchka, E.C. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci. Rep. 2019, 9, 9430. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhang, Q.; Pang, W.; Hou, L.; Liang, Y.; Han, X.; Luo, X.; Wang, P.; Zhang, X.; Li, L.; et al. YTHDC1-mediated augmentation of miR-30d in repressing pancreatic tumorigenesis via attenuation of RUNX1-induced transcriptional activation of Warburg effect. Cell Death Differ. 2021, 28, 3105–3124. [Google Scholar] [CrossRef] [PubMed]
- Rong, L.; Xu, Y.; Zhang, K.; Jin, L.; Liu, X. HNRNPA2B1 inhibited SFRP2 and activated Wnt-β/catenin via m6A-mediated miR-106b-5p processing to aggravate stemness in lung adenocarcinoma. Pathol. Res. Pr. 2022, 233, 153794. [Google Scholar] [CrossRef]
- Yi, D.; Wang, R.; Shi, X.; Xu, L.; Yilihamu, Y.; Sang, J. METTL14 promotes the migration and invasion of breast cancer cells by modulating N6-methyladenosine and hsa-miR-146a-5p expression. Oncol. Rep. 2020, 43, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.; Wang, R.; Shi, X.; Xu, L.; Yilihamu, Y.; Sang, J. METTL14 promotes the migration and invasion of breast cancer cells by modulating N6-methyladenosine and hsa-miR-146a-5p expression. Oncol. Rep. 2020, 43, 1375–1386. [Google Scholar] [CrossRef]
- Gao, C.; Wei, J.; Tang, T.; Huang, Z. Role of microRNA-33a in malignant cells (Review). Oncol. Lett. 2020, 20, 2537–2556. [Google Scholar] [CrossRef]
- Shan, Y.; Liu, Y.; Zhao, L.; Liu, B.; Li, Y.; Jia, L. MicroRNA-33a and let-7e inhibit human colorectal cancer progression by targeting ST8SIA1. Int. J. Biochem. Cell Biol. 2017, 90, 48–58. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Y.; Ding, W.; Lin, Y.; Huang, Z.; Luo, Q. MiR-33a suppresses breast cancer cell proliferation and metastasis by targeting ADAM9 and ROS1. Protein Cell 2015, 6, 881–889. [Google Scholar] [CrossRef]
- Su, X.; Lai, T.; Tao, Y.; Zhang, Y.; Zhao, C.; Zhou, J.; Chen, E.; Zhu, M.; Zhang, S.; Wang, B.; et al. miR-33a-3p regulates METTL3-mediated AREG stability and alters EMT to inhibit pancreatic cancer invasion and metastasis. Sci. Rep. 2023, 13, 13587. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Zhang, Y.; Mao, Y.; Mou, J.; Zhao, J.; Xue, Q.; Wang, D.; Huang, J.; Gao, S.; Gao, Y. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem. Biophys. Res. Commun. 2017, 482, 582–589. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Chen, S.; Ying, Y.; Xie, H.; Li, J.; Ma, X.; Wang, W.; Shen, H.; Wang, X.; Zheng, X.; et al. MicroRNA-501-3p inhibits the proliferation of kidney cancer cells by targeting WTAP. Cancer Med. 2021, 10, 7222–7232. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Gao, X.; Chen, X.; Zhao, N.; Sun, Y.; Zou, Y.; Guan, Y.; Yang, L.; Pei, X.; Wang, G.; et al. miR-139-5p Loss-Mediated WTAP Activation Contributes to Hepatocellular Carcinoma Progression by Promoting the Epithelial to Mesenchymal Transition. Front. Oncol. 2021, 11, 611544. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Xiao, P.; Yu, X.; Zhang, X. A positive feedback loop between AlkB homolog 5 and miR-193a-3p promotes growth and metastasis in esophageal squamous cell carcinoma. Hum. Cell 2020, 34, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Yuan, X.; Wu, S.; Yuan, Y.; Cui, L.; Lin, D.; Peng, X.; Liu, X.; Wang, F. Effects of writers, erasers and readers within miRNA-related m6A modification in cancers. Cell Prolif. 2022, 56, e13340. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Das Mandal, S.; Ray, P.S. Transcriptome-wide analysis reveals spatial correlation between N6-methyladenosine and binding sites of microRNAs and RNA-binding proteins. Genomics 2020, 113, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Kanoria, S.; A Rennie, W.; Carmack, C.S.; Lu, J.; Ding, Y. N 6-methyladenosine enhances post-transcriptional gene regulation by microRNAs. Bioinform. Adv. 2022, 2, vbab046. [Google Scholar] [CrossRef]
- He, X.; Shu, Y. RNA N6-methyladenosine modification participates in miR-660/E2F3 axis-mediated inhibition of cell proliferation in gastric cancer. Pathol. - Res. Pr. 2019, 215, 152393. [Google Scholar] [CrossRef]
- Zhang, M.; Xin, Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J. Hematol. Oncol. 2018, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Yang, Y. Biological functions and applications of circRNA-next generation of RNA-based therapy. J. Mol. Cell Biol. 2023, mjad031. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Chen, R.-X.; Chen, X.; Xia, L.-P.; Zhang, J.-X.; Pan, Z.-Z.; Ma, X.-D.; Han, K.; Chen, J.-W.; Judde, J.-G.; Deas, O.; et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019, 10, 4695. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.-N.; Chen, Z.-Y.; Chen, X.-Y.; Chen, M.; Yi, Y.-C.; Zhu, J.-S.; Zhang, J. METTL14-mediated m6A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol. Cancer 2022, 21, 51. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lan, T.; Li, H.; Xu, L.; Chen, X.; Liao, H.; Chen, X.; Du, J.; Cai, Y.; Wang, J.; et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics 2021, 11, 1396–1411. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yang, H.-Y.; Dai, X.-Y.; Zhang, X.; Huang, Y.-Z.; Shi, L.; Wei, J.-F.; Ding, Q. CircMETTL3, upregulated in a m6A-dependent manner, promotes breast cancer progression. Int. J. Biol. Sci. 2021, 17, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yuan, W.; Zhou, Q.; Shao, B.; Guo, Y.; Wang, W.; Yang, S.; Guo, Y.; Zhao, L.; Dang, Q.; et al. N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics 2021, 11, 4298–4315. [Google Scholar] [CrossRef]
- Ji, F.; Lu, Y.; Chen, S.; Yu, Y.; Lin, X.; Zhu, Y.; Luo, X. IGF2BP2-modified circular RNA circARHGAP12 promotes cervical cancer progression by interacting m6A/FOXM1 manner. Cell Death Discov. 2021, 7, 215. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Tian, K.; Luo, W.; Li, M. m6A-modified circRNA MYO1C participates in the tumor immune surveillance of pancreatic ductal adenocarcinoma through m6A/PD-L1 manner. Cell Death Dis. 2023, 14, 120. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.-L.; Chen, W.; Xie, J.-J.; Zhang, M.-L.; Nie, R.-C.; Liang, H.; Mei, J.; Han, K.; Xiang, Z.-C.; Wang, F.-W.; et al. A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol. Cancer 2022, 21, 93. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yin, X.; Zhao, W.; Xu, W.; Chen, L. Molecular mechanism of m6A methylation of circDLC1 mediated by RNA methyltransferase METTL3 in the malignant proliferation of glioma cells. Cell Death Discov. 2022, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhu, L.; Lu, C.; Wang, C.; Wang, H.; Jin, H.; Ma, X.; Cheng, Z.; Yu, C.; Wang, S.; et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat. Commun. 2021, 12, 295. [Google Scholar] [CrossRef] [PubMed]
- Pisignano, G.; Michael, D.C.; Visal, T.H.; Pirlog, R.; Ladomery, M.; Calin, G.A. Going circular: history, present, and future of circRNAs in cancer. Oncogene 2023, 42, 2783–2800. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2013, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Aprile, M.; Costa, V.; Cimmino, A.; Calin, G.A. Emerging role of oncogenic long noncoding RNA as cancer biomarkers. Int. J. Cancer 2022, 152, 822–834. [Google Scholar] [CrossRef] [PubMed]
- Fonseca-Montaño, M.A.; Vázquez-Santillán, K.I.; Hidalgo-Miranda, A. The current advances of lncRNAs in breast cancer immunobiology research. Front. Immunol. 2023, 14, 1194300. [Google Scholar] [CrossRef]
- Li, J.; Momen-Heravi, F.; Wu, X.; He, K. Mechanism of METTL14 and m6A modification of lncRNA MALAT1 in the proliferation of oral squamous cell carcinoma cells. Oral Dis. 2022, 29, 2012–2026. [Google Scholar] [CrossRef]
- Li, S.; Jiang, F.; Chen, F.; Deng, Y.; Pan, X. Effect of m6A methyltransferase METTL3 -mediated MALAT1/E2F1/AGR2 axis on adriamycin resistance in breast cancer. J. Biochem. Mol. Toxicol. 2021, 36, e22922. [Google Scholar] [CrossRef]
- Lee, J.; Wu, Y.; Harada, B.T.; Li, Y.; Zhao, J.; He, C.; Ma, Y.; Wu, X. N 6 -methyladenosine modification of lncRNA Pvt1 governs epidermal stemness. EMBO J. 2021, 40, e106276. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, L.; Wang, Y. ALKBH5-mediated m6A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020, 20, 34. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Lv, F.; Li, N.; Yuan, X.; Zhang, L.; Zhao, S.; Jin, L.; Qiu, Y. Long noncoding RNAMEG3 inhibits oral squamous cell carcinoma progression via GATA3. FEBS Open Bio 2022, 13, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Gong, Q.; Xiang, X.-D.; Guo, G.; Liu, J.; Zhao, L.; Li, J.; Chen, N.; Li, H.; Zhang, L.-J.; et al. HNRNPA2B1-mediated m6A modification of lncRNA MEG3 facilitates tumorigenesis and metastasis of non-small cell lung cancer by regulating miR-21-5p/PTEN axis. J. Transl. Med. 2023, 21, 382. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Yao, S.; Zhou, Y.; Liu, Y.; Huang, P.; Zhou, A.; Liu, J.; Che, L.; Li, J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol. Cancer 2019, 18, 143. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; He, F.; Hou, Y.; Tu, G.; Li, Q.; Jin, T.; Zeng, H.; Qin, Y.; Wan, X.; Qiao, Y.; et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene 2021, 40, 1609–1627. [Google Scholar] [CrossRef]
- Ma, F.; Liu, X.; Zhou, S.; Li, W.; Liu, C.; Chadwick, M.; Qian, C. Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 2019, 450, 63–75. [Google Scholar] [CrossRef]
- Lan, T.; Li, H.; Zhang, D.; Xu, L.; Liu, H.; Hao, X.; Yan, X.; Liao, H.; Chen, X.; Xie, K.; et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 2019, 18, 186. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.; Su, H.; Zhang, Q.; Wu, W.-Y.; Zeng, Y.; Li, X.-M.; Xiong, J.; Chen, L.-F.; Zhou, Y. Comprehensive analysis of lncRNAs N6-methyladenosine modification in colorectal cancer. Aging 2021, 13, 4182–4198. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Xu, Y.; Xu, S.; Jin, L.; Shen, Y.; Rajan, K.C.; Bhandari, A.; Xia, E. Construction and Analysis of a Colorectal Cancer Prognostic Model Based on N6-Methyladenosine-Related lncRNAs. Front. Cell Dev. Biol. 2021, 9, 698388. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Ren, J.; Yuan, W.; Xiang, R.; Ge, Y.; Fu, T. N6-Methyladenosine-Related lncRNA Signature Predicts the Overall Survival of Colorectal Cancer Patients. Genes 2021, 12, 1375. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Meng, Q.; Ma, B. Characterization of the Prognostic m6A-Related lncRNA Signature in Gastric Cancer. Front. Oncol. 2021, 11, 630260. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.; Wu, L.; Wang, P.; Hu, Q.; Tao, C.; Li, K.; Huang, K.; Zhu, X. N6-Methylandenosine-Related lncRNAs Are Potential Biomarkers for Predicting the Overall Survival of Lower-Grade Glioma Patients. Front. Cell Dev. Biol. 2020, 8, 642. [Google Scholar] [CrossRef] [PubMed]
- Weng, L.; Qiu, K.; Gao, W.; Shi, C.; Shu, F. LncRNA PCGEM1 accelerates non-small cell lung cancer progression via sponging miR-433-3p to upregulate WTAP. BMC Pulm. Med. 2020, 20, 213. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Chang, Y.; Zhu, L.; Shen, C.; Qian, J.; Chang, R. LINC00839/miR-144-3p/WTAP (WT1 Associated protein) axis is involved in regulating hepatocellular carcinoma progression. Bioengineered 2021, 12, 10849–10861. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Liu, M.; Zhang, Y.; Xie, L.; Shi, Z.; Wang, G. SNHG10/miR-141-3p/WTAP axis promotes osteosarcoma proliferation and migration. J. Biochem. Mol. Toxicol. 2022, 36, e23031. [Google Scholar] [CrossRef]
- Huang, T.; Cao, L.; Feng, N.; Xu, B.; Dong, Y.; Wang, M. N6-methyladenosine (m6A)-mediated lncRNA DLGAP1-AS1enhances breast canceradriamycin resistance through miR-299-3p/WTAP feedback loop. Bioengineered 2021, 12, 10935–10944. [Google Scholar] [CrossRef]
- Bedi, R.K.; Huang, D.; Li, Y.; Caflisch, A. Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3. ACS Bio Med Chem Au 2023, 3, 359–370. [Google Scholar] [CrossRef]
- Moroz-Omori, E.V.; Huang, D.; Bedi, R.K.; Cheriyamkunnel, S.J.; Bochenkova, E.; Dolbois, A.; Rzeczkowski, M.D.; Li, Y.; Wiedmer, L.; Caflisch, A. METTL3 Inhibitors for Epitranscriptomic Modulation of Cellular Processes. ChemMedChem 2021, 16, 3035–3043. [Google Scholar] [CrossRef]
- Zhang, L.; Ren, T.; Wang, Z.; Wang, R.; Chang, J. Comparative study of the binding of 3 flavonoids to the fat mass and obesity-associated protein by spectroscopy and molecular modeling. J. Mol. Recognit. 2017, 30, e2606. [Google Scholar] [CrossRef]
- Chen, B.; Ye, F.; Yu, L.; Jia, G.; Huang, X.; Zhang, X.; Peng, S.; Chen, K.; Wang, M.; Gong, S.; et al. Development of Cell-Active N6-Methyladenosine RNA Demethylase FTO Inhibitor. J. Am. Chem. Soc. 2012, 134, 17963–17971. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, M.; Huang, H.; Zhu, J.; Song, H.; Zhu, J.; Park, J.; Ji, S.-J. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 2017, 46, 1412–1423. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yan, J.; Li, Q.; Li, J.; Gong, S.; Zhou, H.; Gan, J.; Jiang, H.; Jia, G.F.; Luo, C.; Yang, C.G. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015, 43, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



