Submitted:
03 October 2023
Posted:
04 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Material and methods
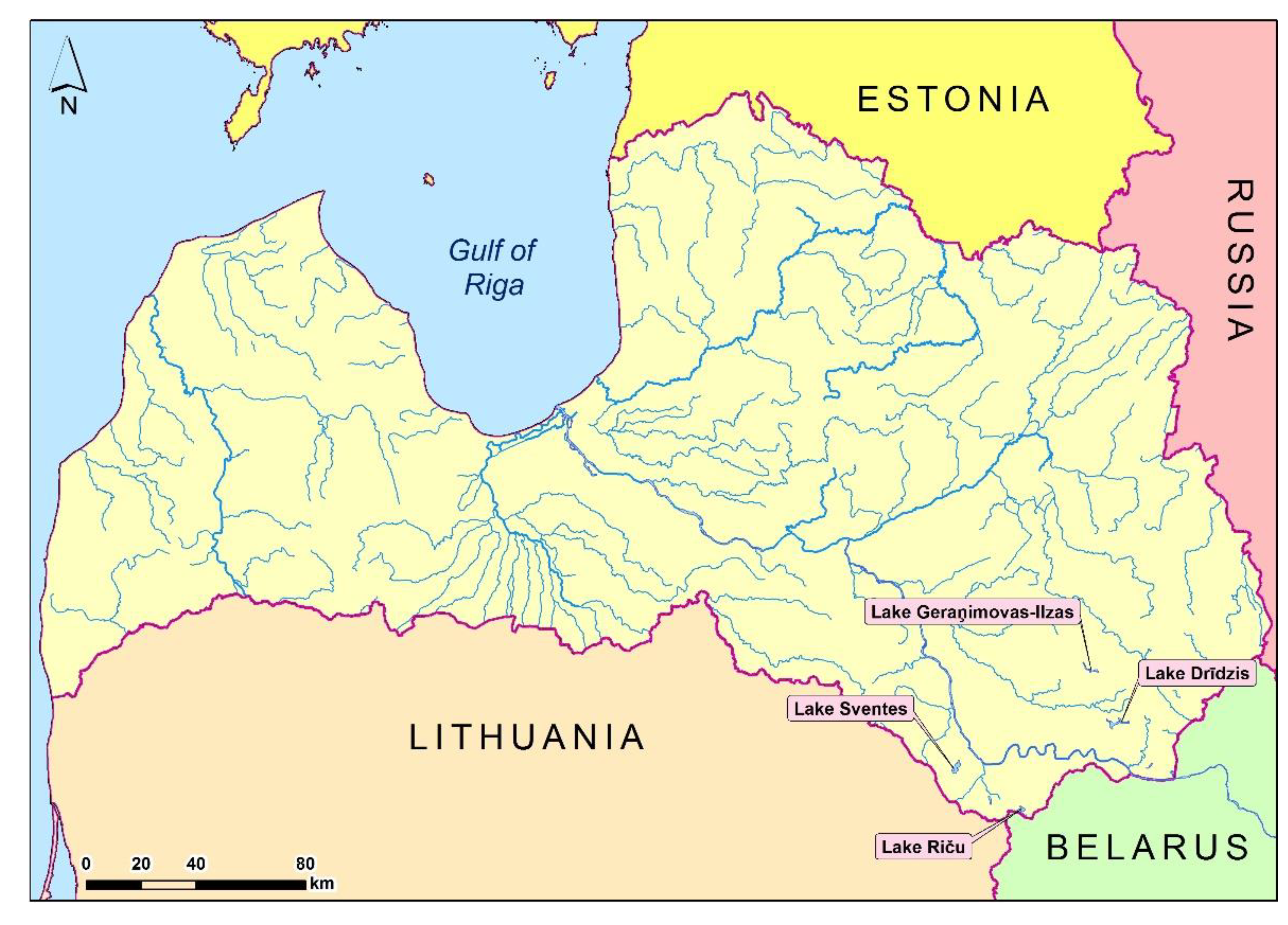
2.1. Sampling sites and material collection of Daphnia cucullata
2.2. Genetic analysis
2.2.1. DNA extraction
2.2.2. Determination of the quantity and quality of isolated DNA
2.2.3. Microsatellites analysis
2.2.4. Statistical processing and analysis of the obtained data
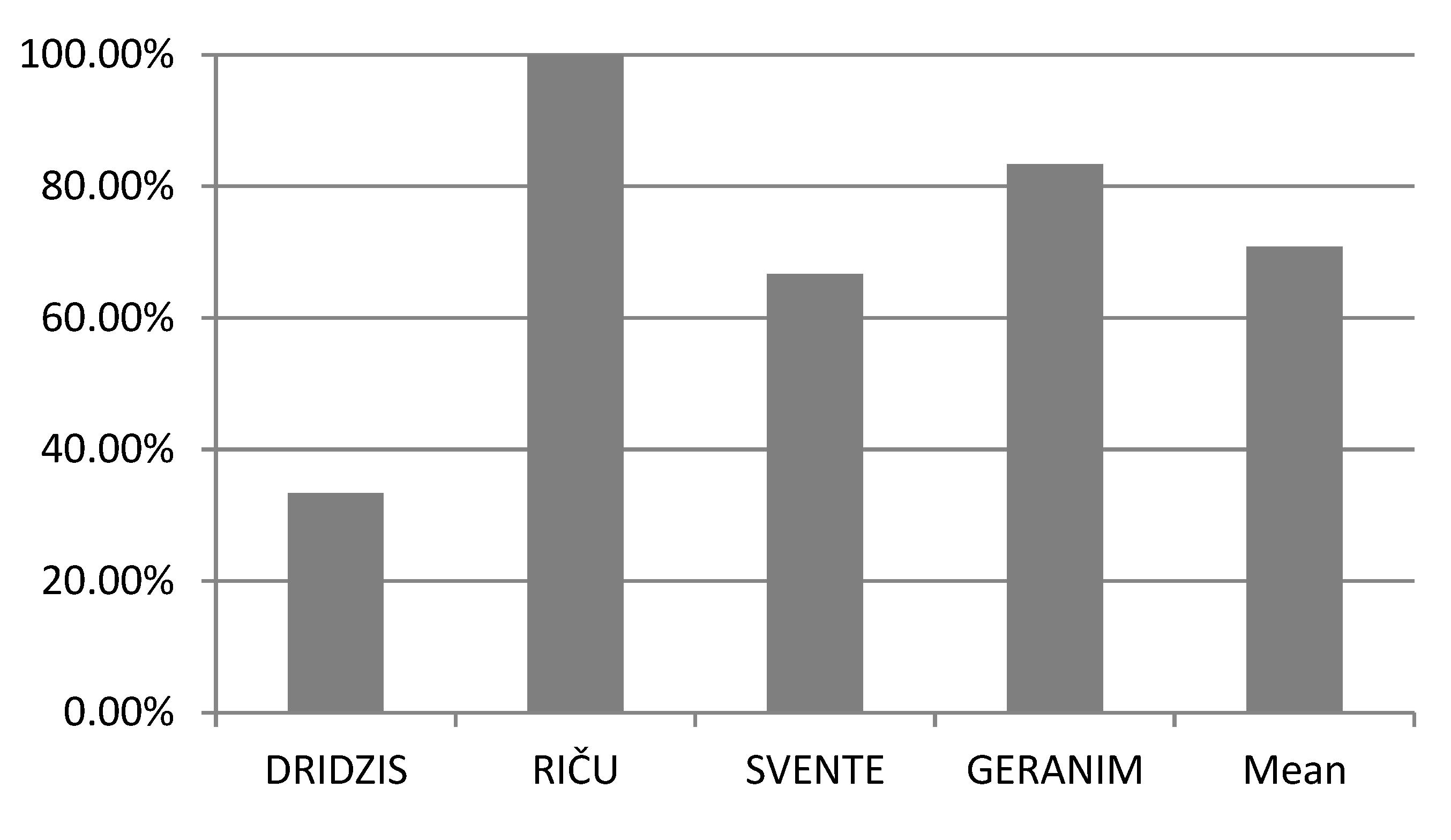
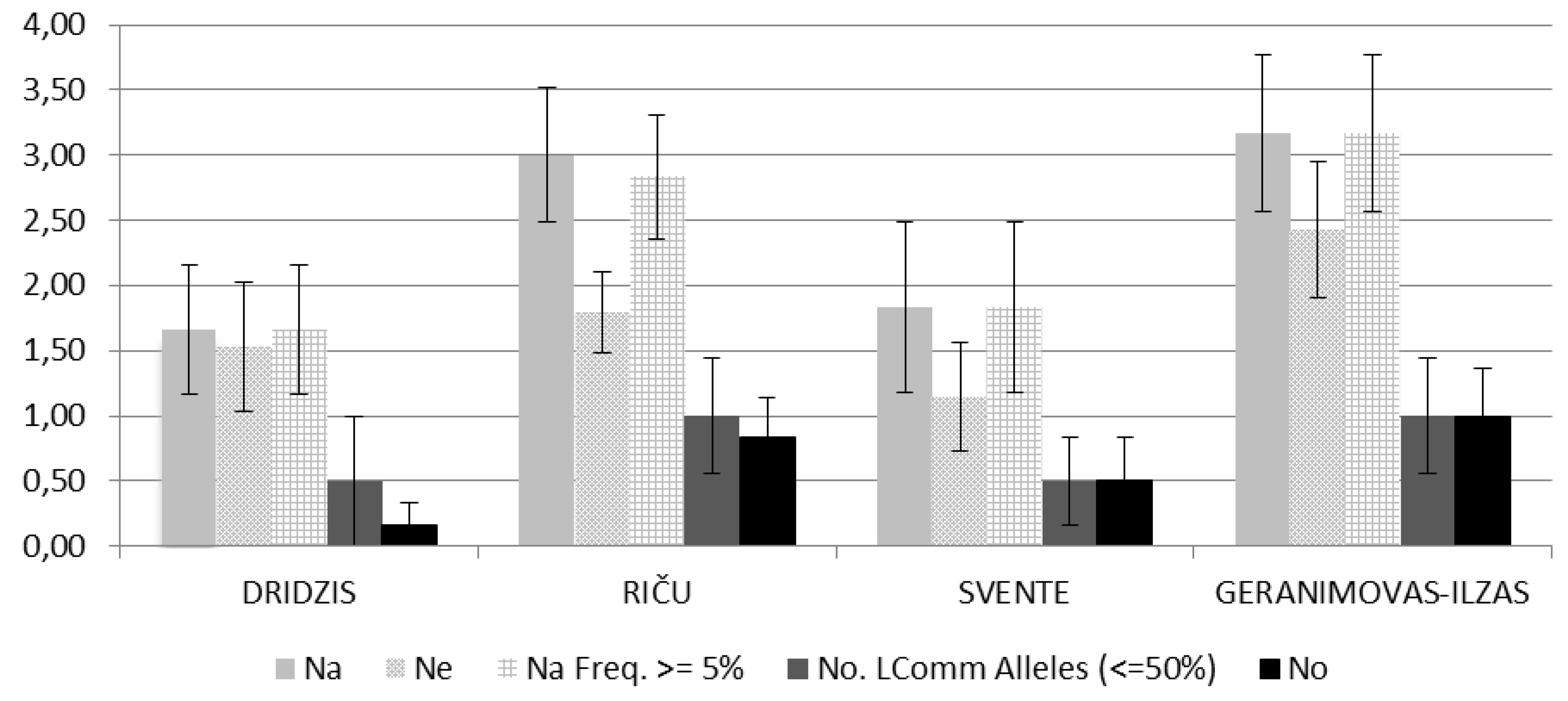
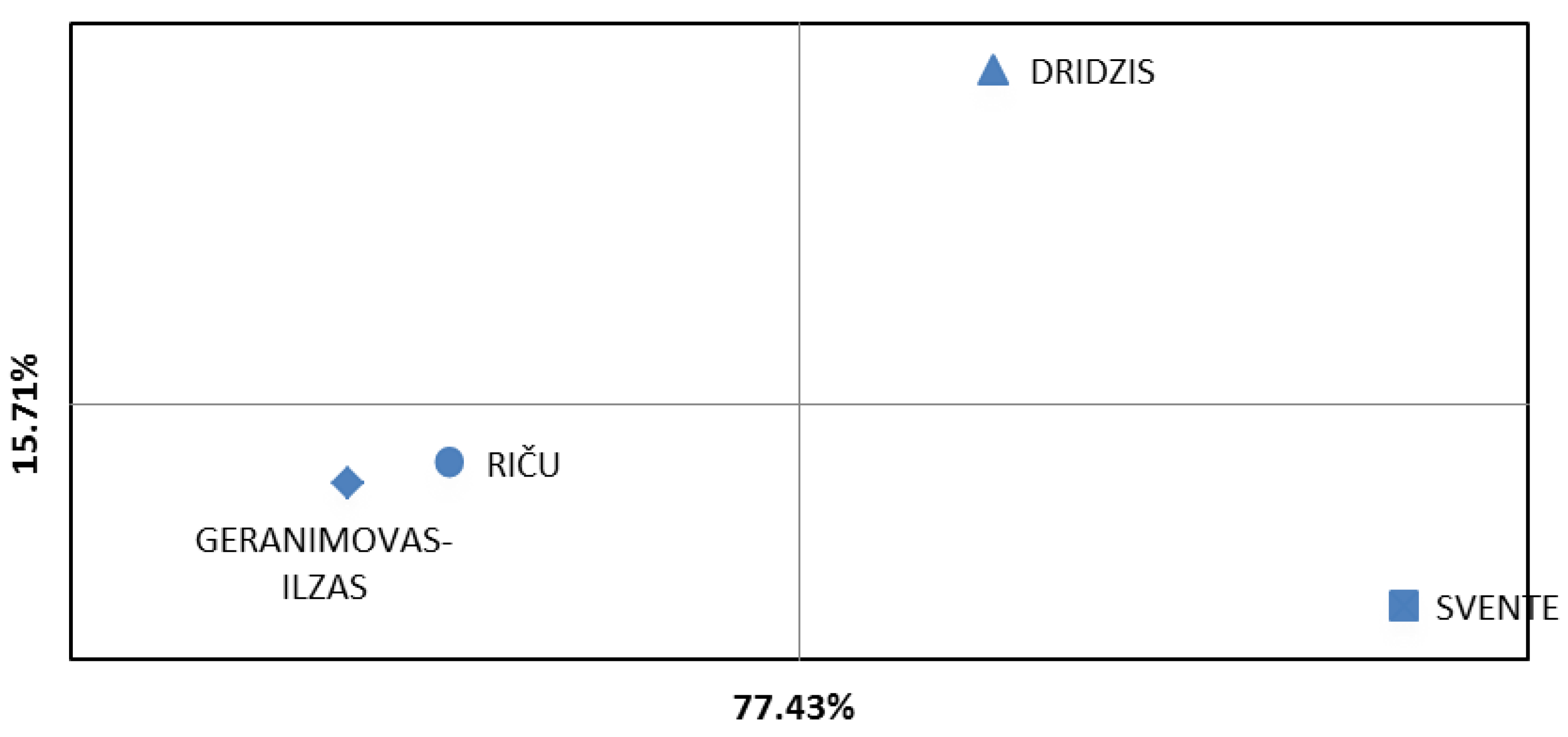
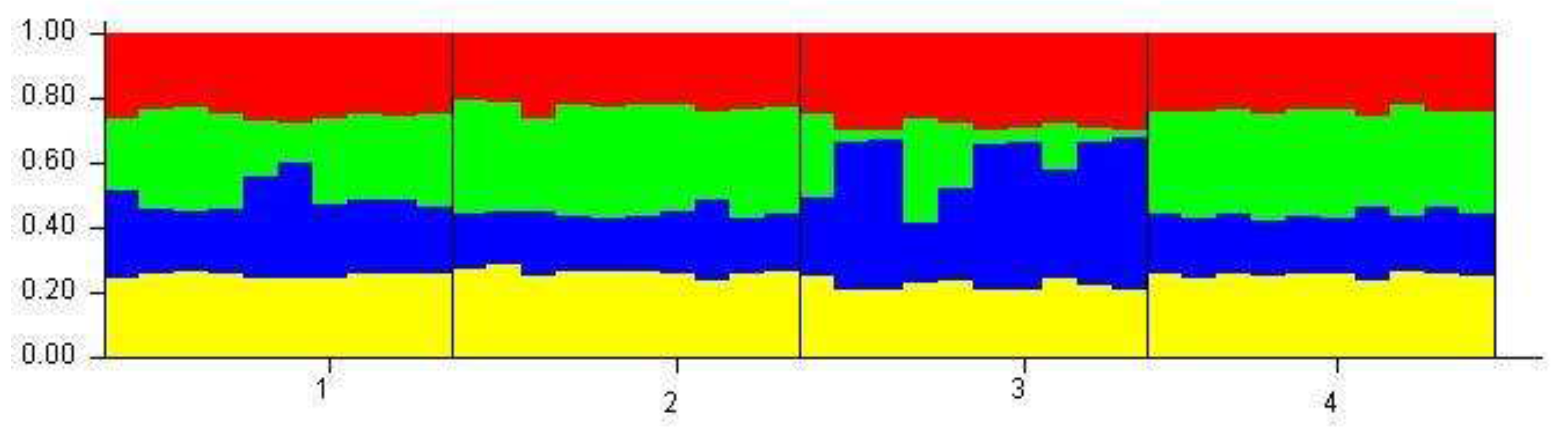
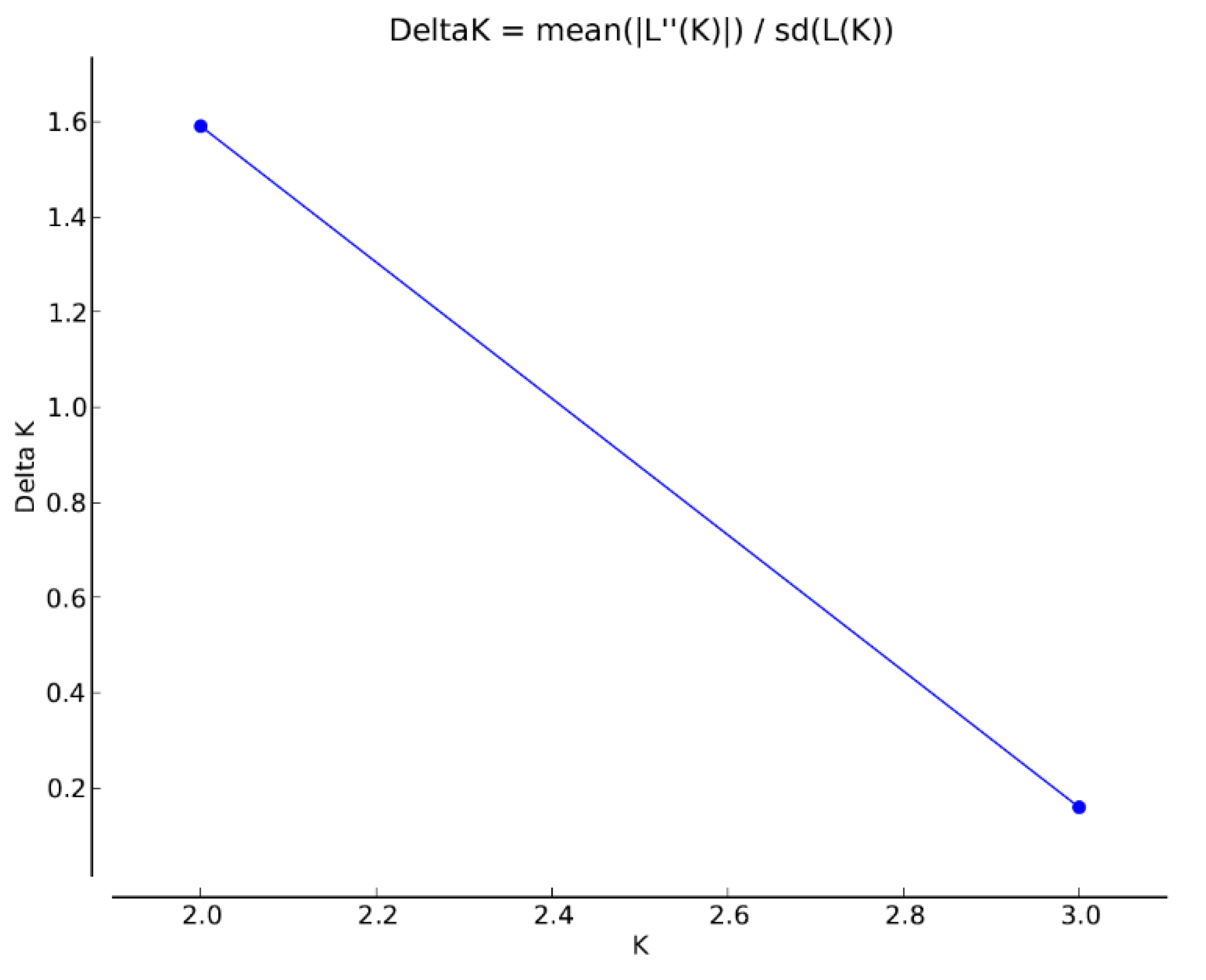
3. Results
- * GERANIM- Lake Geraņimovas-Ilzas
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hebert, P. D. N. Competition in zooplankton communities. Annales Zoologici Fennici 1982, 19, 349–356. [Google Scholar]
- Malone, B.J.; McQueen, D.J. Horizontal patchiness in zooplankton populations in two Ontario kettle lakes. Hydrobiologia 1983, 99, 101–124. [Google Scholar] [CrossRef]
- Pinel-Alloul, B. Spatial heterogeneity as a multiscale characteristic of zooplankton community. Hydrobiologia 1995, 300/301, 17–42. [Google Scholar] [CrossRef]
- Wetzel, R.G. Limnology. Lake and River Ecosystems. Third Edition. Academic Press 2001, 1006. [Google Scholar]
- Adamowicz, S.J.; Hebert, P.D.N.; Marinone, M.C. Species diversity and endemism in the Daphnia of Argentina: a genetic investigation. Zoological Journal of the Linnean Society of London 2004, 140, 171–205. [Google Scholar] [CrossRef]
- Colbourne, J.K.; Hebert, P.D.N. The systematics of North American Daphnia (Crustacea: Anomopoda): a molecularphylogenetic approach. Philosophical Transactions of the Royal Society of London. Series B 1996, 351, 349–360. [Google Scholar]
- Colbourne, J.K.; Wilson, C.C.; Hebert, P.D.N. The systematics of Australian Daphnia and Daphniopsis (Crustacea: Cladocera): A shared phylogenetic history transformed by habitat-specific rates of evolution. Biological Journal of the Linnean Society 2006, 89, 469–488. [Google Scholar] [CrossRef]
- Dussart, B.H.; Defaye, D. Introduction to the Copepoda (2nd edition revised and enlarged). Guides to the Identification of the Microinvertebrates of the Continetal Waters of the World. 16 Backhuys Publishers, Leiden. 2001, 344.
- Harris, K. D. M.; Bartlett, N. J.; Lloyd, V.K. Daphnia as an emerging epigenetic model organism. Genetic Research International. ID 147892 2012, 1–8. [Google Scholar] [CrossRef]
- Lampert, W. Daphnia: model herbivore, predator and prey. Polish Journal of Ecology 2006, 54, 607–620. [Google Scholar]
- Colbourne, J.K.; Hebert, P.D.N.; Taylor, D.J. Evolutionary origins of phenotypic diversity in Daphnia. In Molecular Evolution and Adaptive Radiation. (Givnish, T. J. & Sytsma, K. J. eds), Cambridge University Press, London. 1997, 163 –188.
- Dodson, S.I.; Hanazato, T. Commentary on effects of anthropogenic and natural organic-chemicals on development, swimming behavior and reproduction of Daphnia, a key member of aquatic ecosystems. Environmental Health Perspectives 1995, 103, 7–11. [Google Scholar]
- Brede, N.; Thielsch, A.; Sandrock, C.; Spaak, P.; Keller, B.; Streit, B.; Schwenk, K. Microsatellite markers for European Daphnia. Molecular Ecology Notes 2006, 6, 536–539. [Google Scholar] [CrossRef]
- Cousyn, C.; De Meester, L.; Colbourne, J.K.; Brendonck, L.; Verschuren, D.; Volckaert, F. Rapid, local adaptation of zooplankton behavior to changes in predation pressure in the absence of neutral genetic changes. Proceedings of the National Academy of Sciences of the United States of America 2001, 98, 6256–6260. [Google Scholar] [CrossRef] [PubMed]
- De Meester, L.; Weider, L.J.; Tollrian, R. Alternative antipredator defences and genetic polymorphism in a pelagic predator-prey system. Nature 1995, 378, 483–485. [Google Scholar] [CrossRef]
- Harris, R.P.; Wiebe, P.H.; Lenz, J.; Skjoldal, H.R.; Huntley, M. Zooplankton Methodology Manual. Elsevier Academic Press 2005, 533–570. [Google Scholar]
- Hellsten, M.E.; Sundberg, P. Genetic variation in two sympatric European populations of Bosmina spp. (Cladocera) tested with RAPD markers. Hydrobiologia 2000, 421, 157–164. [Google Scholar] [CrossRef]
- Brakovska, A. Daphnia cucullata Sars, 1862 (CRUSTACEA: CLADOCERA) distribution and location in composition of zooplankton cenosis in Lake Dridzis. Acta Biologica Universitatis Daugavpiliensis 2014, 14, 1–19. [Google Scholar]
- Brakovska, A.; Škute, R. Ecological characterization of zooplankton groups in the deepest lakes of East Latvia. Acta Biologica Universitatis Daugavpiliensis 2007, 7, 165–174. [Google Scholar]
- Brakovska, A.; Škute, R. Ecological evaluation of zooplankton groups in Lake Geranimovas-Ilzas and Lake Garais. Proceedings of the 7th International Scientific and Practical Conference Environment. Technology. Resources 2009, 2, 43–50. [Google Scholar] [CrossRef]
- Brakovska, A.; Škute, R.; Škute, A. Heterogeneity of distribution and community composition of zooplankton in upper layers of Lake Svente. Zoology and Ecology 2012, 22, 172–180. [Google Scholar] [CrossRef]
- Brakovska, A.; Škute, N. Optimisation of DNA extraction and RAPD-PCR amplification for population genetic analysis of Daphnia cucullata Sars, 1862 (Crustacea: Cladocera). Acta Biologica Universitatis Daugavpiliensis 2013, 13, 11–20. [Google Scholar]
- Brakovska, A.; Paidere, J.; Škute, A. Dynamics and factors influencing zooplankton in the lakes Svente, Riča, Dridzis and Geraņimovas-Ilzas (Eastern Latvia). Acta Biologica Universitatis Daugavpiliensis 2020, 20, 71–94. [Google Scholar]
- Selkoe, K. A.; Toonen, R. J. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecology Letters 2006, 9, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Haag, T.; Santos, A. S.; Sana, D. A.; Morato, R. G.; Cullen, L. Jr.; Crawshaw, P. G. Jr.; De Angelo, C.; Di Bitetti, M. S.; Salzano, F. M.; Eizirik, E. The effect of habitat fragmentation on the genetic structure of a top predator: loss of diversity and high differentiation among remnant populations of Atlantic Forest jaguars (Panthera onca). Molecular Ecology 2010, 22, 4906–4921. [Google Scholar] [CrossRef] [PubMed]
- Colbourne, J.K.; Robison, B.; Bogart, K.; Lynch, M. Five hundred and twenty-eight microsatellite markers for ecological genomic investigations using Daphnia. Molecular Ecology Notes 2004, 4, 485–490. [Google Scholar] [CrossRef]
- Frisch, D.; Morton, P. K.; Roy Chowdhury, P.; Culver, B. W.; Colbourne, J. K.; Weider, L. J.; Jeyasingh, P. D. A millennial-scale chronicle of evolutionary responses to cultural eutrophication in Daphnia. Ecology Letters 2014, 17, 360–368. [Google Scholar] [CrossRef]
- Forest, F.; Chase, M. W.; Persson, C.; Crane, P. R.; Hawkins, J. A. The role of biotic and abiotic factors in evolution of ant dispersal in the milkwort family (polygalaceae). Evolution 2007, 61, 1675–1694. [Google Scholar] [CrossRef]
- Pálsson, S. Microsatellite variation in Daphnia pulex from both sides of the Baltic Sea. Molecular Ecology 2000, 9, 1075–1088. [Google Scholar] [CrossRef]
- Ender, A.; Schwenk, K.; Stadler, T.; Streit, B.; Schierwater, B. RAPD identification of microsatellites in Daphnia. Molecular Ecology 1996, 5, 437–441. [Google Scholar] [CrossRef]
- Fox, J.A. New microsatellite primers for Daphnia galeata mendotae. Molecular Ecology Notes 2004, 4, 544–546. [Google Scholar] [CrossRef]
- Lynch, M.; Spitze, K. Evolutionary genetics of Daphnia. In: Ecological Genetics (ed. Real L) Princeton University Press, Princeton, NJ. 1994, 109–128.
- Schwenk, K.; Spaak, P. Evolutionary and ecological consequences of interspecific hybridization in cladocerans. Experientia 1995, 51, 465–481. [Google Scholar] [CrossRef]
- Cousyn, C.; De Meester, L.; Colbourne, J.K.; Brendonck, L.; Verschuren, D.; Volckaert, F. Rapid, local adaptation of zooplankton behavior to changes in predation pressure in the absence of neutral genetic changes. Proceedings of the National Academy of Sciences 2001, 98, 6256–6260. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Pfrender, M.; Spitze, K.; Lehman, N.; Hicks, J.; Allen, D.; Latta, L.; Ottene, M.; Bogue, F.; Colbourne, J. The quantitative and molecular genetic architecture of a subdivided species Evolution 1999, 53, 100–110. 53.
- Mergeay, J.; Verschuren, D.; De Meester, L. Cryptic invasion and dispersal of an American Daphnia in East Africa. Limnol. Oceanogr 2005, 50, 1278–1283. [Google Scholar] [CrossRef]
- Silva R., M.; Pereira, F.; Carneiro, J.; Sobral, O.; Ribeiro, R.; Amorim, A.; Soares A. M. V., M.; Lopes, I. Microevolution in a Natural Population of Daphnia longispina Exposed to Acid Mine Drainage. In Interdisciplinary Studies on Environmental Chemistry — Biological Responses to Contaminants, Eds., N. Hamamura, S. Suzuki, S. Mendo, C. M. Barroso, H. Iwata and S. Tanabe. Terrapub, 2010, 213–218.
- Reid, V. A.; Carvalho, G. R.; George, D. G. Molecular genetic analysis of Daphnia in the English Lake District: species identity, hybridisation and resting egg banks. Freshwater Biology 2020, 44, 247–253. [Google Scholar] [CrossRef]
- Urtāne, L. Cladocera as types and trophic status indicators of Latvian Lakes. Thesis 1998, 168. Not published (In Latvian).
- www.ezeri.lv database. (accessed 24.01.2023).
- Eipurs, I. Lake Geraņimovas-Ilzas. The Nature of Latvia. Riga, Encyclopedia of Latvia. 1995, 2, 106. (In Latvian) [Google Scholar]
- Tidriķis. A. Lake Svente. The Nature of Latvia. Riga, Publishing house. 1998, 5, 180. (In Latvian)
- Tidriķis, A. Lake Riču. The Nature of Latvia. Riga, Publishing house. 1997, 4, 243–244. (In Latvian) [Google Scholar]
- APHA. Standart methods for the examination of waters and wastewater. 21<sup>st</sup> edition. Washington, D.C. APHA. Standart methods for the examination of waters and wastewater. 21st edition. Washington, D.C., American Public Health Association. 2005.
- Wetzel, R.G.; Likens, G.E. Limnological Analyses. New York, Springer Science, Business Media 2000, 429.
- Fitzsimmons, J.M.; Innes, D.M. No evidence of Wolbachia among Great Lakes area populations of Daphnia pulex (Crustacea: Cladocera). Journal of Plankton Research 2005, 27, 121–124. [Google Scholar] [CrossRef]
- Brakovska, A.; Paidere, J.; Škute, R.; Škute, N.; Škute, A. Occurrence of Cladocera and genetic diversity of Daphnia cucullata in pelagic zone of the Latvian salmonid lakes. Estonian Journal of Ecology 2013, 62, 244–264. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Yeh, F. C.; Yang, R. C.; Boyle, T. POPGENE 32-version 1. 31. Population Genetics Software. 1999.
- Nei, M. Analysis of gene diversity in subdivided populations. Proc.Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Tajima, F.; Tateno, Y. Accurancy of estimated phylogenetic trees from molecular data. 2 Genefrequency data. Journal of Molecular Evolution 1983, 19, 153–170. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics. Columbia University Press, New York. 1987, 512.
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, A.J. Interring weak population structure with the assistance of sample group information. Molecular Ecology Resources, 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Nei, M. Estimation of Average Heterozygosity and Genetic Distance from a small number of Individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Biogeographical regions in Europe — European Environment Agency (europa.eu). (accessed 24.01.2023).
- De Meester, L.; Vanoverbeke, J.; De Gelas, K.; Ortells, R.; Spaak, P. Genetic structure of cyclic parthenogenetic zooplankton populations — a conceptual framework. Archiv für Hydrobiologie 2006, 167, 217–244. [Google Scholar] [CrossRef]
- De Meester, L.; Gómez, A.; Simon, J.C. Evolutionary and ecological genetics of cyclical parthenogens. In Moya A, Font E (eds) Evolution from molecules to ecosystems. Oxford University Press, Oxford, 2004, 122–134.
- Kalff, J. Limnology: Inland water ecosystems. Prentice- Hall, Upper Saddle River, NJ. 2002, 592.
- Lampert, W.; Sommer, U. Limnoecology: The Ecology of Lakes and Streams. Oxford University Press, New York Oxford. 1997, 382.
- Haag, C.R.; Riek, M.; Hottinger, J.W.; Pajunen, V.I.; Ebert, D. Genetic diversity and genetic differentiation in Daphnia metapopulations with subpopulations of known age. Genetics 2005, 170, 1809–1820. [Google Scholar] [CrossRef]
- Taylor, D.J; Hebert, P.D.N. Habitat dependent hybrid parentage and differential introgression between neighboringly sympatric Daphnia species. Proceedings of the National Academy of Sciences USA 1993a, 90, 7079–7083. [Google Scholar] [CrossRef]
- Taylor, D.J.; Hebert, P.D.N. A reappraisal of phenotypic variation in Daphnia galeata mendotae: the role of interspecific hybridization. Canadian Journal of Fisheries and Aquatic Sciences 1993b, 50, 2137–2146. [Google Scholar] [CrossRef]
- Green, A.J.; Figuerola, J. Recent advances in the study of longdistance dispersal of aquatic invertebrates via birds. Diversity and Distributions 2005, 11, 149–156. [Google Scholar] [CrossRef]
- Figuerola, J.; Green, A.J.; Santamaria, L. Passive internal transport of aquatic organisms by waterfowl in Doñana, south-west Spain. Global Ecology and Biogeography 2003, 12, 427–436. [Google Scholar] [CrossRef]
- Figuerola, J.; Green, A.J.; Michot, T.C. Invertebrate eggs can fly: evidence of waterfowl-mediated gene flow in aquatic invertebrates. American Naturalist 2005, 165, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Moran, C. Enzyme variability in natural populations of Daphnia carinata King. Heredity 1980, 45, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Crease, T. J.; Lynch, M.; Spitze, K. Hierarchical analysis of population genetic variation in mitochondrial and nuclear genes of Daphnia pulex. Molecular Biology and Evolution 1990, 7, 444–458. [Google Scholar]
- Gomez, A.; Carvalho, G.R. Sex, parthenogenesis and genetic structure of rotifers: microsatellite analysis of contemporary and resting egg bank populations. Molecular Ecology 2000, 9, 203–214. [Google Scholar] [CrossRef]
- Vanoverbeke, J.; De Meester, L. Among-populational genetic differentiation in the cyclical parthenogen Daphnia magna (Crustacea: Anomopoda) and its relation to geographic distance and clonal diversity. Hydrobiologia 1997, 126, 135–142. [Google Scholar] [CrossRef]
- Decaestecker, E.; De Meester, L.; Mergeay, J. Cyclical Parthenogenesis in Daphnia: Sexual Versus Asexual Reproduction. Lost Sex 2009, 295–316. [Google Scholar]
- Hughes, R.N. A Functional Biology of Clonal Animals. Chapman & Hall, London, New York. 1989.
- Hobæk, A.; Larsson, P. Sex determination in Daphnia magna. Ecology 1990, 71, 2255–2268. [Google Scholar] [CrossRef]
- Hebert, P.D.N. Genetics of Daphnia. In: Peters RH, De Bernardi R (eds) Daphnia. MemInst Ital Idrobiol 1987, 45, 439–460. [Google Scholar]
- De Meester, L. Local genetic differentiation and adaptation in freshwater zooplankton populations: patterns and processes. Ecoscience 1996, 3, 385–399. [Google Scholar] [CrossRef]
- Vanoverbeke, J.; De Gelas, K.; De Meester, L. Habitat size and the genetic structure of a cyclical parthenogen, Daphnia magna. Heredity 2007, 98, 419–426. [Google Scholar] [CrossRef]
- Deng, H.W.; Lynch, M. Change of genetic architecture in response to sex. Genetics 1996, 143, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Hairston, N.G.; Holtmeier, C.L.; Lampert, W.; Weider, L. J.; Post, D. M.; Fischer, J. M.; Cáceres, C. E.; Fox, J. A.; Gaedke, U. Natural selection for grazer resistance to toxic cyanobacteria: Evolution of phenotypic plasticity? Evolution 2001, 55, 2203–2214. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Deng, H.W. Genetic slippage in response to sex. American Naturalist 1994, 144, 242–261. [Google Scholar] [CrossRef]
- Michels, E.; Audenaert, E.; Ortells, R.; De Meester, L. Population genetic structure of three pond-inhabiting Daphnia species on a regional scale (Flanders, Belgium). Freshwater Biology 2003, 48, 1825–1839. [Google Scholar] [CrossRef]
- Spaak, P.; Denk, A.; Boersma, M.; Weider, L. J. Spatial and temporal patterns of sexual reproduction in a hybrid Daphnia species complex. Journal of Plankton Research 2004, 26, 625–635. [Google Scholar] [CrossRef]
- Wolf, H. G. Interspecific hybridization between Daphnia hyalina, D. galeata and D. cucullata and seasonal abundances of these species and their hybrids. Hydrobiologia 1987, 145, 213–217. [Google Scholar] [CrossRef]
- Gliwicz, M. Z.; Ślusarczyk, A.; Ślusarczyk, M. Life history synchronization in a long-lifespan single-cohort Daphnia population in a fishless alpine lake. Oecologia, 2001, 128, 368–378. [Google Scholar] [CrossRef]
| Lakes | Coordinates X/Y |
Elevation of lakes above sea level,m | Surface area with island, km2 | Surface area without island, km2 | Max. depth, m* | Mean depth, m* | Catchment basin, km2 | Shore length, km |
|---|---|---|---|---|---|---|---|---|
| Dridzis | 705390.852/ 208462.077 |
159.8 | 7.72 | 7.56 | 64 | 12.8 | 46 | 42 |
| Riča | 670715.594/ 175721.067 |
145.8 | 13.12 | 13.07 | 39 | 9.7 | 123*/130** | 34 |
| Svente | 647412.511/ 192388.091 |
136.9 | 7.06 | 7.03 | 38 | 7.8 | 20 | 26 |
| Geraņimovas-Ilzas | 696251.015/ 228167.042 |
150.7 | 3.17 | 3.17 | 46 | 9.8 | 66 | 24 |
|
Locus |
Primer sequences (5′−3′) |
Repeat unit |
Label dye |
Size range (bp) (our data) |
Size range (bp) (data after Brede et al.) |
Ta(°C) |
|---|---|---|---|---|---|---|
| SwiD1 | F:GCCGTGTTCGAAAGCTAGTC R: AGCCGAACGAAAAACATGC |
(TG)18 | 5'TAM | 122–127 | 116-142 | 59.4 |
| Dgm105 | F:ATGTGAGCGCGCGAGCATTT R:GTCCAGCCGGCCCATTTCAGTT |
(CAG)8AG | 5'FAM | 165–240 | 172-197 | 59.4 |
| Dgm101 | F: TCTTGCTCGAATTCTCTCC R: CCTGTCTCACACGGAGC |
(GA)10AGA | 5'HEX | 165–180 | 162-177 | 54.5 |
| DaB17/17 | F:GAGAACCTTTTATCAGCTTCG R:ACTCATCTGGTGAGATGGATC |
T9 | 5'TAM | 100–106 | 100-109 | 55.9 |
| Dgm109 | F: CCAGCTGTTGACCACCTG R: TGCGCGAGGATTTCCAACAC |
(ACC)7AC | 5'FAM | 250–303 | 247-266 | 58.2 |
| Dp519 | F:AGTCGCGACGACATAAAGC R:GTGGTAGTTGTGGAATCCG |
(TG)6(GA)7 | 5'HEX | 140–142 | 144-160 | 56.7 |
| DaB10/15 | F:AGAGAAGTGTTTGCGTTTC R:TGTTTCCTATATCCCTCGG |
TC6 | 5'TAM | No result | 75–89 | 52.4 |
| Dp512 | F:TTTCGTTCTACCCAGGGAAG R:TTTGCTCGTCTGTGATAGGC |
(TG)4…(GT)8 | 5'HEX | No result | 125-141 | 57.3 |
| DaB17/16 | F: AGGGAACGAGCGGCGATAAG R:TCTTTGGCAGGCCACTGCCAAGG |
GA10 | 5'FAM | No result | 189-195 | 61.4 |
| Locus | Total number of alleles in the locus | Number of private alleles in the locus | Proportion of private alleles (%) | Number of populations in which private alleles have been detected |
|---|---|---|---|---|
| SwiD1 | 5 | 1 | 20 | 1 |
| Dgm105 | 8 | 5 | 62 | 3 |
| Dgm101 | 7 | 4 | 57 | 3 |
| DaB17/17 | 4 | 2 | 50 | 2 |
| Dgm109 | 5 | 3 | 60 | 2 |
| Dp519 | 2 | 0 | 0 | 0 |
| Sample | SwiD1 | Dgm105 | Dgm101 | DaB17/17 | Dgm109 | Dp519 | |
|---|---|---|---|---|---|---|---|
| Dridzis | N | 4 | 4 | 1 | 14 | 4 | 14 |
| Na | 4 | 1 | 1 | 2 | 1 | 1 | |
| No | 0 | 0 | 1 | 0 | 0 | 0 | |
| Ho | 0 | 0 | 0 | 0 | 0 | 0 | |
| He | 0.75 | 0 | 0 | 0.13 | 0 | 0 | |
| Riča | N | 13 | 12 | 12 | 19 | 11 | 19 |
| Na | 2 | 4 | 5 | 2 | 3 | 2 | |
| No | 1 | 1 | 2 | 0 | 1 | 0 | |
| Ho | 0 | 0.25 | 0 | 0 | 0 | 0 | |
| He | 0.14 | 0.51 | 0.68 | 0.46 | 0.31 | 0.1 | |
| Svente | N | 4 | 8 | 0 | 15 | 0 | 16 |
| Na | 3 | 4 | 0 | 2 | 0 | 2 | |
| No | 0 | 2 | 0 | 1 | 0 | 0 | |
| Ho | 0 | 0.25 | 0 | 0 | 0 | 0 | |
| He | 0.62 | 0.33 | 0 | 0.12 | 0 | 0.37 | |
| Geranimovas-Ilzas | N | 7 | 6 | 7 | 14 | 6 | 8 |
| Na | 2 | 5 | 4 | 3 | 4 | 1 | |
| No | 0 | 2 | 1 | 1 | 2 | 0 | |
| Ho | 0 | 0.17 | 0 | 0 | 0.17 | 0 | |
| He | 0.24 | 0.74 | 0.73 | 0.36 | 0.68 | 0 |
| Population/ Microsatellite loci |
SwiD1 | Dgm105 | Dgm101 | DaB17/17 | Dgm109 | Dp519 |
|---|---|---|---|---|---|---|
| Dridzis | ns | M | M | *** | M | M |
| Riča | *** | ns | *** | *** | *** | *** |
| Svente | * | * | M | *** | M | *** |
| Geraņimovas-Ilzas | ** | ns | ** | *** | ns | M |
| Population | Dridzis | Riča | Svente | Geraņimovas-Ilzas |
|---|---|---|---|---|
| Dridzis | 0.29 | 0.45 | 0.37 | |
| Riča | 0.56 | 0.50 | 0.08 | |
| Svente | 0.50 | 1.14 | 0.49 | |
| Geraņimovas-Ilzas | 0.70 | 0.16 | 1.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





