Submitted:
30 September 2023
Posted:
02 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study population
2.2. Linear sequence analysis
2.3. Three-dimensional comparative modelling
2.4. Antigenic prediction
2.5. Search for potential T cell epitopes
3. Results
3.1. Study population
3.1.1. Patient 1
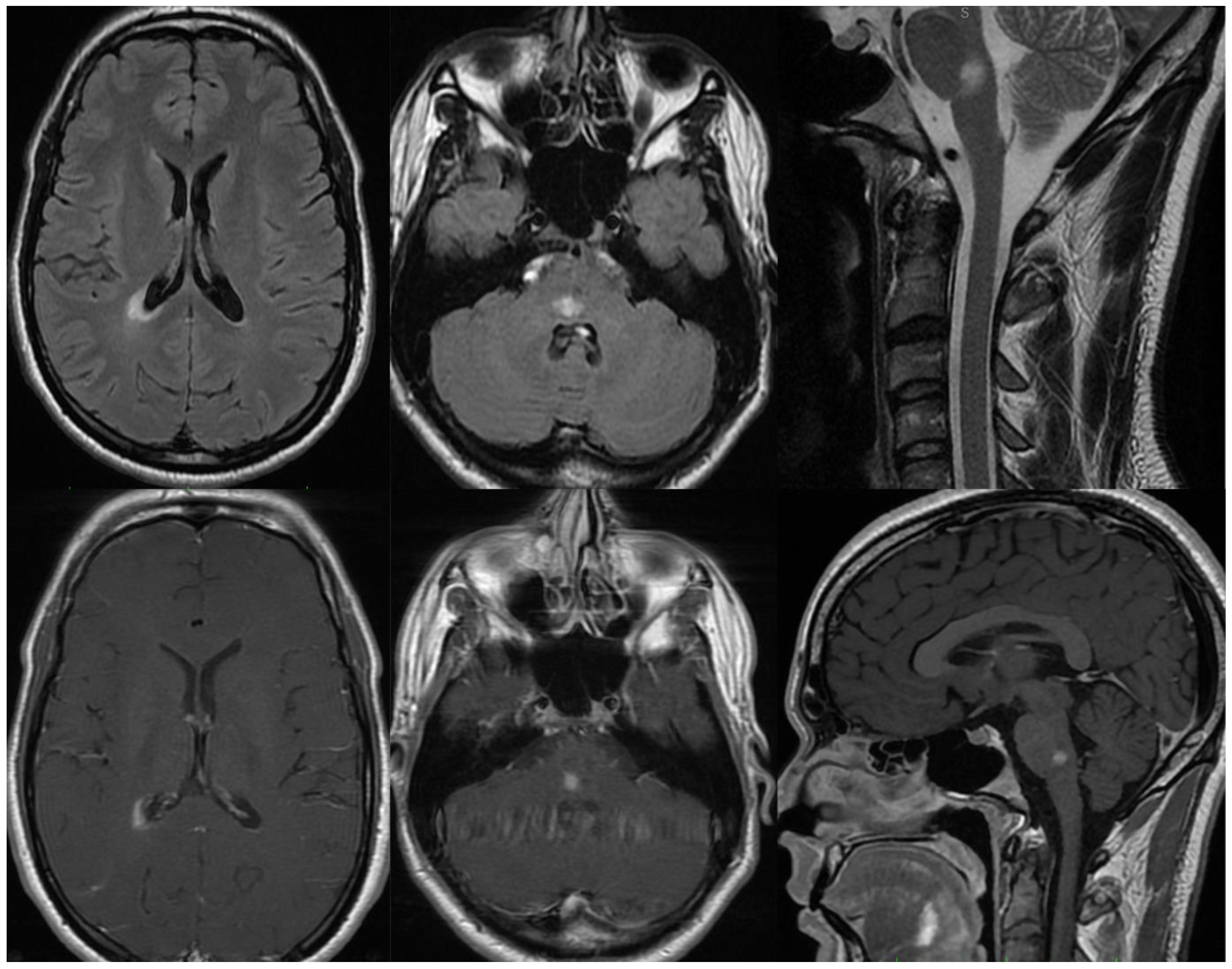
3.1.2. Patient 2
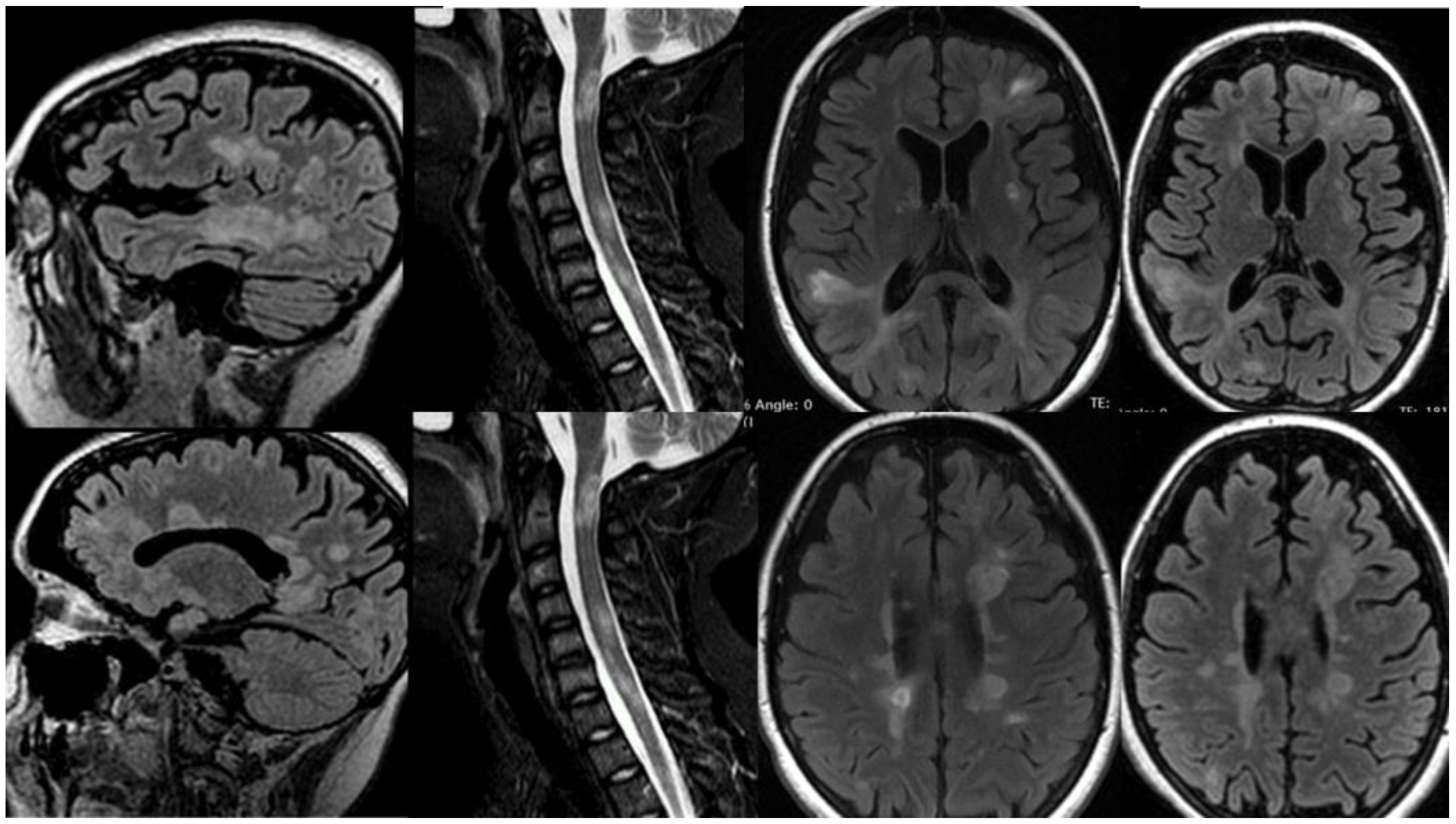
3.1.3. Patient 3
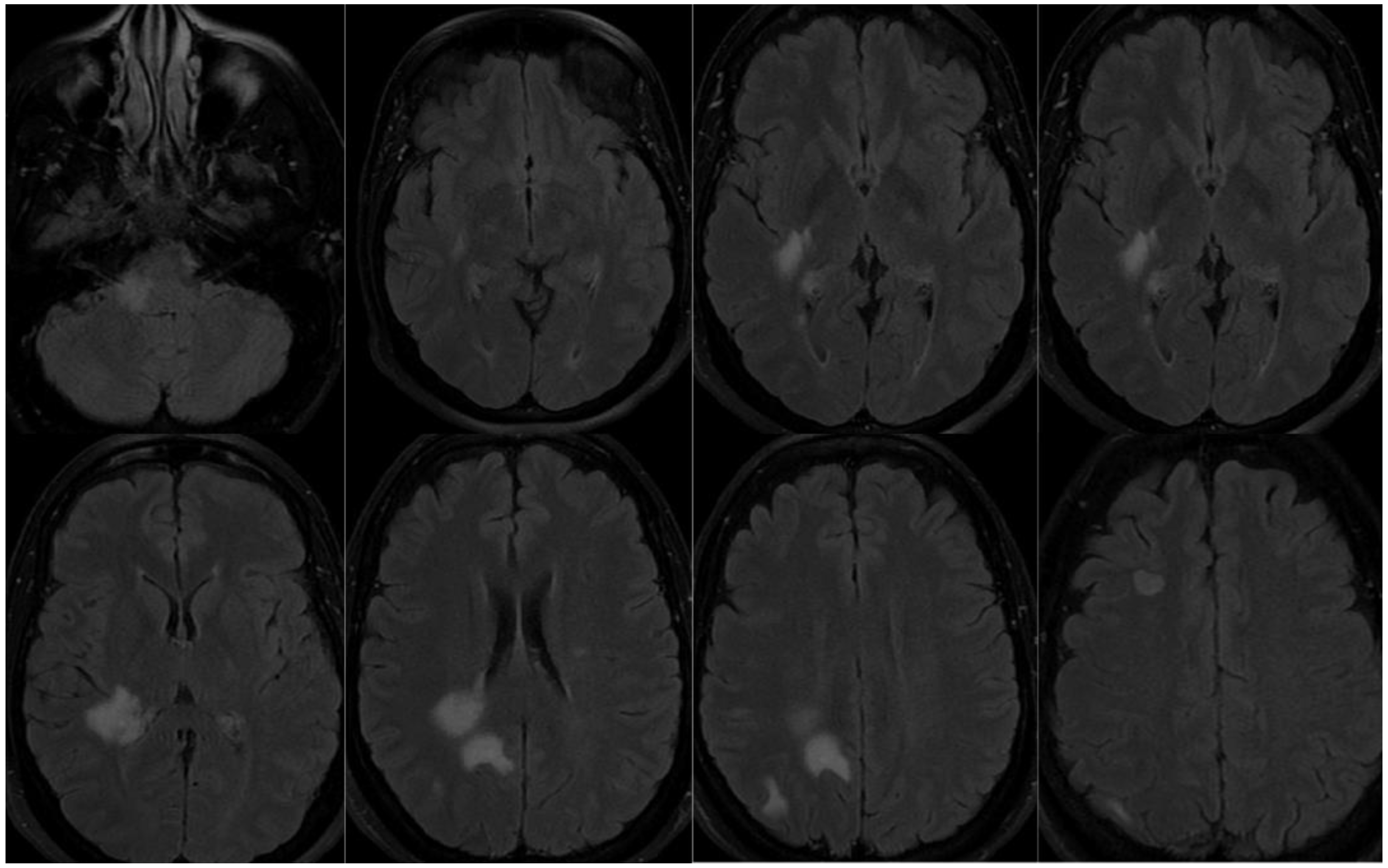
3.2. Sequence identification
| Protein | Number of aminoacids | Gene | NCBI Reference Sequence | Uniprot ID |
|---|---|---|---|---|
| E | 75aa | E | YP_009724392 | P0DTC4 |
| Nsp1 | 180aa | ORF1a | YP_009742608.1 | P0DTD1 |
| M | 222aa | M | YP_009724393.1 | P0DTC5 |
| Nsp2 | 638aa | ORF1a | YP_009742609.1 | P0DTD1 |
| Nsp3 | 1945aa | ORF1a | YP_009742610.1 | P0DTD1 |
| Nsp13 | 601aa | ORF1a | NP_828870.1 | P0DTD1 |
| ORF7a | 121aa | ORF7a | YP_009724395.1 | P0DTC7 |
| S | 1273 aa | S | YP_009724390.1 | P0DTC2 |
| Protein | Number of aminoacids | Gene | NCBI Reference Sequence | Uniprot ID |
|---|---|---|---|---|
| 2’,3’-Cyclic-nucleotide 3’-phosphodiesterase (CNP) | 421aa | CNP | NP_149124.3 | P09543 |
| Aquaporin-4 (AQP4) | 323aa | AQP4 | NP_001641.1 | P55087 |
| Glutamic acid decarboxylase 65-kilodalton isoform (GAD65) | 585aa | GAD2 | NP_001127838.1 | Q05329 |
| Myelin associated glycoprotein (MAG) | 626aa | MAG | NP_002352.1 | P20916 |
| Myelin basic protein (MBP) | 304aa | MBP | NP_001020272.1 | P02686 |
| Myelin oligodendrocyte glycoprotein (MOG) | 247aa | MOG | NP_996532.2 | Q16653 |
| Myelin-associated oligodendrocytic basic protein (MOBP) | 183aa | MOBP | NP_001380633.1 | Q13875 |
| Myeloperoxidase (MPO) | 745aa | MPO | NP_000241.1 | P05164 |
| N-methyl-D-aspartate receptor 1 (NMDAR1) | 938aa | GRIN1 | NP_015566.1 | Q05586 |
| Transaldolase | 337aa | TALDO1 | NP_006746.1 | P37837 |
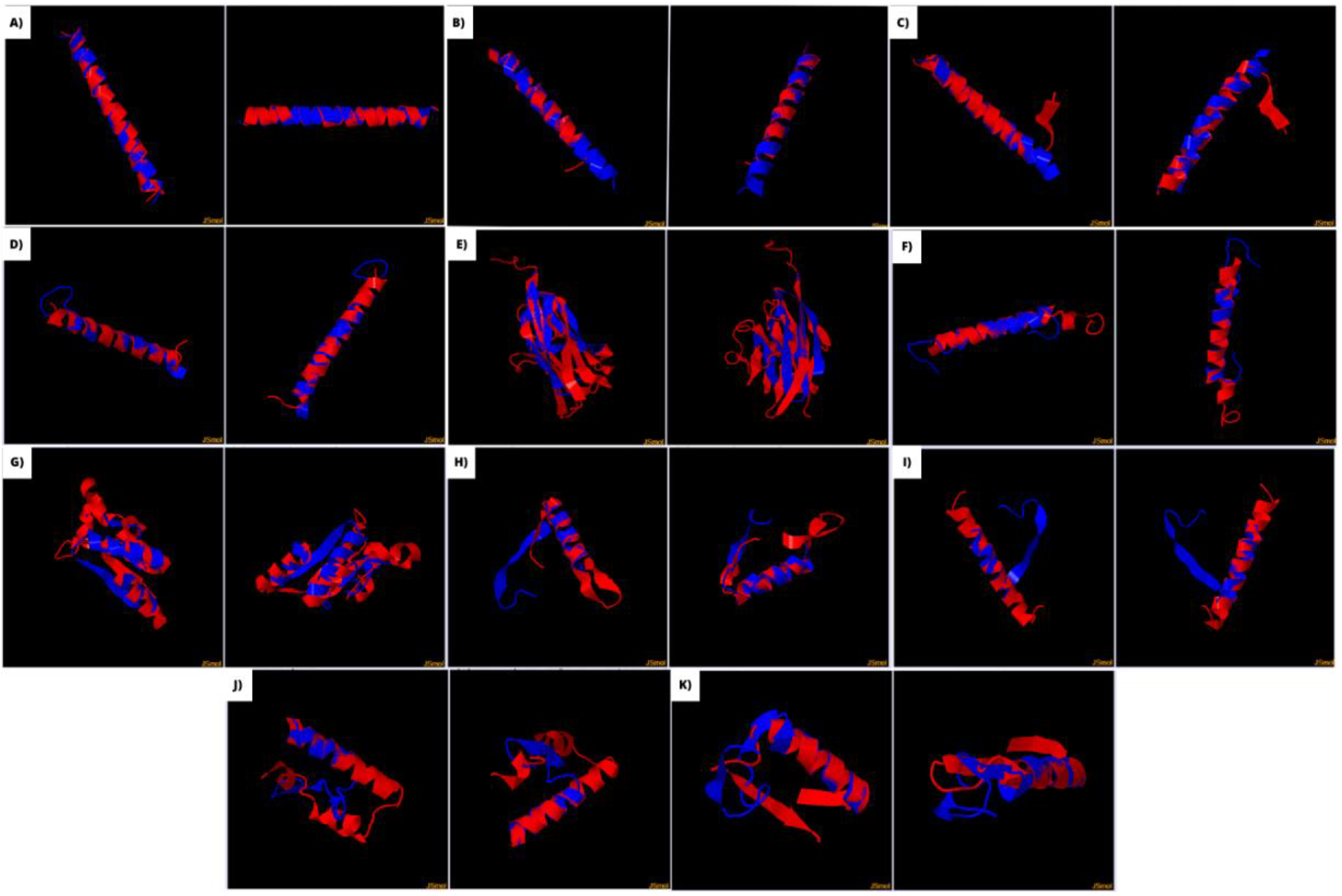
3.3. Linear and three-dimensional analysis
| Sars-Cov-2 antigens | Autoantigens | Region of the SARS-Cov-2 antigen with more identity | Region of the autoantigen with more identity | % Identity | E-value | SWISS MODEL SARS-CoV-2 antigen | SWISS MODEL autoantigen | TM-Score | RMSD | Overall prediction Vaxijen linear | Overall prediction VaxiJen three-dimensional model |
|---|---|---|---|---|---|---|---|---|---|---|---|
| M | NMDAR1 | 54-70 | 562-577 | 100 | 3,00E-04 | 74-106 | 626-658 | 0.89 | 0.55 | 0.9306 | 0.5324 |
| M | MPO | 134-162 | 57-85 | 100 | 1.0 | 76-105 | 67-96 | 0.73 | 1.21 | 0.4740 | 0.5177 |
| Nsp2 | NMDAR1 | 448-465 | 325-344 | 71.43 | 0.038 | 549-584 | 621-650 | 0.69 | 1.35 | 0.5983 | 0.4174 |
| S | MOG | 249-278 | 83-102 | 83.33 | 0.069 | 944-974 | 150-180 | 0.63 | 2.06 | 0.4706 | 0.4059 |
| ORF7a | MOG | 25-32 | 95-102 | 75 | 2,00E-04 | 17-81 | Complete structure | 0.62 | 2.83 | 0.4846 | 0.6598 |
| N | MPO | 227-236 | 149-157 | 70 | 0.002 | 388-419 | 67-98 | 0.59 | 1.91 | 0.4117 | 0.4124 |
| Nsp13 | GAD65 | 466-472 | 439-445 | 66.67 | 3,00E-04 | 290-349 | 312-389 | 0.52 | 3.22 | 0.6555 | 0.4695 |
| Nsp1 | GAD65 | 131-138 | 137-139 | 100 | 0.005 | 32-61 | 302-331 | 0.52 | 1.57 | -0.3299 | 0.6325 |
| Nsp1 | MOG | 103-114 | 210-221 | 71.43 | 0.003 | 33-62 | 204-232 | 0.50 | 1.59 | 0.6013 | 0.6757 |
| Nsp3 | MPO | 903-908 | 613-618 | 83.33 | 2,00E-05 | 180-209 | 71-112 | 0.50 | 2.79 | 1.8236 | 0.8823 |
| S | NMDAR1 | 1020-1027 | 223-230 | 100 | 0.017 | 1020-1050 | 221-250 | 0.50 | 1.47 | 0.8726 | 0.7359 |
| Nsp3 | NMDAR1 | 1800-1809 | 766-774 | 100 | 5,00E-04 | 399-535 | 153-278 | 0.49 | 3.86 | 0.4238 | 0.4044 |
| Nsp13 | PLP | 88-94 | 99-105 | 71.43 | 1,00E-04 | 310-342 | 175-210 | 0.49 | 2.82 | -0.0624 | 0.4685 |
| Nsp1 | PLP | 34-62 | 196-211 | 100 | 0.006 | 33-64 | 240-272 | 0.47 | 1.69 | 0.7645 | 0.5854 |
| Nsp1 | Transaldolase | 84-99 | 139-154 | 100 | 0.001 | 33-62 | 145-176 | 0.46 | 2.18 | 0.6898 | 0.6757 |
| Nsp2 | MPO | 419-425 | 159-165 | 100 | 0.15 | 672-707 | 67-95 | 0.46 | 1.16 | 0.5813 | 0.5890 |
| Nsp3 | PLP | 88-94 | 99-105 | 71.43 | 1,00E-04 | 180-209 | 34-63 | 0.45 | 2.07 | -0.0624 | 0.8823 |
| M | MAG | 132-138 | 369-375 | 100 | 2,00E-04 | 156-186 | 295-325 | 0.43 | 2.59 | 0.4549 | 0.6527 |
| S | Transaldolase | 1110-1117 | 37-44 | 62.50 | 0.002 | 276-305 | 125-156 | 0.42 | 2.10 | 0.8734 | 0.6476 |
| Nsp13 | Transaldolase | 146-151 | 307-312 | 83.33 | 0.001 | 367-396 | 135-164 | 0.41 | 2.60 | 1.0726 | 1.1104 |
| N | CNP | 243-252 | 191 200 | 100 | 0.001 | 195-239 | 322-363 | 0.41 | 2.72 | 0.7056 | 0.6601 |
| Nsp13 | NMDAR1 | 301-306 | 628-636 | 100 | 3,00E-04 | 503-532 | 683-712 | 0.41 | 2.90 | 0.9060 | 0.4818 |
| Nsp2 | Transaldolase | 389-403 | 95-109 | 66.67 | 0.19 | 249-280 | 134-163 | 0.40 | 1.90 | 0.5146 | 0.7524 |
| Nsp2 | CNP | 270-278 | 348-356 | 77.78 | 4,00E-07 | 395-432 | 349-382 | 0.39 | 2.38 | 0.9295 | 0.4387 |
| N | MOBP | 198-209 | 168-178 | 77.78 | 5,00E-06 | 202-233 | 29-74 | 0.38 | 2.38 | 0.4291 | 0.6320 |
| N | PLP | 170-184 | 120-134 | 75 | 0.0003 | 212-241 | 75-98 | 0.37 | 1.44 | 0.4459 | 0.7169 |
| Nsp1 | MBP | 76-102 | 198-116 | 100 | 0.010 | 32-67 | 211-240 | 0.36 | 3.22 | 0.6583 | 0.4770 |
| Nsp3 | AQP4 | 63-77 | 51-65 | 100 | 0.27 | 2667-2697 | 197-227 | 0.35 | 2.36 | 0.4168 | 0.5551 |
| Nsp2 | MBP | 61-73 | 107-119 | 100 | 8,00E-04 | 280-309 | 200-229 | 0.33 | 2.79 | 0.6625 | 0.5624 |
3.3. Search for potential T cell epitopes
| SARS-CoV-2 antigen | Autoantigens | Allele | Potencial SARS-CoV-2 epitope | Corresponding human epitope | IC50 virus peptide | IC50 human peptide |
|---|---|---|---|---|---|---|
| M | NMDAR1 | HLA-DQA1*01:02/DQB1*06:02 | NWITGGIAIAMACLV | VWAGFAMIIVASYTA | 57.00 | 60.00 |
| HLA-DRB1*15:01 | LMWLSYFIASFRLFA | GFAMIIVASYTANLA | 68.00 | 49.00 | ||
| HLA-A*31:01 | LSYFIASFR | LGMVWAGFAM | 12.20 | 262.06 | ||
| M | MPO | HLA-A*31:01 | LSYFIASFR | KQLVDKAYK | 12.20 | 68.78 |
| HLA-B*07:02 | GGIAIAMACLV | RLRSGSASPM | 321.23 | 159.89 | ||
| Nsp2 | NMDAR1 | HLA-DQA1*01:02/DQB1*06:02 | RVLQKAAITILDGIS | VWAGFAMIIVASYTA | 93.00 | 60.00 |
| HLA-DRB1*15:01 | ITILDGISQYSLRLI | VWAGFAMIIVASYTA | 146.00 | 60.00 | ||
| HLA-A*31:01 | RTLETAQNSVR | GAPRSFSAR | 309.40 | 129.86 | ||
| HLA-B*07:02 | SVRVLQKAAI | APRSFSARIL | 357.95 | 29.30 | ||
| S | MOG | HLA-DRB1*15:01 | LNTLVKQLSSNFGAI | GVLVLLAVLPVLLLQ | 89.00 | 58.00 |
| HLA-DQA1*01:02/ DQB1*06:02 | QNAQALNTLVKQLSS | WVSPGVLVLLAVLPV | 319.00 | 229.00 | ||
| ORF7a | MOG | HLA-DQA1*01:02/DQB1*06:02 | YQECVRGTTVLLKEP | YWVSPGVLVLLAVLP | 261.00 | 210.00 |
| N | MPO | HLA-A*31:01 | FSKQLQQSMSS | KQLVDKAYK | 46.72 | 68.78 |
| Nsp13 | GAD65 | HLA-DRB1*15:01 | AIGLALYYPSARIVY | AKQKGFVPFLVSATA | 71.00 | 196.00 |
| HLA-DQA1*01:02/DQB1*06:02 | IVYTACSHAAVDALC | LVSATAGTTVYGAFD | 347.00 | 52.00 | ||
| HLA-A*31:01 | KYLPIDKCSR | KHKWKLSGVER | 18.72 | 35.44 | ||
| HLA-B*07:02 | LPIDKCSR | VPFLVSAT | 187.50 | 69.82 | ||
| Nsp1 | GAD65 | HLA-DRB1*03:01 | SVEEVLSEARQHLKD | RGKMIPSDLERRILE | 489.00 | 448.00 |
| HLA-A*31:01 | HLKDGTCGLVE | KMIPSDLERR | 31.44 | 27.31 | ||
| Nsp1 | MOG | HLA-A*31:01 | HLKDGTCGLVE | CWKITLFVIVP | 31.44 | 284.79 |
| Nsp3 | MPO | HLA-A*31:01 | SYKDWSYSGQS | RLRSGSASPME | 38.77 | 111.56 |
| S | NMDAR1 | HLA-DQA1*01:02/ DQB1*06:02 | ASANLAATKMSECVL | ASEDDAATVYRAAAM | 213.00 | 121.00 |
| HLA-A*31:01 | KMSECVLGQSKR | LSASEDDAATVYR | 297.9 | 495.7 | ||
| HLA-DQA1*01:02/ DQB1*06:02 | AQYTSALLAGTITSG | SRRVLLLAGRLAAQS | 122.00 | 199.00 | ||
| HLA-B*07:02 | MIAQYTSAL | MAAESRRVL | 61.88 | 54.29 |
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- worldometers.info/coronavirus/, “COVID-19 CORONAVIRUS PANDEMIC.”.
- N. I. Nii-Trebi, “Emerging and Neglected Infectious Diseases: Insights, Advances, and Challenges,” Biomed Res Int, vol. 2017, pp. 1–15, 2017, doi: 10.1155/2017/5245021. [CrossRef]
- S. V. Alves-Leon et al., “Zika virus found in brain tissue of a multiple sclerosis patient undergoing an acute disseminated encephalomyelitis-like episode,” Multiple Sclerosis Journal, vol. 25, no. 3, pp. 427–430, Mar. 2019, doi: 10.1177/1352458518781992. [CrossRef]
- F. C. Rueda-Lopes et al., “Clinical and magnetic resonance imaging patterns of extensive Chikungunya virus–associated myelitis,” J Neurovirol, vol. 27, no. 4, pp. 616–625, Aug. 2021, doi: 10.1007/s13365-021-00962-4. [CrossRef]
- S. V. Alves-Leon et al., “Exome-Wide Search for Genes Associated With Central Nervous System Inflammatory Demyelinating Diseases Following CHIKV Infection: The Tip of the Iceberg,” Front Genet, vol. 12, Mar. 2021, doi: 10.3389/fgene.2021.639364. [CrossRef]
- P. Feizi et al., “Central nervous system (CNS) inflammatory demyelinating diseases (IDDs) associated with COVID-19: A case series and review,” J Neuroimmunol, vol. 371, p. 577939, Oct. 2022, doi: 10.1016/j.jneuroim.2022.577939. [CrossRef]
- G. Toscano et al., “Guillain–Barré Syndrome Associated with SARS-CoV-2,” New England Journal of Medicine, vol. 382, no. 26, pp. 2574–2576, Jun. 2020, doi: 10.1056/NEJMc2009191. [CrossRef]
- M. B. A. Oldstone, “Molecular mimicry and immune-mediated diseases,” The FASEB Journal, vol. 12, no. 13, pp. 1255–1265, Oct. 1998, doi: 10.1096/fasebj.12.13.1255. [CrossRef]
- M. Lima et al., “Coronaviruses and their relationship with multiple sclerosis: is the prevalence of multiple sclerosis going to increase after the Covid-19 pandemia?,” Rev Neurosci, vol. 33, no. 7, pp. 703–720, Oct. 2022, doi: 10.1515/revneuro-2021-0148. [CrossRef]
- J. C. Gerdes, I. Klein, B. L. DeVald, and J. S. Burks, “Coronavirus isolates SK and SD from multiple sclerosis patients are serologically related to murine coronaviruses A59 and JHM and human coronavirus OC43, but not to human coronavirus 229E,” J Virol, vol. 38, no. 1, pp. 231–238, Apr. 1981, doi: 10.1128/jvi.38.1.231-238.1981. [CrossRef]
- R. Moody, K. Wilson, K. L. Flanagan, A. Jaworowski, and M. Plebanski, “Adaptive Immunity and the Risk of Autoreactivity in COVID-19,” Int J Mol Sci, vol. 22, no. 16, p. 8965, Aug. 2021, doi: 10.3390/ijms22168965. [CrossRef]
- C. Hammer et al., “Neuropsychiatric disease relevance of circulating anti-NMDA receptor autoantibodies depends on blood–brain barrier integrity,” Mol Psychiatry, vol. 19, no. 10, pp. 1143–1149, Oct. 2014, doi: 10.1038/mp.2013.110. [CrossRef]
- H. Prüss, “Autoantibodies in neurological disease,” Nature Reviews Immunology, vol. 21, no. 12. Nature Research, pp. 798–813, Dec. 01, 2021. doi: 10.1038/s41577-021-00543-w. [CrossRef]
- P. Feizi et al., “Central nervous system (CNS) inflammatory demyelinating diseases (IDDs) associated with COVID-19: A case series and review,” J Neuroimmunol, vol. 371, p. 577939, Oct. 2022, doi: 10.1016/j.jneuroim.2022.577939. [CrossRef]
- H. Wang, “COVID−19, Anti-NMDA Receptor Encephalitis and MicroRNA,” Front Immunol, vol. 13, Mar. 2022, doi: 10.3389/fimmu.2022.825103. [CrossRef]
- M. Salari, B. Zaker Harofteh, and M. Etemadifar, “Autoimmune meningoencephalitis associated with anti-glutamic acid decarboxylase antibody following COVID-19 infection: A case report,” Clin Case Rep, vol. 10, no. 12, Dec. 2022, doi: 10.1002/ccr3.6597. [CrossRef]
- L. Llorente Ayuso, P. Torres Rubio, R. F. Beijinho do Rosário, M. L. Giganto Arroyo, and F. Sierra-Hidalgo, “Bickerstaff encephalitis after COVID-19,” J Neurol, vol. 268, no. 6, pp. 2035–2037, Jun. 2021, doi: 10.1007/s00415-020-10201-1. [CrossRef]
- M. Gaughan, S. Connolly, S. O’Riordan, N. Tubridy, C. McGuigan, and J. A. Kinsella, “Pediatric Parainfectious Encephalitis Associated With COVID-19,” Neurology, vol. 96, no. 11, pp. 541–544, Mar. 2021, doi: 10.1212/WNL.0000000000011476. [CrossRef]
- N. Ahsan, S. Jafarpour, and J. D. Santoro, “Myelin oligodendrocyte glycoprotein antibody encephalitis following severe acute respiratory syndrome coronavirus 2 in a pediatric patient,” Clin Exp Pediatr, vol. 64, no. 6, pp. 310–312, Jun. 2021, doi: 10.3345/cep.2020.01963. [CrossRef]
- A. Guilmot et al., “Immune-mediated neurological syndromes in SARS-CoV-2-infected patients,” J Neurol, vol. 268, no. 3, pp. 751–757, Mar. 2021, doi: 10.1007/s00415-020-10108-x. [CrossRef]
- Marino Gammazza et al., “Human molecular chaperones share with SARS-CoV-2 antigenic epitopes potentially capable of eliciting autoimmunity against endothelial cells: possible role of molecular mimicry in COVID-19,” Cell Stress Chaperones, vol. 25, no. 5, pp. 737–741, Sep. 2020, doi: 10.1007/s12192-020-01148-3. [CrossRef]
- D. Kanduc, “From Anti-SARS-CoV-2 Immune Responses to COVID-19 via Molecular Mimicry,” Antibodies, vol. 9, no. 3, p. 33, Jul. 2020, doi: 10.3390/antib9030033. [CrossRef]
- E. Gusev, A. Sarapultsev, L. Solomatina, and V. Chereshnev, “SARS-CoV-2-Specific Immune Response and the Pathogenesis of COVID-19,” Int J Mol Sci, vol. 23, no. 3, p. 1716, Feb. 2022, doi: 10.3390/ijms23031716. [CrossRef]
- Sharma and J. Bayry, “High risk of autoimmune diseases after COVID-19,” Nat Rev Rheumatol, vol. 19, no. 7, pp. 399–400, Jul. 2023, doi: 10.1038/s41584-023-00964-y. [CrossRef]
- A. Marino Gammazza et al., “Human molecular chaperones share with SARS-CoV-2 antigenic epitopes potentially capable of eliciting autoimmunity against endothelial cells: possible role of molecular mimicry in COVID-19,” Cell Stress Chaperones, vol. 25, no. 5, pp. 737–741, Sep. 2020, doi: 10.1007/s12192-020-01148-3. [CrossRef]
- G. Lucchese and A. Flöel, “SARS-CoV-2 and Guillain-Barré syndrome: molecular mimicry with human heat shock proteins as potential pathogenic mechanism,” Cell Stress Chaperones, vol. 25, no. 5, pp. 731–735, Sep. 2020, doi: 10.1007/s12192-020-01145-6. [CrossRef]
- A. Felipe Cuspoca, P. Isaac Estrada, and A. Velez-van-Meerbeke, “Molecular Mimicry of SARS-CoV-2 Spike Protein in the Nervous System: A Bioinformatics Approach,” Comput Struct Biotechnol J, vol. 20, pp. 6041–6054, 2022, doi: 10.1016/j.csbj.2022.10.022. [CrossRef]
- F. Madeira et al., “Search and sequence analysis tools services from EMBL-EBI in 2022,” Nucleic Acids Res, vol. 50, no. W1, pp. W276–W279, Jul. 2022, doi: 10.1093/nar/gkac240. [CrossRef]
- S. F. Altschul, W. Gish, W. Miller, E. W. Myers, and D. J. Lipman, “Basic local alignment search tool,” J Mol Biol, vol. 215, no. 3, pp. 403–410, Oct. 1990, doi: 10.1016/S0022-2836(05)80360-2. [CrossRef]
- S. F. Altschul et al., “Protein database searches using compositionally adjusted substitution matrices,” FEBS Journal, vol. 272, no. 20, pp. 5101–5109, Oct. 2005, doi: 10.1111/j.1742-4658.2005.04945.x. [CrossRef]
- Y. Zhang, “TM-align: a protein structure alignment algorithm based on the TM-score,” Nucleic Acids Res, vol. 33, no. 7, pp. 2302–2309, Apr. 2005, doi: 10.1093/nar/gki524. [CrossRef]
- M. Nielsen, C. Lundegaard, and O. Lund, “Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method,” BMC Bioinformatics, vol. 8, no. 1, p. 238, Dec. 2007, doi: 10.1186/1471-2105-8-238. [CrossRef]
- M. Mohkhedkar, S. S. K. Venigalla, and V. Janakiraman, “Untangling COVID-19 and autoimmunity: Identification of plausible targets suggests multi organ involvement,” Mol Immunol, vol. 137, pp. 105–113, Sep. 2021, doi: 10.1016/j.molimm.2021.06.021. [CrossRef]
- C. P. Figueiredo et al., “Zika virus replicates in adult human brain tissue and impairs synapses and memory in mice,” Nat Commun, vol. 10, no. 1, Dec. 2019, doi: 10.1038/s41467-019-11866-7. [CrossRef]
- U. K. Ranawaka, “Emerging tropical neurological infections,” Clinical Medicine, vol. 22, no. 1, pp. 18–20, Jan. 2022, doi: 10.7861/clinmed.2021-0799. [CrossRef]
- L. Zerboni, M. Reichelt, and A. Arvin, “Varicella-Zoster Virus Neurotropism in SCID Mouse–Human Dorsal Root Ganglia Xenografts,” 2010, pp. 255–276. doi: 10.1007/82_2009_8. [CrossRef]
- M. K. Das, A. Sarma, and T. Chakraborty, “Nano-ART and NeuroAIDS,” Drug Deliv Transl Res, vol. 6, no. 5, pp. 452–472, Oct. 2016, doi: 10.1007/s13346-016-0293-z. [CrossRef]
- V. da Silva, F. L. Fontes-Dantas, T. V. Dantas, A. Dutra, O. J. M. Nascimento, and S. V. Alves-Leon, “Shared Molecular Signatures Across Zika Virus Infection and Multiple Sclerosis Highlight AP-1 Transcription Factor as a Potential Player in Post-ZIKV MS-Like Phenotypes,” Mol Neurobiol, vol. 60, no. 8, pp. 4184–4205, Aug. 2023, doi: 10.1007/s12035-023-03305-y. [CrossRef]
- L. C. França et al., “Mimetismo molecular entre o vírus Zika e os distúrbios inflamatórios desmielinizantes do sistema nervoso central: o papel do epítopo NS5 do vírus Zika e dos autoantígenos PLP,” Arq Neuropsiquiatr, vol. 81, no. 4, pp. 357–368, Apr. 2023, doi: 10.1055/s-0043-1768698. [CrossRef]
- L. T. de F. Cavalcante et al., “Buffy Coat Transcriptomic Analysis Reveals Alterations in Host Cell Protein Synthesis and Cell Cycle in Severe COVID-19 Patients,” Int J Mol Sci, vol. 23, no. 21, p. 13588, Nov. 2022, doi: 10.3390/ijms232113588. [CrossRef]
- L. Yan et al., “Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis,” Cell, vol. 184, no. 1, pp. 184-193.e10, Jan. 2021, doi: 10.1016/j.cell.2020.11.016. [CrossRef]
- J. Almqvist et al., “Neurological manifestations of coronavirus infections – a systematic review,” Ann Clin Transl Neurol, vol. 7, no. 10, pp. 2057–2071, Oct. 2020, doi: 10.1002/acn3.51166. [CrossRef]
- R. Chang, T. Yen-Ting Chen, S.-I. Wang, Y.-M. Hung, H.-Y. Chen, and C.-C. J. Wei, “Risk of autoimmune diseases in patients with COVID-19: a retrospective cohort study,” EClinicalMedicine, vol. 56, p. 101783, Feb. 2023, doi: 10.1016/j.eclinm.2022.101783. [CrossRef]
- Tesch et al., “Incident autoimmune diseases in association with a SARS-CoV-2 infection: A matched cohort study”, doi: 10.1101/2023.01.25.23285014. [CrossRef]
- D. G. Corrêa, F. C. de Souza Lima, D. da Cruz Bezerra, A. C. Coutinho, and L. C. Hygino da Cruz, “COVID-19 associated with encephalomyeloradiculitis and positive anti-aquaporin-4 antibodies: Cause or coincidence?,” Multiple Sclerosis Journal, vol. 27, no. 6, pp. 973–976, May 2021, doi: 10.1177/1352458520949988. [CrossRef]
- Lotan, S. Nishiyama, G. S. Manzano, M. Lydston, and M. Levy, “COVID-19 and the risk of CNS demyelinating diseases: A systematic review,” Front Neurol, vol. 13, Sep. 2022, doi: 10.3389/fneur.2022.970383. [CrossRef]
- C. Galeotti and J. Bayry, “Autoimmune and inflammatory diseases following COVID-19,” Nat Rev Rheumatol, vol. 16, no. 8, pp. 413–414, Aug. 2020, doi: 10.1038/s41584-020-0448-7. [CrossRef]
- A. Schattner and B. Rager-Zisman, “Virus-Induced Autoimmunity,” Clinical Infectious Diseases, vol. 12, no. 2, pp. 204–222, Mar. 1990, doi: 10.1093/clinids/12.2.204. [CrossRef]
- M. B. A. Oldstone, “Molecular mimicry and immune-mediated diseases,” The FASEB Journal, vol. 12, no. 13, pp. 1255–1265, Oct. 1998, doi: 10.1096/fasebj.12.13.1255. [CrossRef]
- S. Morsy and A. Morsy, “Epitope mimicry analysis of SARS-COV-2 surface proteins and human lung proteins,” J Mol Graph Model, vol. 105, Jun. 2021, doi: 10.1016/j.jmgm.2021.107836. [CrossRef]
- V. A. Petrenko, J. W. Gillespie, L. M. De Plano, and M. A. Shokhen, “Phage-Displayed Mimotopes of SARS-CoV-2 Spike Protein Targeted to Authentic and Alternative Cellular Receptors,” Viruses, vol. 14, no. 2, Feb. 2022, doi: 10.3390/v14020384. [CrossRef]
- K. Karagöz, M. R. Munk, M. Kaya, R. Rückert, M. Yıldırım, and L. Karabaş, “Using bioinformatic protein sequence similarity to investigate if SARS CoV-2 infection could cause an ocular autoimmune inflammatory reactions?,” Exp Eye Res, vol. 203, Feb. 2021, doi: 10.1016/j.exer.2020.108433. [CrossRef]
- Lucchese and A. Flöel, “Molecular mimicry between SARS-CoV-2 and respiratory pacemaker neurons,” Autoimmun Rev, vol. 19, no. 7, p. 102556, Jul. 2020, doi: 10.1016/j.autrev.2020.102556. [CrossRef]
- Y. Lavi et al., “Dysregulated Levels of Circulating Autoantibodies against Neuronal and Nervous System Autoantigens in COVID-19 Patients,” Diagnostics, vol. 13, no. 4, p. 687, Feb. 2023, doi: 10.3390/diagnostics13040687. [CrossRef]
- Giovannoni and G. Ebers, “Multiple sclerosis: the environment and causation,” Curr Opin Neurol, vol. 20, no. 3, pp. 261–268, Jun. 2007, doi: 10.1097/WCO.0b013e32815610c2. [CrossRef]
- T. Olsson, L. F. Barcellos, and L. Alfredsson, “Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis,” Nat Rev Neurol, vol. 13, no. 1, pp. 25–36, Jan. 2017, doi: 10.1038/nrneurol.2016.187. [CrossRef]
- N. Kaushansky, M. Eisenstein, R. Zilkha-Falb, and A. Ben-Nun, “The myelin-associated oligodendrocytic basic protein (MOBP) as a relevant primary target autoantigen in multiple sclerosis,” Autoimmun Rev, vol. 9, no. 4, pp. 233–236, Feb. 2010, doi: 10.1016/j.autrev.2009.08.002. [CrossRef]
- F. Mohammadi-Milasi, K. Mahnam, and M. Shakhsi-Niaei, “In silico study of the association of the HLA-A*31:01 allele (human leucocyte antigen allele 31:01) with neuroantigenic epitopes of PLP (proteolipid protein), MBP (myelin basic protein) and MOG proteins (myelin oligodendrocyte glycoprotein) for studying the multiple sclerosis disease pathogenesis,” J Biomol Struct Dyn, vol. 39, no. 7, pp. 2526–2542, May 2021, doi: 10.1080/07391102.2020.1751291. [CrossRef]
- M. Minohara et al., “Upregulation of myeloperoxidase in patients with opticospinal multiple sclerosis: Positive correlation with disease severity,” J Neuroimmunol, vol. 178, no. 1–2, pp. 156–160, Sep. 2006, doi: 10.1016/j.jneuroim.2006.05.026. [CrossRef]
- E. Gray, T. L. Thomas, S. Betmouni, N. Scolding, and S. Love, “Elevated Activity and Microglial Expression of Myeloperoxidase in Demyelinated Cerebral Cortex in Multiple Sclerosis,” Brain Pathology, vol. 18, no. 1, pp. 86–95, Jan. 2008, doi: 10.1111/j.1750-3639.2007.00110.x. [CrossRef]
- R. M. Nagra et al., “Immunohistochemical and genetic evidence of myeloperoxidase involvement in multiple sclerosis,” J Neuroimmunol, vol. 78, no. 1–2, pp. 97–107, Sep. 1997, doi: 10.1016/S0165-5728(97)00089-1. [CrossRef]
- Lotan, S. Nishiyama, G. S. Manzano, M. Lydston, and M. Levy, “COVID-19 and the risk of CNS demyelinating diseases: A systematic review,” Front Neurol, vol. 13, Sep. 2022, doi: 10.3389/fneur.2022.970383. [CrossRef]
- S. Sedighi et al., “Comprehensive Investigations Relationship Between Viral Infections and Multiple Sclerosis Pathogenesis,” Curr Microbiol, vol. 80, no. 1, p. 15, Jan. 2023, doi: 10.1007/s00284-022-03112-z. [CrossRef]
- S. Mariotto et al., “Clinical spectrum and IgG subclass analysis of anti-myelin oligodendrocyte glycoprotein antibody-associated syndromes: a multicenter study,” J Neurol, vol. 264, no. 12, pp. 2420–2430, Dec. 2017, doi: 10.1007/s00415-017-8635-4. [CrossRef]
- A. Cobo-Calvo et al., “Clinical spectrum and prognostic value of CNS MOG autoimmunity in adults,” Neurology, vol. 90, no. 21, pp. e1858–e1869, May 2018, doi: 10.1212/WNL.0000000000005560. [CrossRef]
- V. Sehgal, P. Bansal, S. Arora, S. Kapila, and G. S. Bedi, “Myelin Oligodendrocyte Glycoprotein Antibody Disease After COVID-19 Vaccination - Causal or Incidental?,” Cureus, Jul. 2022, doi: 10.7759/cureus.27024. [CrossRef]
- M. Netravathi et al., “COVID-19 vaccine associated demyelination & its association with MOG antibody,” Mult Scler Relat Disord, vol. 60, p. 103739, Apr. 2022, doi: 10.1016/j.msard.2022.103739. [CrossRef]
- Lambe et al., “Myelin oligodendrocyte glycoprotein-IgG associated disorders (MOGAD) following SARS-CoV-2 infection: A case series,” J Neuroimmunol, vol. 370, p. 577933, Sep. 2022, doi: 10.1016/j.jneuroim.2022.577933. [CrossRef]
- V. Vasilevska, P. C. Guest, H. G. Bernstein, M. L. Schroeter, C. Geis, and J. Steiner, “Molecular mimicry of NMDA receptors may contribute to neuropsychiatric symptoms in severe COVID-19 cases,” Journal of Neuroinflammation, vol. 18, no. 1. BioMed Central Ltd, Dec. 01, 2021. doi: 10.1186/s12974-021-02293-x. [CrossRef]
- M. Etemadifar, H. Nouri, M. Salari, and N. Sedaghat, “Detection of anti-NMDA receptor antibodies following BBIBP-CorV COVID-19 vaccination in a rituximab-treated person with multiple sclerosis presenting with manifestations of an acute relapse,” Hum Vaccin Immunother, vol. 18, no. 1, Jan. 2022, doi: 10.1080/21645515.2022.2033540. [CrossRef]
- Valadez-Calderon, A. Ordinola Navarro, E. Rodriguez-Chavez, and O. Vera-Lastra, “Co-expression of anti-NMDAR and anti-GAD65 antibodies. A case of autoimmune encephalitis in a post-COVID-19 patient,” Neurología, vol. 37, no. 6, pp. 503–504, Jul. 2022, doi: 10.1016/j.nrl.2021.09.003. [CrossRef]
- M. Salari, B. Zaker Harofteh, and M. Etemadifar, “Autoimmune meningoencephalitis associated with anti-glutamic acid decarboxylase antibody following COVID-19 infection: A case report,” Clin Case Rep, vol. 10, no. 12, Dec. 2022, doi: 10.1002/ccr3.6597. [CrossRef]
- F. Assi, R. Abdallah, A. Mecheik, H. H. Rahhal, and J. Wazne, “Acute Disseminated Encephalomyelitis Following COVID-19 Infection,” Cureus, Jan. 2023, doi: 10.7759/cureus.33365. [CrossRef]
- Mumoli, V. Vescio, D. Pirritano, E. Russo, and D. Bosco, “ADEM anti-MOG antibody-positive after SARS-CoV2 vaccination,” Neurological Sciences, vol. 43, no. 2, pp. 763–766, Feb. 2022, doi: 10.1007/s10072-021-05761-7. [CrossRef]
- Y. Wang, Y. Wang, L. Huo, Q. Li, J. Chen, and H. Wang, “SARS-CoV-2-associated acute disseminated encephalomyelitis: a systematic review of the literature,” J Neurol, vol. 269, no. 3, pp. 1071–1092, Mar. 2022, doi: 10.1007/s00415-021-10771-8. [CrossRef]
- Miyamoto, J. Koh, M. Takahashi, M. Niwa, and H. Ito, “A case of anti-MOG antibody-positive ADEM following COVID-19 mRNA vaccination,” Neurological Sciences, vol. 43, no. 6, pp. 3513–3514, Jun. 2022, doi: 10.1007/s10072-022-06019-6. [CrossRef]
- A. A. Pinto, L. S. Carroll, V. Nar, A. Varatharaj, and I. Galea, “CNS inflammatory vasculopathy with antimyelin oligodendrocyte glycoprotein antibodies in COVID-19,” Neurology - Neuroimmunology Neuroinflammation, vol. 7, no. 5, p. e813, Sep. 2020, doi: 10.1212/NXI.0000000000000813. [CrossRef]
- Naidu and R. Tayler, “Anti N-Methyl-D-Aspartate receptor antibody associated Acute Demyelinating Encephalomyelitis in a patient with COVID-19: a case report,” J Med Case Rep, vol. 17, no. 1, p. 247, Jun. 2023, doi: 10.1186/s13256-023-03979-x. [CrossRef]
- H. K. Al-Hakeim, H. T. Al-Rubaye, D. S. Al-Hadrawi, A. F. Almulla, and M. Maes, “Long-COVID post-viral chronic fatigue and affective symptoms are associated with oxidative damage, lowered antioxidant defenses and inflammation: a proof of concept and mechanism study,” Mol Psychiatry, vol. 28, no. 2, pp. 564–578, Feb. 2023, doi: 10.1038/s41380-022-01836-9. [CrossRef]
- H. K. Al-Hakeim, H. T. Al-Rubaye, A. F. Almulla, D. S. Al-Hadrawi, and M. Maes, “Chronic Fatigue, Depression and Anxiety Symptoms in Long COVID Are Strongly Predicted by Neuroimmune and Neuro-Oxidative Pathways Which Are Caused by the Inflammation during Acute Infection,” J Clin Med, vol. 12, no. 2, p. 511, Jan. 2023, doi: 10.3390/jcm12020511. [CrossRef]
- F. Li and J. Z. Tsien, “Memory and the NMDA Receptors,” New England Journal of Medicine, vol. 361, no. 3, pp. 302–303, Jul. 2009, doi: 10.1056/NEJMcibr0902052. [CrossRef]
- C. Franke et al., “Association of cerebrospinal fluid brain-binding autoantibodies with cognitive impairment in post-COVID-19 syndrome,” Brain Behav Immun, vol. 109, pp. 139–143, Mar. 2023, doi: 10.1016/j.bbi.2023.01.006. [CrossRef]
- S. H. Mueller et al., “Genetic predisposition in anti-LGI1 and anti-NMDA receptor encephalitis,” Ann Neurol, vol. 83, no. 4, pp. 863–869, Apr. 2018, doi: 10.1002/ana.25216. [CrossRef]
- “Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis,” Nature, vol. 476, no. 7359, pp. 214–219, Aug. 2011, doi: 10.1038/nature10251. [CrossRef]
- S. Vieira Alves-Leon et al., “ACUTE DISSEMINATED ENCEPHALOMYELITIS Clinical features, HLA DRB1*1501, HLA DRB1*1503, HLA DQA1*0102, HLA DQB1*0602, and HLA DPA1*0301 allelic association study,” 2009.
- A. De Silvestri et al., “The Involvement of HLA Class II Alleles in Multiple Sclerosis: A Systematic Review with Meta-analysis,” Dis Markers, vol. 2019, pp. 1–7, Nov. 2019, doi: 10.1155/2019/1409069. [CrossRef]
- F. Barcellos et al., “Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis,” Hum Mol Genet, vol. 15, no. 18, pp. 2813–2824, Sep. 2006, doi: 10.1093/hmg/ddl223. [CrossRef]
- F. L. Fontes-Dantas et al., “SARS-CoV-2 spike protein induces TLR4-mediated long-term cognitive dysfunction recapitulating post-COVID syndrome in mice,” Cell Rep, p. 112189, Feb. 2023, doi: 10.1016/j.celrep.2023.112189. [CrossRef]
- I. Ismail and S. Salama, “A systematic review of cases of CNS demyelination following COVID-19 vaccination,” J Neuroimmunol, vol. 362, p. 577765, Jan. 2022, doi: 10.1016/j.jneuroim.2021.577765. [CrossRef]
- P. Flannery, I. Yang, M. Keyvani, and G. Sakoulas, “Acute Psychosis Due to Anti-N-Methyl D-Aspartate Receptor Encephalitis Following COVID-19 Vaccination: A Case Report,” Front Neurol, vol. 12, Nov. 2021, doi: 10.3389/fneur.2021.764197. [CrossRef]
- H. C. Lee et al., “Anti-N-Methyl-D-Aspartate Receptor Encephalitis after BNT162b2 COVID-19 Vaccination,” J Epilepsy Res, vol. 12, no. 2, pp. 71–73, Dec. 2022, doi: 10.14581/jer.22013. [CrossRef]
- S. Jarius, N. Bieber, J. Haas, and B. Wildemann, “MOG encephalomyelitis after vaccination against severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2): case report and comprehensive review of the literature,” J Neurol, vol. 269, no. 10, pp. 5198–5212, Oct. 2022, doi: 10.1007/s00415-022-11194-9. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





