Submitted:
29 September 2023
Posted:
30 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Phenotypic Analysis
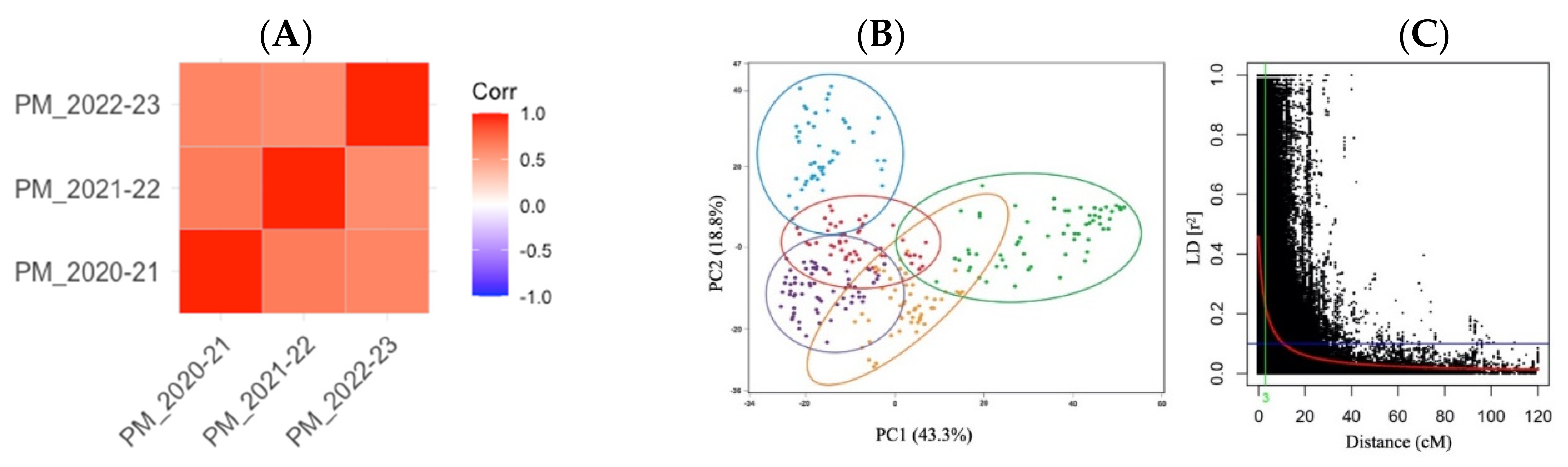
2.2. Population Structure Analysis
2.3. Genome-wide Marker Trait Association (MTA) Analysis
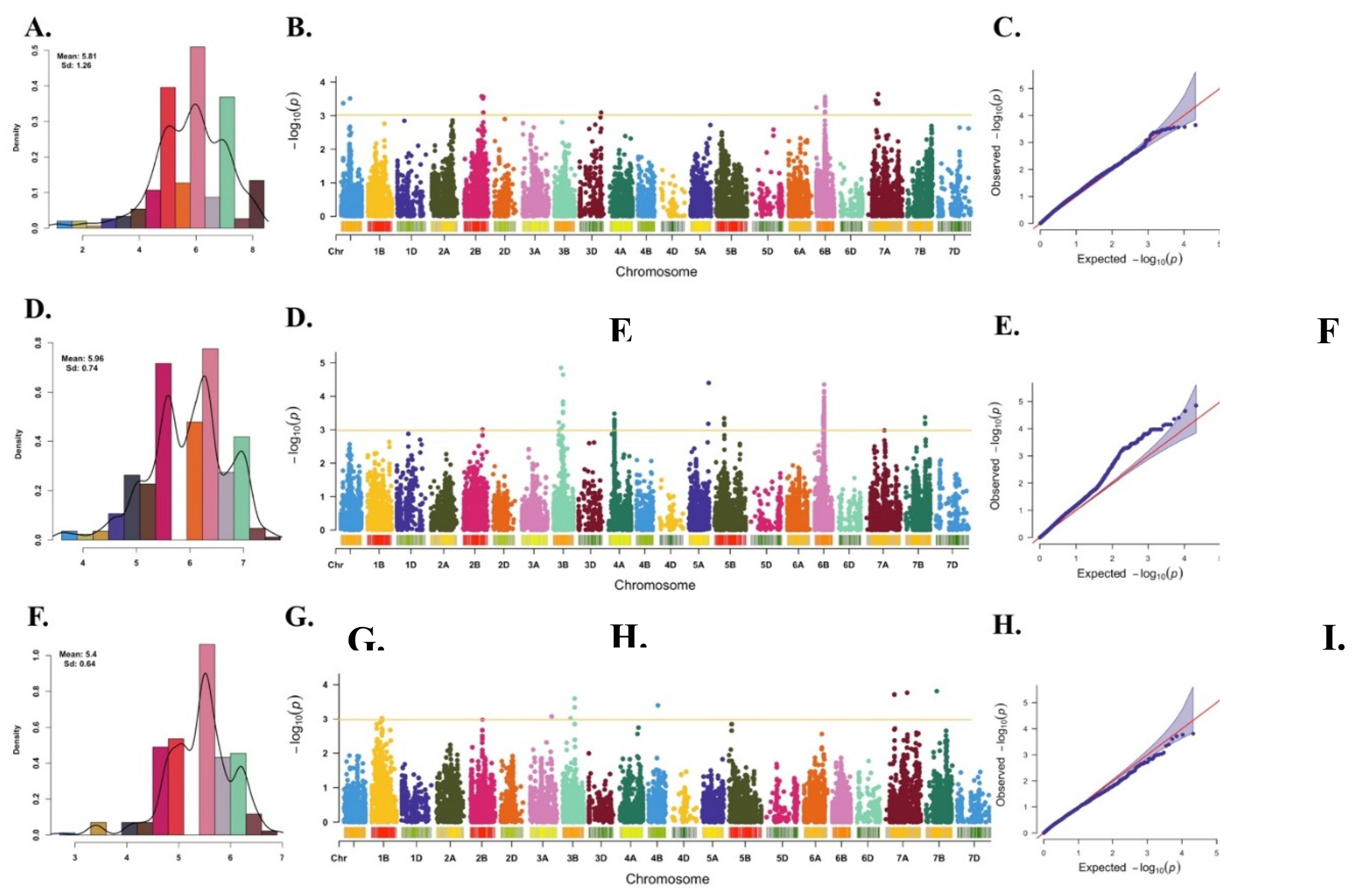
| Marker | Chr | Pos (cM)1 | Pos (bp)2 | Effect | p value |
|---|---|---|---|---|---|
| 2020-21 | |||||
| RAC875_c1710_376$ | 1A | 27.0 | 9103721 | -0.55 | 0.0004 |
| Kukri_c35200_895$ | 1A | 74.0 | 459514896 | -0.59 | 0.0003 |
| BS00080318_51$ | 2B | 132.0 | 763842555 | 0.32 | 0.0003 |
| Excalibur_c5438_274$ | 2B | 142.0 | 774958099 | 0.30 | 0.0003 |
| Excalibur_rep_c109577_698$ | 2B | 144.0 | 775368259 | -0.27 | 0.0008 |
| RAC875_rep_c83950_222$ | 2B | 144.0 | 775053184 | -0.29 | 0.0003 |
| Ra_c22493_190$ | 6B | 0.0 | 6196794 | -0.48 | 0.0006 |
| Excalibur_rep_c106789_271 | 6B | 59.0 | 153957925 | -0.33 | 0.0004 |
| RAC875_c25489_1208 | 6B | 59.0 | 153959656 | -0.31 | 0.0008 |
| wsnp_Ex_c39304_46635517 | 6B | 59.0 | 151609970 | -0.34 | 0.0003 |
| wsnp_Ex_rep_c102044_87296690 | 6B | 59.0 | 153959656 | -0.32 | 0.0005 |
| wsnp_Ex_rep_c102044_87297599 | 6B | 59.0 | 153957925 | -0.34 | 0.0003 |
| wsnp_Ra_c48999_54089942 | 6B | 59.0 | 153956896 | -0.31 | 0.0008 |
| Excalibur_c3795_198$ | 6B | 60.0 | 156266046 | -0.31 | 0.0005 |
| Ra_c14852_1487 | 6B | 60.0 | 156266570 | -0.32 | 0.0003 |
| RAC875_c66770_208 | 7A | 57.0 | 26879337 | -0.58 | 0.0004 |
| Excalibur_c7538_2718$ | 7A | 65.0 | 35602223 | -0.48 | 0.0004 |
| wsnp_Ex_c35_77935$ | 7A | 73.0 | 41960564 | -0.36 | 0.0002 |
| BS00023055_51 | 7A | 74.0 | 41959666 | -0.32 | 0.0004 |
| 2021-22 | |||||
| Excalibur_c7971_1573 | 2B | 144.0 | 775371388 | 0.04 | 0.0009 |
| BS00011728_51$ | 3B | 57.0 | 59064340 | -0.28 | 0.0000 |
| BS00022741_51$ | 3B | 61.0 | 66953509 | -0.26 | 0.0009 |
| Excalibur_c21372_142 | 3B | 61.0 | 66954023 | -0.26 | 0.0009 |
| Tdurum_contig51993_52$ | 3B | 61.0 | 66953509 | -0.26 | 0.0009 |
| BS00011869_51 | 3B | 71.0 | 426452313 | -0.23 | 0.0003 |
| Excalibur_c80041_400$ | 3B | 71.0 | 429610611 | -0.26 | 0.0000 |
| Kukri_c21818_519$ | 3B | 71.0 | 556059780 | -0.23 | 0.0002 |
| RAC875_c58159_989 | 3B | 71.0 | 569425136 | -0.19 | 0.0008 |
| wsnp_Ku_c21818_31604716$ | 3B | 71.0 | 556059565 | -0.24 | 0.0001 |
| BobWhite_c27944_234$ | 4A | 48.0 | 109443802 | -0.23 | 0.0007 |
| Ex_c17894_1159$ | 4A | 48.0 | 654418148 | -0.22 | 0.0005 |
| Excalibur_rep_c66815_273 | 4A | 48.0 | 104375678 | -0.22 | 0.0005 |
| GENE-2637_94$ | 4A | 48.0 | 164633479 | -0.23 | 0.0008 |
| IAAV3906$ | 4A | 48.0 | 113854919 | -0.22 | 0.0006 |
| IAAV8784 | 4A | 48.0 | 135358615 | -0.22 | 0.0005 |
| IACX1896$ | 4A | 48.0 | 109431746 | -0.22 | 0.0007 |
| Kukri_c44469_1240 | 4A | 48.0 | 136432865 | -0.22 | 0.0005 |
| Kukri_c48155_158$ | 4A | 48.0 | 120605031 | -0.22 | 0.0005 |
| RAC875_c22562_429$ | 4A | 48.0 | 111292238 | -0.23 | 0.0003 |
| RAC875_rep_c74695_101$ | 4A | 48.0 | 164632278 | -0.23 | 0.0008 |
| tplb0035b22_184 | 4A | 48.0 | 135614180 | -0.22 | 0.0005 |
| wsnp_BE442869A_Ta_2_1 | 4A | 48.0 | 140134106 | -0.22 | 0.0005 |
| wsnp_Ex_c10527_17198865$ | 4A | 48.0 | 115570013 | -0.22 | 0.0005 |
| wsnp_Ex_c1387_2659020$ | 4A | 48.0 | 115912754 | -0.22 | 0.0005 |
| wsnp_Ex_c14529_22547438 | 4A | 48.0 | 115913618 | -0.22 | 0.0005 |
| wsnp_Ex_c1865_3515470$ | 4A | 48.0 | 136894495 | -0.22 | 0.0005 |
| wsnp_Ex_c36141_44153175$ | 4A | 48.0 | 111091261 | -0.23 | 0.0007 |
| wsnp_Ex_c4286_7734046 | 4A | 48.0 | 114744375 | -0.22 | 0.0006 |
| wsnp_Ex_c43734_49968808$ | 4A | 48.0 | 211000047 | -0.24 | 0.0005 |
| wsnp_Ex_rep_c69890_68851948$ | 4A | 48.0 | 105249095 | -0.24 | 0.0005 |
| Tdurum_contig51134_191$ | 5A | 144.0 | 698501228 | -0.36 | 0.0000 |
| RAC875_c19099_308$ | 5B | 68.0 | 519153210 | 0.24 | 0.0007 |
| Tdurum_contig53926_455$ | 5B | 68.0 | 516469300 | 0.26 | 0.0006 |
| BS00046963_51 | 6B | 59.0 | 150665070 | -0.19 | 0.0006 |
| Excalibur_rep_c106789_271 | 6B | 59.0 | 153957925 | -0.20 | 0.0005 |
| Kukri_c38732_225$ | 6B | 59.0 | 151131387 | -0.18 | 0.0007 |
| wsnp_Ex_rep_c102044_87296690 | 6B | 59.0 | 153959656 | -0.19 | 0.0007 |
| wsnp_Ex_rep_c102044_87297599 | 6B | 59.0 | 153957925 | -0.20 | 0.0004 |
| Ra_c14852_1487$ | 6B | 60.0 | 156266570 | -0.18 | 0.0006 |
| BS00040868_51$ | 6B | 63.0 | 228977167 | -0.24 | 0.0001 |
| BobWhite_c34920_228 | 6B | 64.0 | 249498169 | -0.21 | 0.0006 |
| BS00003955_51$ | 6B | 64.0 | 260548319 | -0.24 | 0.0001 |
| BS00021686_51$ | 6B | 64.0 | 174566651 | -0.24 | 0.0003 |
| BS00045761_51$ | 6B | 64.0 | 227070547 | -0.24 | 0.0002 |
| BS00067871_51$ | 6B | 64.0 | 261585651 | -0.24 | 0.0002 |
| BS00067873_51$ | 6B | 64.0 | 261585682 | -0.23 | 0.0003 |
| BS00080544_51$ | 6B | 64.0 | 234560063 | -0.22 | 0.0007 |
| Ex_c49055_617$ | 6B | 64.0 | 260548488 | -0.22 | 0.0005 |
| Excalibur_c20503_382$ | 6B | 64.0 | 257568556 | -0.24 | 0.0001 |
| Excalibur_c47738_334 | 6B | 64.0 | 262473204 | -0.22 | 0.0004 |
| Excalibur_c5136_2314$ | 6B | 64.0 | 231677537 | -0.22 | 0.0003 |
| Excalibur_c53834_416$ | 6B | 64.0 | 257993802 | -0.25 | 0.0000 |
| Excalibur_c79066_165$ | 6B | 64.0 | 232168465 | -0.24 | 0.0002 |
| GENE-2606_197$ | 6B | 64.0 | 257567774 | -0.25 | 0.0001 |
| Kukri_c25377_106$ | 6B | 64.0 | 174567929 | -0.23 | 0.0005 |
| Kukri_c38058_532 | 6B | 64.0 | 257567774 | -0.22 | 0.0009 |
| Kukri_c52515_442 | 6B | 64.0 | 259877872 | -0.21 | 0.0005 |
| Ra_c77985_260$ | 6B | 64.0 | 214089373 | -0.22 | 0.0008 |
| RAC875_c12805_908$ | 6B | 64.0 | 234559907 | -0.23 | 0.0005 |
| RAC875_c24962_1326 | 6B | 64.0 | 229280173 | -0.23 | 0.0003 |
| RFL_Contig311_951 | 6B | 64.0 | 231354317 | -0.23 | 0.0003 |
| TA005139-0719$ | 6B | 64.0 | 234559694 | -0.24 | 0.0004 |
| wsnp_BQ161448B_Ta_2_1$ | 6B | 64.0 | 261530574 | -0.23 | 0.0002 |
| wsnp_Ex_c1603_3056226 | 6B | 64.0 | 221813724 | -0.23 | 0.0004 |
| wsnp_Ex_c23474_32717535$ | 6B | 64.0 | 276221225 | -0.25 | 0.0001 |
| wsnp_Ex_c27934_37093614 | 6B | 64.0 | 259881611 | -0.24 | 0.0002 |
| wsnp_Ex_c42372_48966781 | 6B | 64.0 | 229280173 | -0.21 | 0.0007 |
| wsnp_Ex_c46160_51746546 | 6B | 64.0 | 229278919 | -0.23 | 0.0002 |
| wsnp_Ex_rep_c103466_88415738 | 6B | 64.0 | 274209042 | -0.24 | 0.0002 |
| wsnp_Ex_rep_c103466_88415994 | 6B | 64.0 | 274208315 | -0.22 | 0.0006 |
| wsnp_Ex_rep_c103497_88437811$ | 6B | 64.0 | 277165618 | -0.24 | 0.0001 |
| wsnp_Ex_rep_c68480_67305954 | 6B | 64.0 | 259292578 | -0.24 | 0.0001 |
| wsnp_JD_c6448_7610859 | 6B | 64.0 | 257993802 | -0.24 | 0.0001 |
| wsnp_Ku_c27423_37369145 | 6B | 64.0 | 259883491 | -0.21 | 0.0005 |
| wsnp_Ra_c16850_25605248 | 6B | 64.0 | 262473278 | -0.23 | 0.0002 |
| wsnp_Ra_c33358_42248399$ | 6B | 64.0 | 257993727 | -0.22 | 0.0004 |
| wsnp_Ra_rep_c111161_93528347$ | 6B | 64.0 | 277163269 | -0.21 | 0.0005 |
| wsnp_Ra_rep_c73725_71801179 | 6B | 64.0 | 274207889 | -0.21 | 0.0006 |
| wsnp_Ra_rep_c73725_71801237 | 6B | 64.0 | 274207831 | -0.21 | 0.0005 |
| wsnp_Ra_rep_c73731_71807419 | 6B | 64.0 | 257996714 | -0.24 | 0.0001 |
| BobWhite_c32911_243$ | 6B | 65.0 | 255268901 | -0.22 | 0.0003 |
| Kukri_c24148_254$ | 7B | 136.0 | 705270901 | -0.16 | 0.0007 |
| TA005284-0990$ | 7B | 136.0 | 704271881 | -0.16 | 0.0006 |
| wsnp_JD_c13673_13606066$ | 7B | 136.0 | 704271648 | -0.17 | 0.0004 |
| 2022-23 | |||||
| IACX3595$ | 1B | 76.0 | 539562016 | 0.17 | 0.0009 |
| BS00030652_51 | 3A | 158.0 | 693002205 | -0.25 | 0.0008 |
| Excalibur_c8284_580$ | 3B | 57.0 | 736672449 | -0.20 | 0.0009 |
| IAAV6566 | 3B | 84.0 | 736712583 | -0.17 | 0.0003 |
| Tdurum_contig8365_433 | 3B | 84.0 | 736672399 | -0.17 | 0.0005 |
| Kukri_c2526_1375$ | 4B | 66.0 | 523447728 | -0.28 | 0.0004 |
| Excalibur_c25471_225 | 7A | 64.0 | 34539197 | 0.17 | 0.0002 |
| BS00094965_51$ | 7A | 150.0 | 669729091 | -0.30 | 0.0002 |
| Kukri_c30836_582 | 7B | 73.0 | 699427808 | 0.16 | 0.0002 |
2.4. Candidate Genes
3. Discussion
3.1. Investigating the Concordance among Significant MTAs and Previously Mapped QTL/Pm Resistance Genes
3.2. Candidate Genes for PM Resistance
4. Materials and Methods
4.1. Plant Material
4.2. Experimental Design and Trait Evaluation
4.3. DNA Extraction and Genotyping
4.4. Descriptive Statistics Analyses
4.5. Population Structure, Kinship Matrix and Principal Components Analyses
4.6. Linkage Disequilibrium and Genome-Wide Association Analyses
4.7. Identification of Candidate Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grote, U.; Fasse, A.; Nguyen, T.T.; Erenstein, O. Food Security and the Dynamics of Wheat and Maize Value Chains in Africa and Asia. Frontiers in Sustainable Food Systems 2021, 4. [CrossRef]
- Shahbandeh, M. Total wheat consumption worldwide 2022/23. Available online: https://www.statista.com/statistics/1094056/total-global-rice-consumption/ (accessed on August 8).
- Alam, M.A., Mandal, M.S.N., Wang, C. and Wanquan, J. Chromosomal location and SSR markers of a powdery mildew resistance gene in common wheat line N0308. Afr. J. Microbiol Res. 2013, 7, 477-482.
- Mwale, V.M., Chilembwe H.C., Uluko H.C. Wheat powdery mildew (Blumeria graminis f. sp. tritici): Damage effects and genetic resistance developed in wheat (Triticum aestivum). Journal of Plant Science 2014, 5, 1-16.
- Te Beest, D.E.; Paveley, N.D.; Shaw, M.W.; van den Bosch, F. Disease-weather relationships for powdery mildew and yellow rust on winter wheat. Phytopathology 2008, 98, 609-617. [CrossRef]
- Mehta, A.B.A.K.B.D.K.B.D. Effect of weather parameters on powdery mildew development of wheat at different location in Himachal Pradesh. Indian Phytopathology 2018, 71, 349-353. [CrossRef]
- Hartl, L.; Weiss, H.; Stephan, U.; Zeller, F.J.; Jahoor, A. Molecular identification of powdery mildew resistance genes in common wheat (Triticum aestivum L.). Theoretical and Applied Genetics 1995, 90, 601-606. [CrossRef]
- Keller, M.; Keller, B.; Schachermayr, G.; Winzeler, M.; Schmid, J.E.; Stamp, P.; Messmer, M.M. Quantitative trait loci for resistance against powdery mildew in a segregating wheat×spelt population. Theoretical and Applied Genetics 1999, 98, 903-912. [CrossRef]
- Hysing, S.-C.; Merker, A.; Liljeroth, E.; Koebner, R.M.D.; Zeller, F.J.; Hsam, S.L.K. Powdery mildew resistance in 155 Nordic bread wheat cultivars and landraces. Hereditas 2007, 144, 102-119. [CrossRef]
- Hua, W.; Liu, Z.; Zhu, J.; Xie, C.; Yang, T.; Zhou, Y.; Duan, X.; Sun, Q.; Liu, Z. Identification and genetic mapping of pm42, a new recessive wheat powdery mildew resistance gene derived from wild emmer (Triticum turgidum var. dicoccoides). Theoretical and Applied Genetics 2009, 119, 223-230. [CrossRef]
- Hao, Y.; Parks, R.; Cowger, C.; Chen, Z.; Wang, Y.; Bland, D.; Murphy, J.P.; Guedira, M.; Brown-Guedira, G.; Johnson, J. Molecular characterization of a new powdery mildew resistance gene Pm54 in soft red winter wheat. Theoretical and Applied Genetics 2015, 128, 465-476. [CrossRef]
- Liu, N.; Bai, G.; Lin, M.; Xu, X.; Zheng, W. Genome-wide Association Analysis of Powdery Mildew Resistance in U.S. Winter Wheat. Scientific Reports 2017, 7, 11743. [CrossRef]
- Li, G.; Cowger, C.; Wang, X.; Carver, B.F.; Xu, X. Characterization of Pm65, a new powdery mildew resistance gene on chromosome 2AL of a facultative wheat cultivar. Theoretical and Applied Genetics 2019, 132, 2625-2632. [CrossRef]
- Li, G.; Xu, X.; Tan, C.; Carver, B.F.; Bai, G.; Wang, X.; Bonman, J.M.; Wu, Y.; Hunger, R.; Cowger, C. Identification of powdery mildew resistance loci in wheat by integrating genome-wide association study (GWAS) and linkage mapping. The Crop Journal 2019, 7, 294-306. [CrossRef]
- Jia, M.; Xu, H.; Liu, C.; Mao, R.; Li, H.; Liu, J.; Du, W.; Wang, W.; Zhang, X.; Han, R.; et al. Characterization of the Powdery Mildew Resistance Gene in the Elite Wheat Cultivar Jimai 23 and Its Application in Marker-Assisted Selection. Frontiers in Genetics 2020, 11. [CrossRef]
- Kang, Y.; Barry, K.; Cao, F.; Zhou, M. Genome-wide association mapping for adult resistance to powdery mildew in common wheat. Molecular Biology Reports 2020, 47, 1241-1256. [CrossRef]
- Leonova, I.N. Genome-Wide Association Study of Powdery Mildew Resistance in Russian Spring Wheat (T. aestivum L.) Varieties. Russian Journal of Genetics 2019, 55, 1360-1374. [CrossRef]
- McIntosh R. A., D.J., Rogers W. J., Xia X. C., Raupp W. J. Catalogue of gene symbols for wheat: 2020 supplement. In Proceedings of the Annual Wheat Newsletter, Manhattan, USA, 2020; pp. 98–113.
- McIntosh, R.A.; Hart, G.E.; Devos, K.M.; Gale, M.D.; Rogers, W.J. Catalogue of Gene Symbols for Wheat. 1998. https://wheat.pw.usda.gov/ggpages/wgc/98/.
- Simeone, R.; Piarulli, L.; Nigro, D.; Signorile, M.A.; Blanco, E.; Mangini, G.; Blanco, A. Mapping Powdery Mildew (Blumeria graminis f. sp. tritici) Resistance in Wild and Cultivated Tetraploid Wheats. Int J Mol Sci 2020, 21. [CrossRef]
- Pang, Y.; Wu, Y.; Liu, C.; Li, W.; St. Amand, P.; Bernardo, A.; Wang, D.; Dong, L.; Yuan, X.; Zhang, H.; et al. High-resolution genome-wide association study and genomic prediction for disease resistance and cold tolerance in wheat. Theoretical and Applied Genetics 2021, 134, 2857-2873. [CrossRef]
- Du, X.; Xu, W.; Peng, C.; Li, C.; Zhang, Y.; Hu, L. Identification and validation of a novel locus, Qpm-3BL, for adult plant resistance to powdery mildew in wheat using multilocus GWAS. BMC Plant Biology 2021, 21, 357. [CrossRef]
- He, H.; Liu, R.; Ma, P.; Du, H.; Zhang, H.; Wu, Q.; Yang, L.; Gong, S.; Liu, T.; Huo, N.; et al. Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theoretical and Applied Genetics 2021, 134, 53-62. [CrossRef]
- Hinterberger, V.; Douchkov, D.; Lück, S.; Kale, S.; Mascher, M.; Stein, N.; Reif, J.C.; Schulthess, A.W. Mining for New Sources of Resistance to Powdery Mildew in Genetic Resources of Winter Wheat. Frontiers in Plant Science 2022, 13. [CrossRef]
- Jin, P.; Guo, X.; Guo, M.; Li, R.; Li, Q.; Cheng, P.; Wang, B. Genome-Wide Association Mapping of Resistance to Powdery Mildew in Regional Trials of Wheat Mainly from China. Plant Dis 2022, 106, 2701-2710. [CrossRef]
- Spielmeyer, W.; McIntosh, R.A.; Kolmer, J.; Lagudah, E.S. Powdery mildew resistance and Lr34/Yr18 genes for durable resistance to leaf and stripe rust cosegregate at a locus on the short arm of chromosome 7D of wheat. Theor Appl Genet 2005, 111, 731-735. [CrossRef]
- Lillemo, M.; Asalf, B.; Singh, R.P.; Huerta-Espino, J.; Chen, X.M.; He, Z.H.; Bjørnstad, Å. The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar. Theoretical and Applied Genetics 2008, 116, 1155-1166. [CrossRef]
- Herrera-Foessel, S.A.; Singh, R.P.; Lillemo, M.; Huerta-Espino, J.; Bhavani, S.; Singh, S.; Lan, C.; Calvo-Salazar, V.; Lagudah, E.S. Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor Appl Genet 2014, 127, 781-789. [CrossRef]
- Marone, D.; Russo, M.A.; Laidò, G.; De Vita, P.; Papa, R.; Blanco, A.; Gadaleta, A.; Rubiales, D.; Mastrangelo, A.M. Genetic basis of qualitative and quantitative resistance to powdery mildew in wheat: from consensus regions to candidate genes. BMC Genomics 2013, 14, 562. [CrossRef]
- Lee, J.H.; Graybosch, R.A.; Peterson, C.J. Quality and biochemical effects of a IBL/IRS wheat-rye translocation in wheat. Theoretical and Applied Genetics 1995, 90, 105-112. [CrossRef]
- Zeng, F.-s.; Yang, L.-j.; Gong, S.-j.; Shi, W.-q.; Zhang, X.-j.; Wang, H.; Xiang, L.-b.; Xue, M.-F.; Yu, D.-z. Virulence and Diversity of Blumeria graminis f. sp. tritici Populations in China. Journal of Integrative Agriculture 2014, 13, 2424-2437. [CrossRef]
- Bapela, T.; Shimelis, H.; Terefe, T.; Bourras, S.; Sánchez-Martín, J.; Douchkov, D.; Desiderio, F.; Tsilo, T.J. Breeding Wheat for Powdery Mildew Resistance: Genetic Resources and Methodologies—A Review. Agronomy 2023, 13. [CrossRef]
- Dhariwal, R.; Randhawa, H.S. Mapping Quantitative Trait Loci in Wheat: Historic Perspective, Tools, and Methods for Analysis. In Accelerated Breeding of Cereal Crops, Bilichak, A., Laurie, J.D., Eds.; Springer Protocols Handbooks; Springer US: New York, NY, 2022; pp. 31-75.
- Bartoli, C.; Roux, F. Genome-Wide Association Studies In Plant Pathosystems: Toward an Ecological Genomics Approach. Frontiers in Plant Science 2017, 8. [CrossRef]
- Vagndorf, N.; Nielsen, N.H.; Edriss, V.; Andersen, J.R.; Orabi, J.; Jørgensen, L.N.; Jahoor, A. Genomewide association study reveals novel quantitative trait loci associated with resistance towards Septoria tritici blotch in North European winter wheat. Plant Breeding 2017, 136, 474-482. [CrossRef]
- Juliana, P.; Singh, R.P.; Poland, J.; Mondal, S.; Crossa, J.; Montesinos-López, O.A.; Dreisigacker, S.; Pérez-Rodríguez, P.; Huerta-Espino, J.; Crespo-Herrera, L.; et al. Prospects and Challenges of Applied Genomic Selection—A New Paradigm in Breeding for Grain Yield in Bread Wheat. The Plant Genome 2018, 11, 180017. [CrossRef]
- Muqaddasi, Q.H.; Zhao, Y.; Rodemann, B.; Plieske, J.; Ganal, M.W.; Röder, M.S. Genome-wide Association Mapping and Prediction of Adult Stage Septoria tritici Blotch Infection in European Winter Wheat via High-Density Marker Arrays. The Plant Genome 2019, 12, 180029. [CrossRef]
- Stadlmeier, M.; Jørgensen, L.N.; Corsi, B.; Cockram, J.; Hartl, L.; Mohler, V. Genetic Dissection of Resistance to the Three Fungal Plant Pathogens Blumeria graminis, Zymoseptoria tritici, and Pyrenophora tritici-repentis Using a Multiparental Winter Wheat Population. G3 Genes|Genomes|Genetics 2019, 9, 1745-1757. [CrossRef]
- Huang, B.E.; Verbyla, K.L.; Verbyla, A.P.; Raghavan, C.; Singh, V.K.; Gaur, P.; Leung, H.; Varshney, R.K.; Cavanagh, C.R. MAGIC populations in crops: current status and future prospects. Theor Appl Genet 2015, 128, 999-1017. [CrossRef]
- Buckler, E.S.t.; Thornsberry, J.M. Plant molecular diversity and applications to genomics. Curr Opin Plant Biol 2002, 5, 107-111. [CrossRef]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and Prospects of Association Mapping in Plants. 2008, 1, 5-20. [CrossRef]
- Nelson, R.; Wiesner-Hanks, T.; Wisser, R.; Balint-Kurti, P. Navigating complexity to breed disease-resistant crops. Nat Rev Genet 2018, 19, 21-33. [CrossRef]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnology Journal 2014, 12, 787-796. [CrossRef]
- Bolser, D.; Staines, D.M.; Pritchard, E.; Kersey, P. Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomics Data. Methods in molecular biology (Clifton, N.J.) 2016, 1374, 115-140. [CrossRef]
- Hill, W.G.; Weir, B.S. Variances and covariances of squared linkage disequilibria in finite populations. Theor Popul Biol 1988, 33, 54-78. [CrossRef]
- Alemu, A.; Brazauskas, G.; Gaikpa, D.S.; Henriksson, T.; Islamov, B.; Jørgensen, L.N.; Koppel, M.; Koppel, R.; Liatukas, Ž.; Svensson, J.T.; et al. Genome-Wide Association Analysis and Genomic Prediction for Adult-Plant Resistance to Septoria Tritici Blotch and Powdery Mildew in Winter Wheat. Frontiers in Genetics 2021, 12. [CrossRef]
- Lopes, M.S.; Dreisigacker, S.; Peña, R.J.; Sukumaran, S.; Reynolds, M.P. Genetic characterization of the wheat association mapping initiative (WAMI) panel for dissection of complex traits in spring wheat. Theoretical and Applied Genetics 2015, 128, 453-464. [CrossRef]
- Zeller, F.J.; Lutz, J.; Stephan, U. Chromosome location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L.) 1. Mlk and other alleles at the Pm3 locus. Euphytica 1993, 68, 223-229. [CrossRef]
- Li, G.; Carver, B.F.; Cowger, C.; Bai, G.; Xu, X. Pm223899, a new recessive powdery mildew resistance gene identified in Afghanistan landrace PI 223899. Theor Appl Genet 2018, 131, 2775-2783. [CrossRef]
- Chen, Y.; Hunger, R.M.; Carver, B.F.; Zhang, H.; Yan, L. Genetic characterization of powdery mildew resistance in U.S. hard winter wheat. Mol Breeding 2009, 24, 141-152. [CrossRef]
- Liang, S.S.; Suenaga, K.; He, Z.H.; Wang, Z.L.; Liu, H.Y.; Wang, D.S.; Singh, R.P.; Sourdille, P.; Xia, X.C. Quantitative trait Loci mapping for adult-plant resistance to powdery mildew in bread wheat. Phytopathology 2006, 96, 784-789. [CrossRef]
- Huang, Q.; Ruilian, J.; Xinyuan, W. QTL mapping for adult-plant resistance to powdery mildew in common wheat. Scientia Agricultura Sinica 2008.
- Ma, P.; Xu, H.; Han, G.; Luo, Q.; Xu, Y.; Zhang, X.; An, D.; Li, L.; Sun, Y. Characterization of a Segregation Distortion Locus with Powdery Mildew Resistance in a Wheat-Thinopyrum intermedium Introgression Line WE99. Plant Dis 2016, 100, 1541-1547. [CrossRef]
- Bougot, Y.; Lemoine, J.; Pavoine, M.T.; Guyomar'ch, H.; Gautier, V.; Muranty, H.; Barloy, D. A major QTL effect controlling resistance to powdery mildew in winter wheat at the adult plant stage. Plant Breeding 2006, 125, 550-556. [CrossRef]
- Jia, A.; Ren, Y.; Gao, F.; Yin, G.; Liu, J.; Guo, L.; Zheng, J.; He, Z.; Xia, X. Mapping and validation of a new QTL for adult-plant resistance to powdery mildew in Chinese elite bread wheat line Zhou8425B. Theoretical and Applied Genetics 2018, 131, 1063-1071. [CrossRef]
- Xu, H.; Yao, G.; Xiong, L.; Yang, L.; Jiang, Y.; Fu, B.; Zhao, W.; Zhang, Z.; Zhang, C.; Ma, Z. Identification and mapping of pm2026: a recessive powdery mildew resistance gene in an einkorn (Triticum monococcum L.) accession. Theor Appl Genet 2008, 117, 471-477. [CrossRef]
- Chen, X.M.; Luo, Y.H.; Xia, X.C.; Xia, L.Q.; Chen, X.; Ren, Z.L.; He, Z.H.; Jia, J.Z. Chromosomal location of powdery mildew resistance gene Pm16 in wheat using SSR marker analysis. Plant Breeding 2005, 124, 225-228. [CrossRef]
- Lan, C.; Liang, S.; Wang, Z.; Yan, J.; Zhang, Y.; Xia, X.; He, Z. Quantitative Trait Loci Mapping for Adult-Plant Resistance to Powdery Mildew in Chinese Wheat Cultivar Bainong 64. Phytopathology® 2009, 99, 1121-1126. [CrossRef]
- Ma, Q.; Luo, P.; Ren, Z.; Jiang, H.; Yang, Z. Genetic analysis and chromosomal location of two new genes for resistance to powdery mildew in wheat (Triticum aestivum L.). Acta Agronomica Sinica 2010, 33, 1-8.
- Xiao, M.; Song, F.; Jiao, J.; Wang, X.; Xu, H.; Li, H. Identification of the gene Pm47 on chromosome 7BS conferring resistance to powdery mildew in the Chinese wheat landrace Hongyanglazi. Theor Appl Genet 2013, 126, 1397-1403. [CrossRef]
- Zhong, S.; Ma, L.; Fatima, S.A.; Yang, J.; Chen, W.; Liu, T.; Hu, Y.; Li, Q.; Guo, J.; Zhang, M.; et al. Collinearity Analysis and High-Density Genetic Mapping of the Wheat Powdery Mildew Resistance Gene Pm40 in PI 672538. PLOS ONE 2016, 11, e0164815. [CrossRef]
- Dellaporta, S.L.W., J.; Hicks, J.B. Isolation of DNA from higher plants. Plant Mol Biol Rep 1983, 4, 19–21.
- van Ooijen, G.; Mayr, G.; Kasiem, M.M.; Albrecht, M.; Cornelissen, B.J.; Takken, F.L. Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot 2008, 59, 1383-1397. [CrossRef]
- Faris, J.D.; Zhang, Z.; Lu, H.; Lu, S.; Reddy, L.; Cloutier, S.; Fellers, J.P.; Meinhardt, S.W.; Rasmussen, J.B.; Xu, S.S.; et al. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proceedings of the National Academy of Sciences 2010, 107, 13544-13549. [CrossRef]
- Krishnan, P.; Ma, X.; McDonald, B.A.; Brunner, P.C. Widespread signatures of selection for secreted peptidases in a fungal plant pathogen. BMC Evolutionary Biology 2018, 18, 7. [CrossRef]
- Saintenac, C.; Lee, W.-S.; Cambon, F.; Rudd, J.J.; King, R.C.; Marande, W.; Powers, S.J.; Bergès, H.; Phillips, A.L.; Uauy, C.; et al. Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nature Genetics 2018, 50, 368-374. [CrossRef]
- Gupta, P.K.; Chand, R.; Vasistha, N.K.; Pandey, S.P.; Kumar, U.; Mishra, V.K.; Joshi, A.K. Spot blotch disease of wheat: the current status of research on genetics and breeding. Plant Pathology 2018, 67, 508-531. [CrossRef]
- Noman, A.; Aqeel, M.; Khalid, N.; Islam, W.; Sanaullah, T.; Anwar, M.; Khan, S.; Ye, W.; Lou, Y. Zinc finger protein transcription factors: Integrated line of action for plant antimicrobial activity. Microbial Pathogenesis 2019, 132, 141-149. [CrossRef]
- Dmochowska-Boguta, M.; Kloc, Y.; Zielezinski, A.; Werecki, P.; Nadolska-Orczyk, A.; Karlowski, W.M.; Orczyk, W. TaWAK6 encoding wall-associated kinase is involved in wheat resistance to leaf rust similar to adult plant resistance. PLOS ONE 2020, 15, e0227713. [CrossRef]
- Yates, S.; Mikaberidze, A.; Krattinger, S.G.; Abrouk, M.; Hund, A.; Yu, K.; Studer, B.; Fouche, S.; Meile, L.; Pereira, D.; et al. Precision Phenotyping Reveals Novel Loci for Quantitative Resistance to Septoria Tritici Blotch. Plant Phenomics 2019. [CrossRef]
- Mahmoudi, Z.; Taliei, F.; Ahangar, L.; Kheyrgoo, M. Assessment of salicylic acid-induced resistance against Septoria tritici blotch disease on wheat using real-time PCR. mdrsjrns 2021, 10, 151-165.
- Lin, P.-C.; Pomeranz, M.C.; Jikumaru, Y.; Kang, S.G.; Hah, C.; Fujioka, S.; Kamiya, Y.; Jang, J.-C. The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. The Plant Journal 2011, 65, 253-268. [CrossRef]
- Peng, F.Y.; Yang, R.C. Prediction and analysis of three gene families related to leaf rust (Puccinia triticina) resistance in wheat (Triticum aestivum L.). BMC Plant Biol 2017, 17, 108. [CrossRef]
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta-Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 323, 1360-1363. [CrossRef]
- Krattinger, S.G.; Lagudah, E.S.; Wicker, T.; Risk, J.M.; Ashton, A.R.; Selter, L.L.; Matsumoto, T.; Keller, B. Lr34 multi-pathogen resistance ABC transporter: molecular analysis of homoeologous and orthologous genes in hexaploid wheat and other grass species. The Plant journal : for cell and molecular biology 2011, 65, 392-403. [CrossRef]
- Kolodziej, M.C.; Singla, J.; Sánchez-Martín, J.; Zbinden, H.; Šimková, H.; Karafiátová, M.; Doležel, J.; Gronnier, J.; Poretti, M.; Glauser, G.; et al. A membrane-bound ankyrin repeat protein confers race-specific leaf rust disease resistance in wheat. Nature Communications 2021, 12, 956. [CrossRef]
- Wang, H.; Zou, S.; Li, Y.; Lin, F.; Tang, D. An ankyrin-repeat and WRKY-domain-containing immune receptor confers stripe rust resistance in wheat. Nature Communications 2020, 11, 1353. [CrossRef]
- Yang, Y.; Yu, Y.; Bi, C.; Kang, Z. Quantitative Proteomics Reveals the Defense Response of Wheat against Puccinia striiformis f. sp. tritici. Scientific Reports 2016, 6, 34261. [CrossRef]
- Kim, H.S.; Delaney, T.P. Arabidopsis SON1 Is an F-Box Protein That Regulates a Novel Induced Defense Response Independent of Both Salicylic Acid and Systemic Acquired Resistance. The Plant Cell 2002, 14, 1469-1482. [CrossRef]
- Sharma, M.; Kaur, S.; Saluja, M.; Chhuneja, P. Mapping and characterization of powdery mildew resistance gene in synthetic wheat. Czech Journal of Genetics and Plant Breeding 2016, 52, 120-123.
- Panse, V.G.S., P.V. . Statistical methods for Agricultural workers. 1985.
- Gautam, A.K. Studies on some powdery mildew of Himachal Pradesh, India. Australasian Mycologist 2015, 32, 10-13.
- Bennett, F.G.A.; Westcott, B. Field assessment of resistance to powdery mildew in mature wheat plants. Plant Pathology 1982, 31, 261-268. [CrossRef]
- Sukumaran, S.; Crossa, J.; Jarquín, D.; Lopes, M.; Reynolds, M.P. Genomic and pedigree prediction with genotype × environment interaction in spring wheat grown in South and Western Asia, North Africa, and Mexico. 2016, hdl:11529/10714. [CrossRef]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences 1984, 81, 8014-8018. [CrossRef]
- Sukumaran, S.; Lopes, M.; Dreisigacker, S.; Reynolds, M. Genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theoretical and Applied Genetics 2018, 131, 985-998. [CrossRef]
- Ahirwar, R.N.; Mishra, V.K.; Chand, R.; Budhlakoti, N.; Mishra, D.C.; Kumar, S.; Singh, S.; Joshi, A.K. Genome-wide association mapping of spot blotch resistance in wheat association mapping initiative (WAMI) panel of spring wheat (Triticum aestivum L.). PLOS ONE 2018, 13, e0208196. [CrossRef]
- R Core Team R: A language and environment for statistical computing., R Foundation for Statistical Computing: Vienna, Austria., 2013.
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. Journal of Statistical Software 2015, 67, 1-48.
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics 2006, 38, 904-909. [CrossRef]
- Schwarz, G. Estimating the Dimension of a Model. The Annals of Statistics 1978, 6, 461-464. [CrossRef]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J Dairy Sci 2008, 91, 4414-4423. [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated Tool for Genome-wide Association Study. Genomics, Proteomics & Bioinformatics 2021, 19, 619-628. [CrossRef]
- Marroni, F.; Pinosio, S.; Zaina, G.; Fogolari, F.; Felice, N.; Cattonaro, F.; Morgante, M. Nucleotide diversity and linkage disequilibrium in Populus nigra cinnamyl alcohol dehydrogenase (CAD4) gene. Tree Genetics & Genomes 2011, 7, 1011-1023. [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics 2006, 38, 203-208. [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLOS Genetics 2016, 12, e1005767. [CrossRef]
- Maccaferri, M.; El-Feki, W.; Nazemi, G.; Salvi, S.; Canè, M.A.; Colalongo, M.C.; Stefanelli, S.; Tuberosa, R. Prioritizing quantitative trait loci for root system architecture in tetraploid wheat. J Exp Bot 2016, 67, 1161-1178. [CrossRef]
- Singh, S.; Gaurav, S.S.; Vasistha, N.K.; Kumar, U.; Joshi, A.K.; Mishra, V.K.; Chand, R.; Gupta, P.K. Genetics of spot blotch resistance in bread wheat (Triticum aestivum L.) using five models for GWAS. Frontiers in Plant Science 2023, 13. [CrossRef]
| Source | 2020-21 | 2021-22 | 2022-23 |
|---|---|---|---|
| Min | 1 | 2.5 | 1.5 |
| Max | 8.5 | 8.3 | 9 |
| Mean | 5.3 | 5.1 | 5.3 |
| LSD | 1.2 | 1.4 | 1.3 |
| CV | 8.4 | 12.2 | 9.0 |
| Heritability | 0.89 | 0.65 | 0.81 |
| Source of variation | Df | MSS |
|---|---|---|
| Genotypes | 285 | 4.9 ** |
| Environments (years) | 2 | 52.7 ** |
| Replications | 1 | 8.5 |
| Genotype × Environment | 570 | 1.22 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





