Submitted:
27 September 2023
Posted:
28 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Isolation and selection of antagonistic rhizospheric bacterial strains
2.2. Molecular identification of best-performing bacterial strains
2.3. Screening of potential agonist/s of tomato receptor-like kinases SlLYK12
2.3.1. Preparation of protein structure and quality analysis
2.3.2. Model refinement and validation
2.3.3. Virtual screening by molecular docking
2.4. In-vivo effect of the synthetic elicitor and consortium of Bacillus strains on Fusarium wilt disease development and growth of tomato plants
2.5. Analysis of the biochemical basis of induced defense responses in tomato plants against Fusarium wilt disease
2.5.1. Analysis of total phenolics and plant defense-related enzymes
2.5.2. Analysis of the metabolomic profile of tomato plants
2.6. Statistical analysis
3. Results
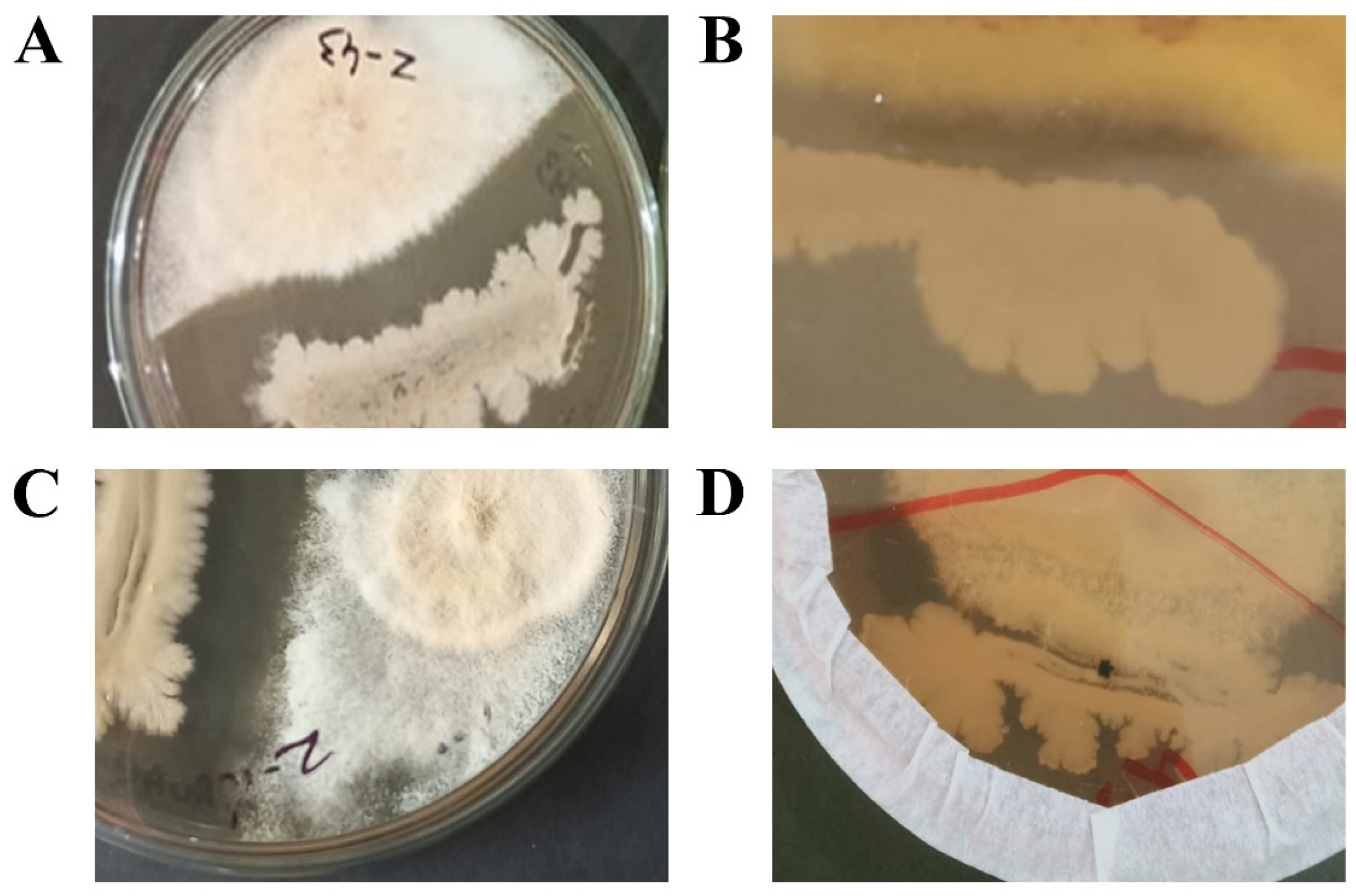
3.1. Selection of antagonistic rhizospheric bacterial strains
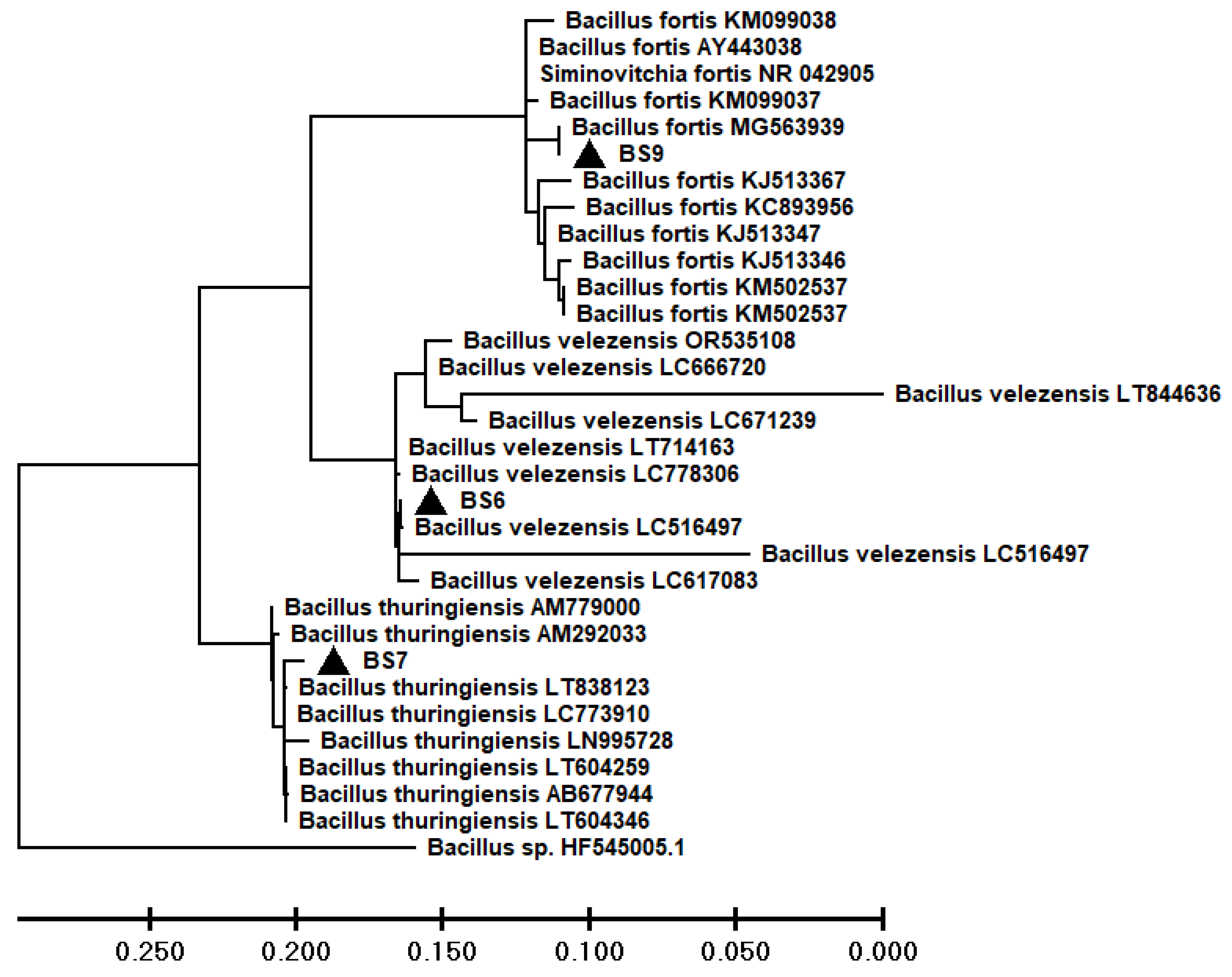
3.2. Molecular identification of bacterial strains
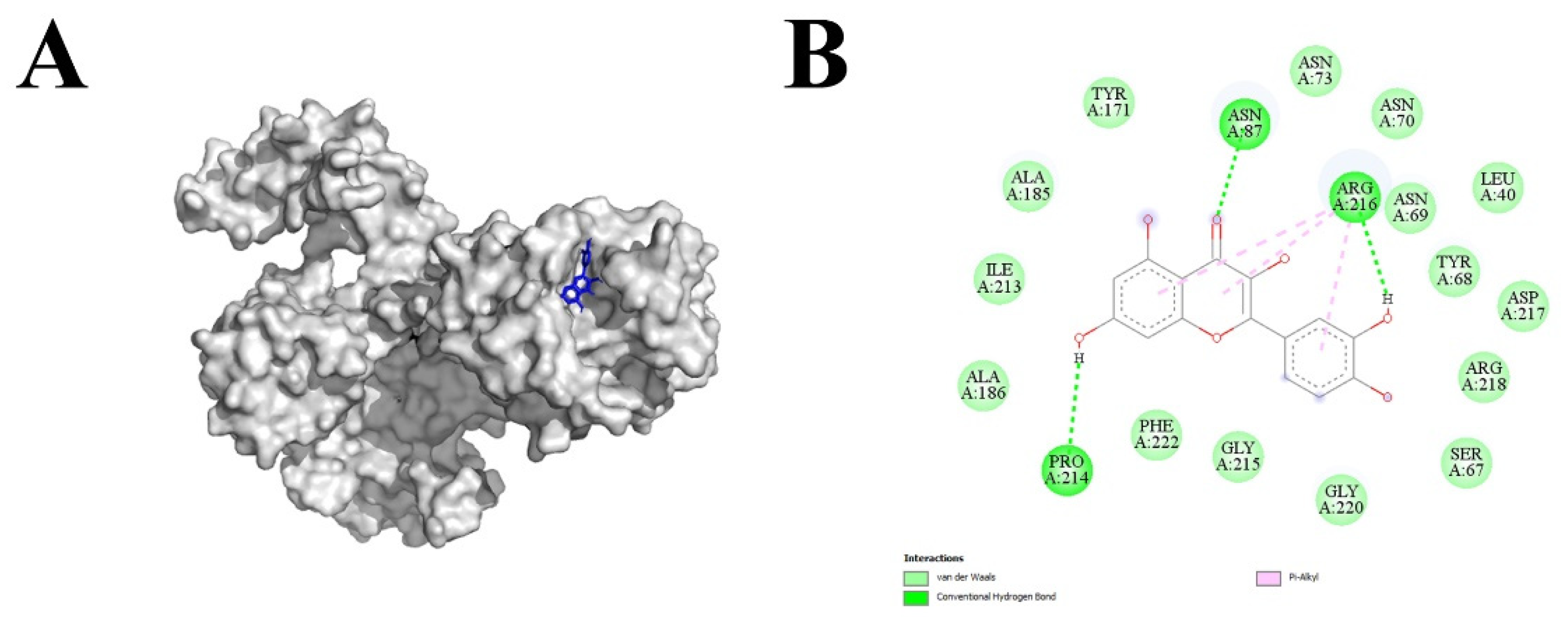
3.3. Screening of potential agonist/s of tomato receptor-like kinases SlLYK12
3.4. In-vivo effect of the synthetic elicitor and consortium of Bacillus strains on Fusarium wilt disease development and growth of tomato plants
3.5. Analysis of the biochemical basis of induced defense responses in tomato plants against Fusarium wilt disease
3.5.1. Analysis of total phenolics and defense-related enzymes
3.5.2. Non-targeted metabolomic analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hussain, I.; Alam, S.S.; Khan, I.; Shah, B.; Naeem, A.; Khan, N.; Ullah, W.; Iqbal, B.; Adnan, M.; Junaid, K. Study on the biological control of fusarium wilt of tomato. J Entomol Zool Studies 2016, 4, 525–528. [Google Scholar]
- Pathak, T.B.; Stoddard, C.S. Climate change effects on the processing tomato growing season in California using growing degree day model. Modeling Earth Systems and Environment 2018, 4, 765–775. [Google Scholar] [CrossRef]
- Venbrux, M.; Crauwels, S.; Rediers, H. Current and emerging trends in techniques for plant pathogen detection. Frontiers in Plant Science 2023, 14, 1120968. [Google Scholar] [CrossRef]
- Brahimi, M.; Boukhalfa, K.; Moussaoui, A. Deep learning for tomato diseases: classification and symptoms visualization. Applied Artificial Intelligence 2017, 31, 299–315. [Google Scholar] [CrossRef]
- Chacón, C.; Bojórquez-Quintal, E.; Caamal-Chan, G.; Ruíz-Valdiviezo, V.M.; Montes-Molina, J.A.; Garrido-Ramírez, E.R.; Rojas-Abarca, L.M.; Ruiz-Lau, N. In vitro antifungal activity and chemical composition of Piper auritum Kunth essential oil against Fusarium oxysporum and Fusarium equiseti. Agronomy 2021, 11, 1098. [Google Scholar] [CrossRef]
- Asif, M.; Haider, M.S.; Akhter, A. Impact of Biochar on Fusarium Wilt of Cotton and the Dynamics of Soil Microbial Community. Sustainability 2023, 15, 12936. [Google Scholar] [CrossRef]
- Chitwood-Brown, J.; Vallad, G.E.; Lee, T.G.; Hutton, S.F. Breeding for resistance to Fusarium wilt of tomato: A review. Genes 2021, 12, 1673. [Google Scholar] [CrossRef] [PubMed]
- Ju, W.; Liu, L.; Jin, X.; Duan, C.; Cui, Y.; Wang, J.; Ma, D.; Zhao, W.; Wang, Y.; Fang, L. Co-inoculation effect of plant-growth-promoting rhizobacteria and rhizobium on EDDS assisted phytoremediation of Cu contaminated soils. Chemosphere 2020, 254, 126724. [Google Scholar] [CrossRef] [PubMed]
- Heimpel, G.E.; Mills, N.J. Biological control; Cambridge University Press: 2017.
- Anckaert, A.; Arguelles Arias, A.; Hoff, G.; Calonne-Salmon, M.; Declerck, S.; Ongena, M. The use of Bacillus spp. as bacterial biocontrol agents to control plant diseases. 2021. [Google Scholar]
- Wang, X.; Zhao, D.; Shen, L.; Jing, C.; Zhang, C. Application and mechanisms of Bacillus subtilis in biological control of plant disease. Role of rhizospheric microbes in soil: Volume 1: Stress management and agricultural sustainability 2018, 225–250. [Google Scholar] [CrossRef]
- Cawoy, H.; Bettiol, W.; Fickers, P.; Ongena, M. Bacillus-based biological control of plant diseases. Pesticides in the modern world-pesticides use and management 2011, 273–302. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Y.; Liu, Y.; Zhang, R. Research progress in biocontrol of Bacillus spp. against plant diseases. Jiangsu Journal of Agricultural Sciences 2012, 28, 999–1006. [Google Scholar]
- Shafi, J.; Tian, H.; Ji, M. Bacillus species as versatile weapons for plant pathogens: a review. Biotechnology & Biotechnological Equipment 2017, 31, 446–459. [Google Scholar] [CrossRef]
- Patel, S.; Saraf, M. Interaction of root colonizing biocontrol agents demonstrates the antagonistic effect against Fusarium oxysporum f. sp. lycopersici on tomato. European Journal of plant pathology 2017, 149, 425–433. [Google Scholar] [CrossRef]
- Smith, D.L.; Praslickova, D.; Ilangumaran, G. Inter-organismal signaling and management of the phytomicrobiome. Frontiers in plant science 2015, 6, 722. [Google Scholar] [CrossRef]
- Bukhat, S.; Imran, A.; Javaid, S.; Shahid, M.; Majeed, A.; Naqqash, T. Communication of plants with microbial world: Exploring the regulatory networks for PGPR mediated defense signaling. Microbiological research 2020, 238, 126486. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; He, S.Y. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Ibanez, S.; Ntoukakis, V.; Rathjen, J.P. The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant signaling & behavior 2009, 4, 539–541. [Google Scholar] [CrossRef]
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proceedings of the National Academy of Sciences 2007, 104, 19613–19618. [Google Scholar] [CrossRef] [PubMed]
- Shirron, N.; Yaron, S. Active suppression of early immune response in tobacco by the human pathogen Salmonella Typhimurium. PLoS One 2011, 6, e18855. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Ibanez, S.; Hann, D.R.; Ntoukakis, V.; Petutschnig, E.; Lipka, V.; Rathjen, J.P. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Current biology 2009, 19, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. elife 2014, 3, e03766. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Plant pattern-recognition receptors. Trends in immunology 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Sun, X.; Wang, N.; Song, F.; Liang, Y. Tomato LysM receptor-like kinase SlLYK12 is involved in arbuscular mycorrhizal symbiosis. Frontiers in Plant Science 2018, 9, 1004. [Google Scholar] [CrossRef]
- Sulistiyani, T.R.; Kusmiati, M.; Putri, G.A. The 16S rRNA analysis and enzyme screening of Bacillus from rhizosphere soil of Lombok Island. Jurnal Ilmu Pertanian Indonesia 2021, 26, 582–590. [Google Scholar] [CrossRef]
- Anith, K.; Radhakrishnan, N.; Manomohandas, T. Screening of antagonistic bacteria for biological control of nursery wilt of black pepper (Piper nigrum). Microbiological Research 2003, 158, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Naik, S.; Palys, S.; Di Falco, M.; Tsang, A.; Périnet, P.; Ramanan, U.S.; Dayanandan, S. Isolation and Characterization of Bacillus velezensis EB14, an Endophytic Bacterial Strain Antagonistic to Poplar Stem Canker Pathogen Sphaerulina musiva and Its Interactions with the Endophytic Fungal Microbiome in Poplar. PhytoFrontiers™ 2021, 1, 229–238. [Google Scholar] [CrossRef]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Applied and environmental microbiology 1997, 63, 3233–3241. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular biology and evolution 2018, 35, 1547. [Google Scholar] [CrossRef]
- Karunarathna, K.H.T.; Senathilake, N.; Mewan, K.M.; Weerasena, O.; Perera, S. In silico structural homology modelling of EST073 motif coding protein of tea Camellia sinensis (L). Journal of Genetic Engineering and Biotechnology 2020, 18, 1–10. [Google Scholar] [CrossRef]
- Vriend, G. WHAT IF: a molecular modeling and drug design program. Journal of molecular graphics 1990, 8, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Vollan, H.S.; Caugant, D.A.; Eldholm, V.; Alfsnes, K.; Debech, N.; Brynildsrud, O. Naturally occurring Neisseria gonorrhoeae can have large deletions in housekeeping gene abcZ, making them untypable with multilocus sequence typing. Microbial Genomics 2022, 8. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.; Janjanam, J.; Kumar, S.; Kaushik, J.K.; Mohanty, A.K. Structural and functional characterization of buffalo oviduct-specific glycoprotein (OVGP1) expressed during estrous cycle. Bioscience Reports 2019, 39. [Google Scholar] [CrossRef]
- Mora Lagares, L.; Minovski, N.; Caballero Alfonso, A.Y.; Benfenati, E.; Wellens, S.; Culot, M.; Gosselet, F.; Novič, M. Homology modeling of the human p-glycoprotein (Abcb1) and insights into ligand binding through molecular docking studies. International journal of molecular sciences 2020, 21, 4058. [Google Scholar] [CrossRef] [PubMed]
- Forli, S.; Olson, A.J. A force field with discrete displaceable waters and desolvation entropy for hydrated ligand docking. Journal of medicinal chemistry 2012, 55, 623–638. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved protein–ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Research 2022, 50, W159–W164. [Google Scholar] [CrossRef]
- Meng, X. Y, Zhang H-X, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr. Comp. Aided Drug Design 2011, 7, 146–157. [Google Scholar] [CrossRef] [PubMed]
- PURWATI, R.D.; Hidayah, N. Inoculation methods and conidial densities of Fusarium oxysporum f. sp. cubense in Abaca. HAYATI Journal of Biosciences 2008, 15, 1–7. [Google Scholar] [CrossRef]
- Abbasi, S.; Safaie, N.; Sadeghi, A.; Shamsbakhsh, M. Streptomyces strains induce resistance to Fusarium oxysporum f. sp. lycopersici race 3 in tomato through different molecular mechanisms. Frontiers in Microbiology 2019, 10, 1505. [Google Scholar] [CrossRef] [PubMed]
- Hua, G.K.H.; Timper, P.; Ji, P. Meloidogyne incognita intensifies the severity of Fusarium wilt on watermelon caused by Fusarium oxysporum f. sp. niveum. Canadian Journal of Plant Pathology 2019, 41, 261–269. [Google Scholar] [CrossRef]
- Wei, L.; Jiangchun, H.; Shujin, W. Growth-promotion and biocontrol of cucumber fusarium wilt by marine Bacillus subtilis 3512A. Journal of Shenyang Agricultural University 2008. [Google Scholar]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant physiology 1949, 24, 1. [Google Scholar] [CrossRef]
- Sundaramoorthy, S.; Raguchander, T.; Ragupathi, N.; Samiyappan, R. Combinatorial effect of endophytic and plant growth promoting rhizobacteria against wilt disease of Capsicum annum L. caused by Fusarium solani. Biological Control 2012, 60, 59–67. [Google Scholar] [CrossRef]
- Dalisay, R.; Kuć, J. Persistence of reduced penetration by Colletotrichum lagenarium into cucumber leaves with induced systemic resistance and its relation to enhanced peroxidase and chitinase activities. Physiological and Molecular Plant Pathology 1995, 47, 329–338. [Google Scholar] [CrossRef]
- Mozzetti, C.; Ferraris, L.; Tamietti, G.; Matta, A. Variation in enzyme activities in leaves and cell suspensions as markers of incompatibility in different Phytophthora-pepper interactions. Physiological and Molecular Plant Pathology 1995, 46, 95–107. [Google Scholar] [CrossRef]
- Cahill, D.M.; McComb, J.A. A comparison of changes in phenylalanine ammonia-lyase activity, lignin and phenolic synthesis in the roots of Eucalyptus calophylla (field resistant) and E. marginata (susceptible) when infected with Phytophthora cinnamomi. Physiological and Molecular Plant Pathology 1992, 40, 315–332. [Google Scholar] [CrossRef]
- Jo, H.E.; Song, K.; Kim, J.-G.; Lee, C.H. Non-targeted metabolomic analysis for the comparative evaluation of volatile organic compounds in 20 globally representative cucumber lines. Frontiers in Plant Science 2022, 13, 1028735. [Google Scholar] [CrossRef]
- Li, X.; Wang, P.; Tong, Y.; Liu, J.; Shu, G. UHPLC-Q-Exactive orbitrap MS/MS-Based untargeted metabolomics and molecular networking reveal the differential chemical constituents of the bulbs and flowers of Fritillaria thunbergii. Molecules 2022, 27, 6944. [Google Scholar] [CrossRef]
- Onofri, A. Routine statistical analyses of field experiments by using an Excel extension. In Proceedings of the Proceedings 6th National Conference Italian Biometric Society:“La statistica nelle scienze della vita e dell’ambiente”, Pisa, 2007; pp. 93–96. [Google Scholar]
- Mari, F.; Memon, R.; Lohano, H. Measuring returns to scale for onion, tomato and chillies production in Sindh province of Pakistan. International Journal of Agriculture and Biology (Pakistan) 2007. [Google Scholar]
- Deng, X.; Zhang, N.; Shen, Z.; Zhu, C.; Li, R.; Salles, J.F.; Shen, Q. Rhizosphere bacteria assembly derived from fumigation and organic amendment triggers the direct and indirect suppression of tomato bacterial wilt disease. Applied soil ecology 2020, 147, 103364. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P.; Touré, Y.; Destain, J.; Jabrane, A.; Thonart, P. Involvement of fengycin-type lipopeptides in the multifaceted biocontrol potential of Bacillus subtilis. Applied microbiology and biotechnology 2005, 69, 29–38. [Google Scholar] [CrossRef]
- Adrees, H.; Haider, M.S.; Anjum, T.; Akram, W. Inducing systemic resistance in cotton plants against charcoal root rot pathogen using indigenous rhizospheric bacterial strains and chemical elicitors. Crop protection 2019, 115, 75–83. [Google Scholar] [CrossRef]
- Mojica-Marín, V.; Luna-Olvera, H.A.; Sandoval-Coronado, C.F.; Pereyra-Alférez, B.; Morales-Ramos, L.H.; Hernández-Luna, C.E.; Alvarado-Gomez, O.G. Antagonistic activity of selected strains of Bacillus thuringiensis against Rhizoctonia solani of chili pepper. African Journal of Biotechnology 2008, 7. [Google Scholar]
- Yan, H.; Qiu, Y.; Yang, S.; Wang, Y.; Wang, K.; Jiang, L.; Wang, H. Antagonistic activity of Bacillus velezensis SDTB038 against Phytophthora infestans in Potato. Plant Disease 2021, 105, 1738–1747. [Google Scholar] [CrossRef] [PubMed]
- Rayavarapu, V.B.; Padmavathi, T. Bacillus sp. as potential plant growth promoting rhizobacteria. Int J Adv Life Sci 2016, 9, 29–36. [Google Scholar]
- Bisen, K.; Singh, V.; Keswani, C.; Ray, S.; Sarma, B.K.; Singh, H. Use of Biocontrol Agents for the Management of Seed-Borne Diseases. Seed-Borne Diseases of Agricultural Crops: Detection, Diagnosis & Management 2020, 651–663. [Google Scholar]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annual review of phytopathology 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Gond, S.K.; Bergen, M.S.; Torres, M.S.; White Jr, J.F. Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiological research 2015, 172, 79–87. [Google Scholar] [CrossRef]
- Sun, Y.; Qiao, Z.; Muchero, W.; Chen, J.-G. Lectin receptor-like kinases: the sensor and mediator at the plant cell surface. Frontiers in plant science 2020, 11, 596301. [Google Scholar] [CrossRef] [PubMed]
- Scardino, V.; Di Filippo, J.I.; Cavasotto, C.N. How good are AlphaFold models for docking-based virtual screening? Iscience 2023, 26. [Google Scholar] [CrossRef] [PubMed]
- Oltrogge, L.M.; Boxer, S.G. Short hydrogen bonds and proton delocalization in green fluorescent protein (GFP). ACS Central Science 2015, 1, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Fu, H.; Fryar, K.L.; Landua, J.; Trevino, S.R.; Shirley, B.A.; Hendricks, M.M.; Iimura, S.; Gajiwala, K.; Scholtz, J.M.; et al. Contribution of Hydrophobic Interactions to Protein Stability. Journal of Molecular Biology 2011, 408, 514–528. [Google Scholar] [CrossRef] [PubMed]
- Hafez, E.; Omara, A.E.D.; Ahmed, A. The coupling effects of plant growth promoting rhizobacteria and salicylic acid on physiological modifications, yield traits, and productivity of wheat under water deficient conditions. Agronomy 2019, 9, 524. [Google Scholar] [CrossRef]
- Mufti, R.; Bano, A.; Munis, M.F.H.; Andleeb, T.; Quraishi, U.M.; Khan, N. Integrated Application of Salicylic Acid and PGPRs to Control Fusarium Wilt of Chickpea. Frontiers in Bioscience-Landmark 2023, 28, 20. [Google Scholar] [CrossRef]
- Chittoor, J.M.; Leach, J.E.; White, F.F. Induction of peroxidase during defense against pathogens. Pathogenesis-related proteins in plants 1999, 171–193. [Google Scholar]
- Vanitha, S.C.; Niranjana, S.R.; Umesha, S. Role of phenylalanine ammonia lyase and polyphenol oxidase in host resistance to bacterial wilt of tomato. Journal of Phytopathology 2009, 157, 552–557. [Google Scholar] [CrossRef]
- Nikoo, F.S.; Sahebani, N.; Aminian, H.; Mokhtarnejad, L.; Ghaderi, R. Induction of systemic resistance and defense-related enzymes in tomato plants using Pseudomonas fluorescens CHAO and salicylic acid against root-knot nematode Meloidogyne javanica. Journal of Plant Protection Research 2014. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Metabolomic evaluation of tissue-specific defense responses in tomato plants modulated by PGPR-priming against Phytophthora capsici infection. Plants 2021, 10, 1530. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Metabolic profiling of PGPR-treated tomato plants reveal priming-related adaptations of secondary metabolites and aromatic amino acids. Metabolites 2020, 10, 210. [Google Scholar] [CrossRef]
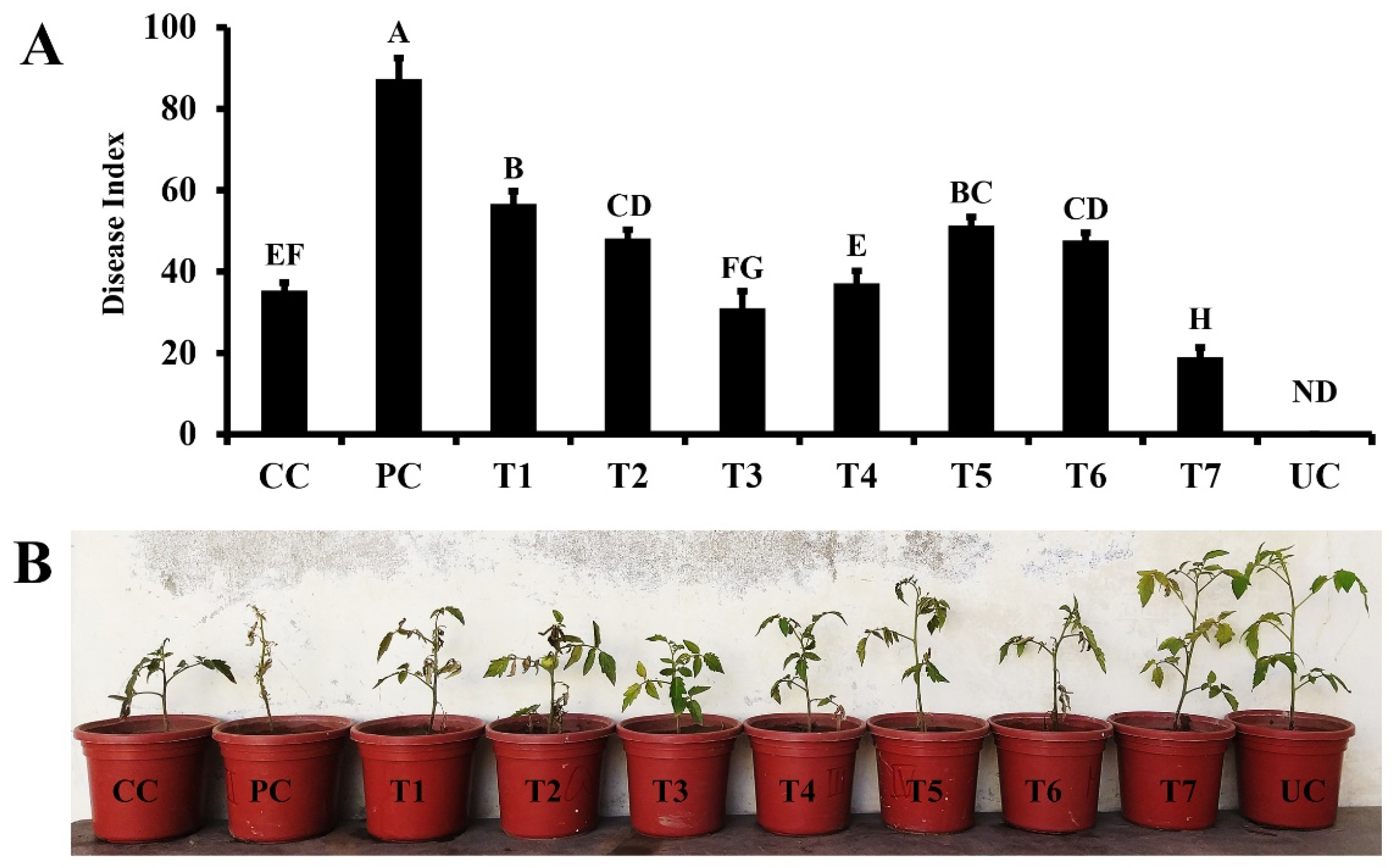
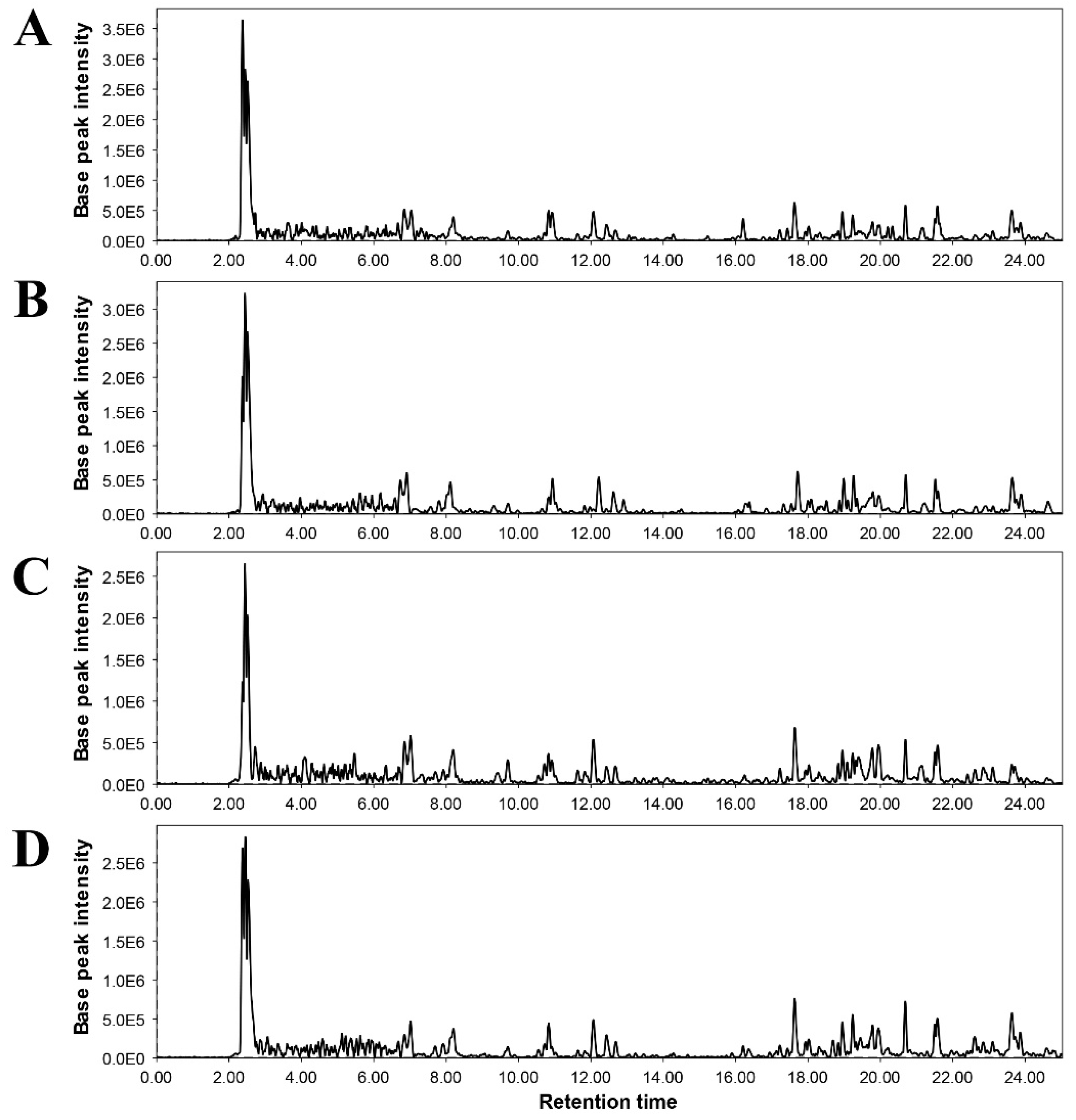
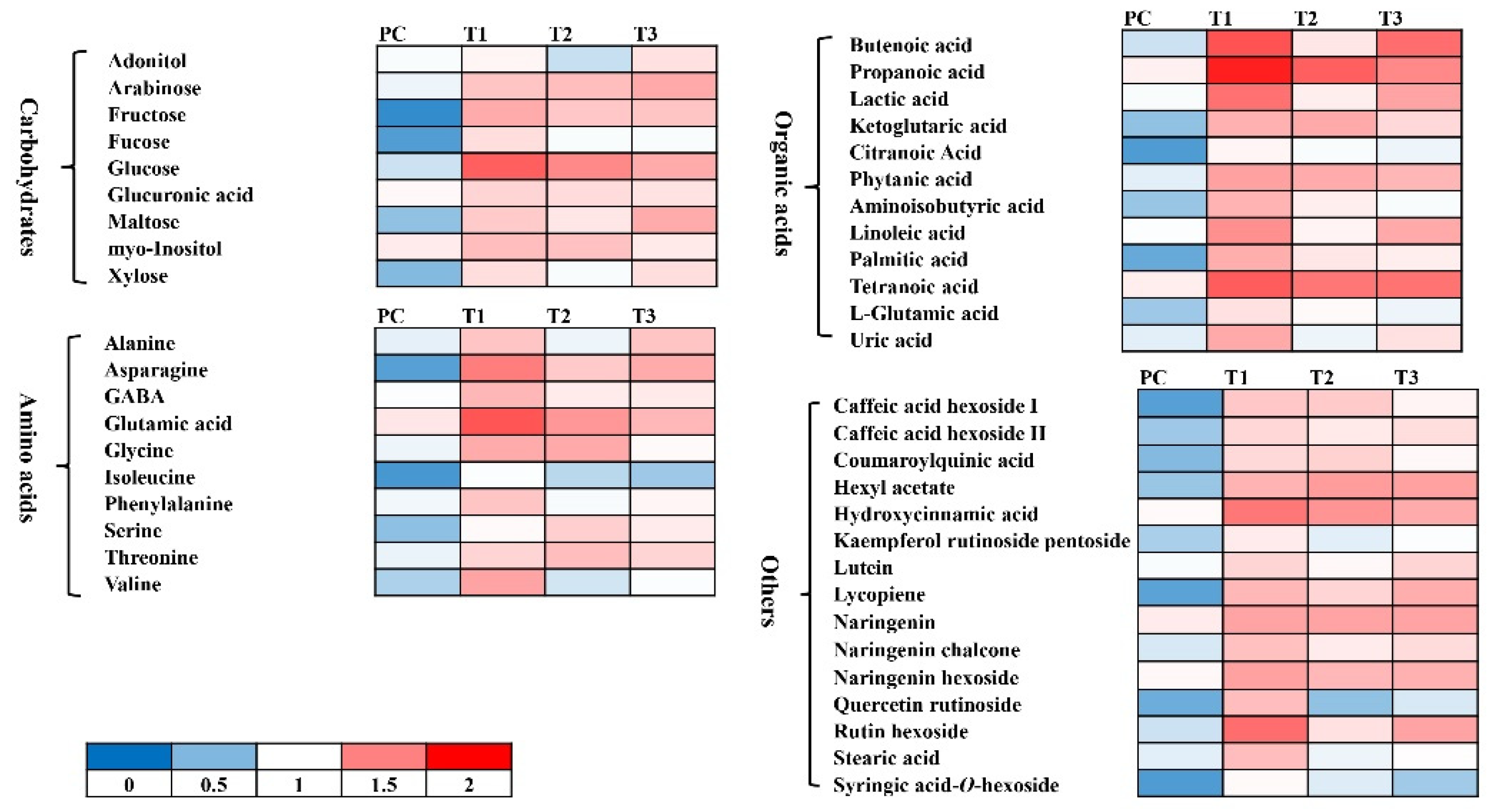
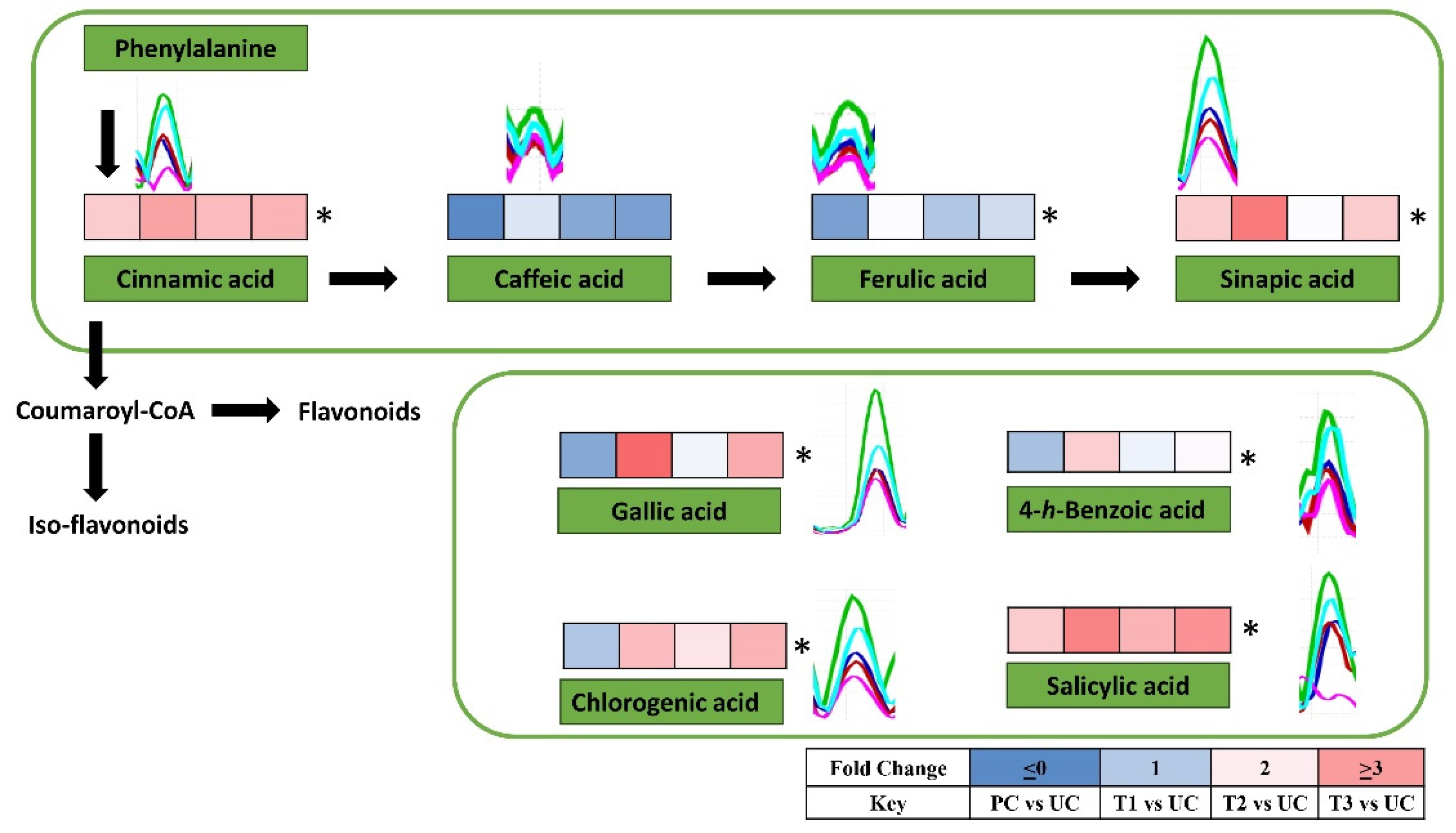
| Treatment | Description |
|---|---|
| CC | Carbendazim control |
| PC | Pathogen control (FOL) |
| T1 | Quercetin (1.0 mM) + FOL |
| T2 | Quercetin (0.1 mM) + FOL |
| T3 | Quercetin (0.01 mM) + FOL |
| T4 | Consortium + FOL |
| T5 | Quercetin (1.0 mM) + Consortium + FOL |
| T6 | Quercetin (0.1 mM) + Consortium + FOL |
| T7 | Quercetin (0.01 mM) + Consortium + FOL |
| UC | Untreated control |
| No | Code | Gram staining | Antagonistic phenotype |
|---|---|---|---|
| 1 | BS1 | Positive | NA |
| 2 | BS2 | Positive | CGI |
| 3 | BS3 | Positive | NA |
| 4 | BS4 | Positive | NA |
| 5 | BS5 | Positive | NA |
| 6 | BS6 | Positive | ZGI |
| 7 | BS7 | Positive | ZGI |
| 8 | BS8 | Positive | CGI |
| 9 | BS9 | Positive | ZGI |
| 10 | BS10 | Positive | NA |
| 11 | BS11 | Positive | NA |
| Parameter | Value |
|---|---|
| PROCHECK | |
| Errors | 5 |
| Warning | 2 |
| Pass | 2 |
| ERRAT | 97.03 |
| Ramachandran plot | |
| FR | 91.8 |
| AR | 8.1 |
| GR | 0.2 |
| DR | 0.00 |
| Verify3D | 62.4 |
| Compound | Formula | Molecular weight | Binding Affinity (kcal/mol) |
|---|---|---|---|
| Quercetin | C15H10O7 | 302.2 | -8.7 |
| Galbacin | C20H20O5 | 340.4 | -8.7 |
| Traumatin | C12H20O3 | 212.2 | -8.3 |
| 7-Oxotyphasterol | C28H48O5 | 464.7 | -8.2 |
| Treatment | Shoot length (cm) | Root length (cm) | Shoot biomass (g) | Root Biomass (g) | Total Chlorophyll (mg/g fw) |
|---|---|---|---|---|---|
| CC | 08.01±0.5e-g | 08.62±0.5ef | 1.82±0.07de | 0.31±0.02gh | 11.23±0.76e-g |
| PC | 09.28±0.8ef | 07.23±0.5e-g | 1.58±0.09ef | 0.24±0.02i | 07.17±0.04h |
| T1 | 11.12±0.9de | 09.26±0.5de | 2.06±0.08b-d | 0.43±0.03ef | 10.87±0.76fg |
| T2 | 10.38±1.4de | 10.71±0.7cd | 1.93±0.12cd | 0.51±0.04c-e | 15.91±1.27cd |
| T3 | 13.66±1.5cd | 11.96±0.7b-d | 2.24±0.13bc | 0.57±0.02cd | 16.08±1.27cd |
| T4 | 14.26±1.3b-d | 12.81±0.9bc | 2.59±0.14bc | 0.65±0.03c | 16.53±0.98c |
| T5 | 17.05±1.0bc | 12.54±1.1bc | 2.66±0.24ab | 0.59±0.04cd | 19.58±1.13a |
| T6 | 19.17±1.5ab | 15.97±1.4ab | 2.89±0.10a | 0.81±0.05ab | 18.32±1.64ab |
| T7 | 21.09±1.1a | 17.26±1.2a | 3.07±0.18a | 0.87±0.06a | 19.53±1.25a |
| Con | 23.18±1.6a | 15.62±1.1ab | 3.24±0.21a | 0.92±0.05a | 21.35±1.57a |
| Treatment | Total Phenolics (mg g-1FW) | PO (ΔOD min-1 g-1 FW) | PPO (ΔOD min-1 g-1 FW) | PAL (ΔOD min-1 g-1 FW) |
|---|---|---|---|---|
| Con | 2.03 ± 0.14e | 0.09±0.00e | 0.72±0.03c | 0.18 ± 0.01d |
| PC | 4.93 ± 0.51b-d | 0.68±0.04c | 1.12±0.25b | 0.49 ± 0.02bc |
| T1 | 7.18 ± 0.37a | 1.03±0.25a | 2.68±0.09a | 0.66 ± 0.05a |
| T2 | 5.39±0.20bc | 0.87±0.03b | 2.17±0.15a | 0.52±0.03ab |
| T3 | 6.28±0.37ab | 0.56±0.02cd | 1.32±0.07b | 0.63±0.04a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





