Submitted:
22 September 2023
Posted:
26 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
2.1. Seed Germination of Chenopodium quinoa UDEC2 and UDEC4 Accessions in Response to Salt Stress
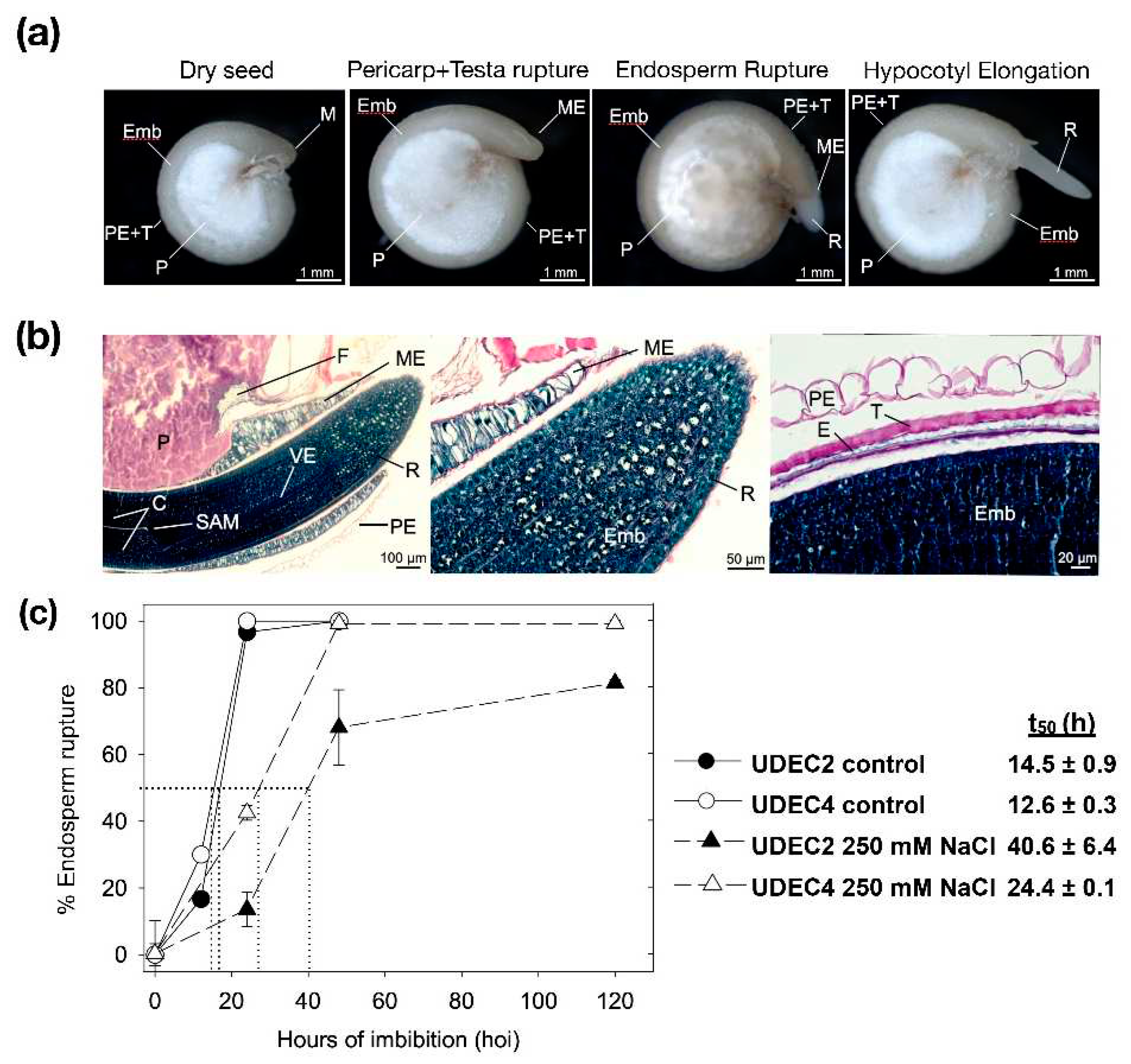
2.1.1. Anatomical and Histological Structure of quinoa Imbibed Seeds
2.1.2. Germination sensu stricto in Chenopodium Quinoa Seeds under Salt Stress
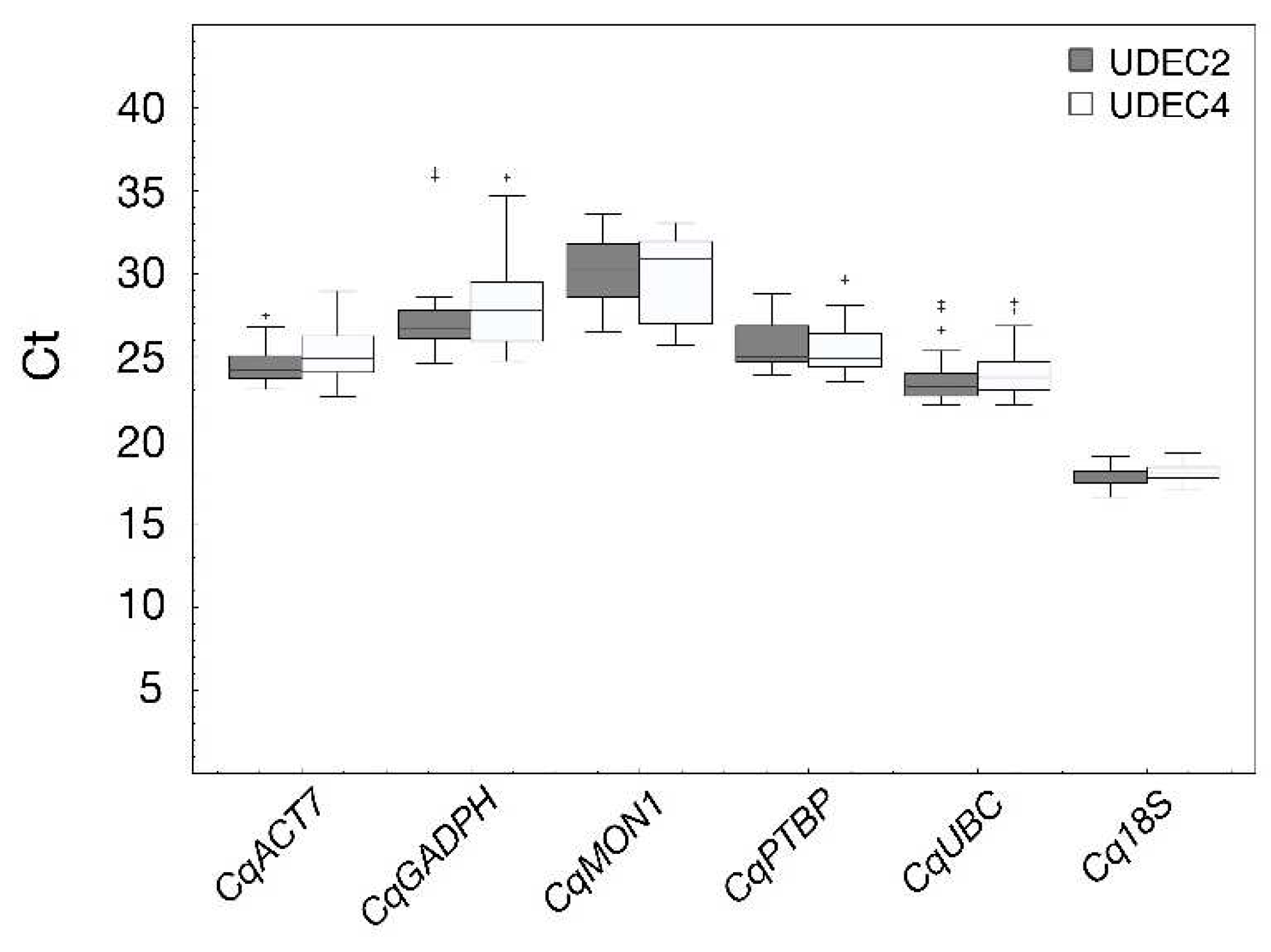
2.2. Selection of Candidate Reference Genes
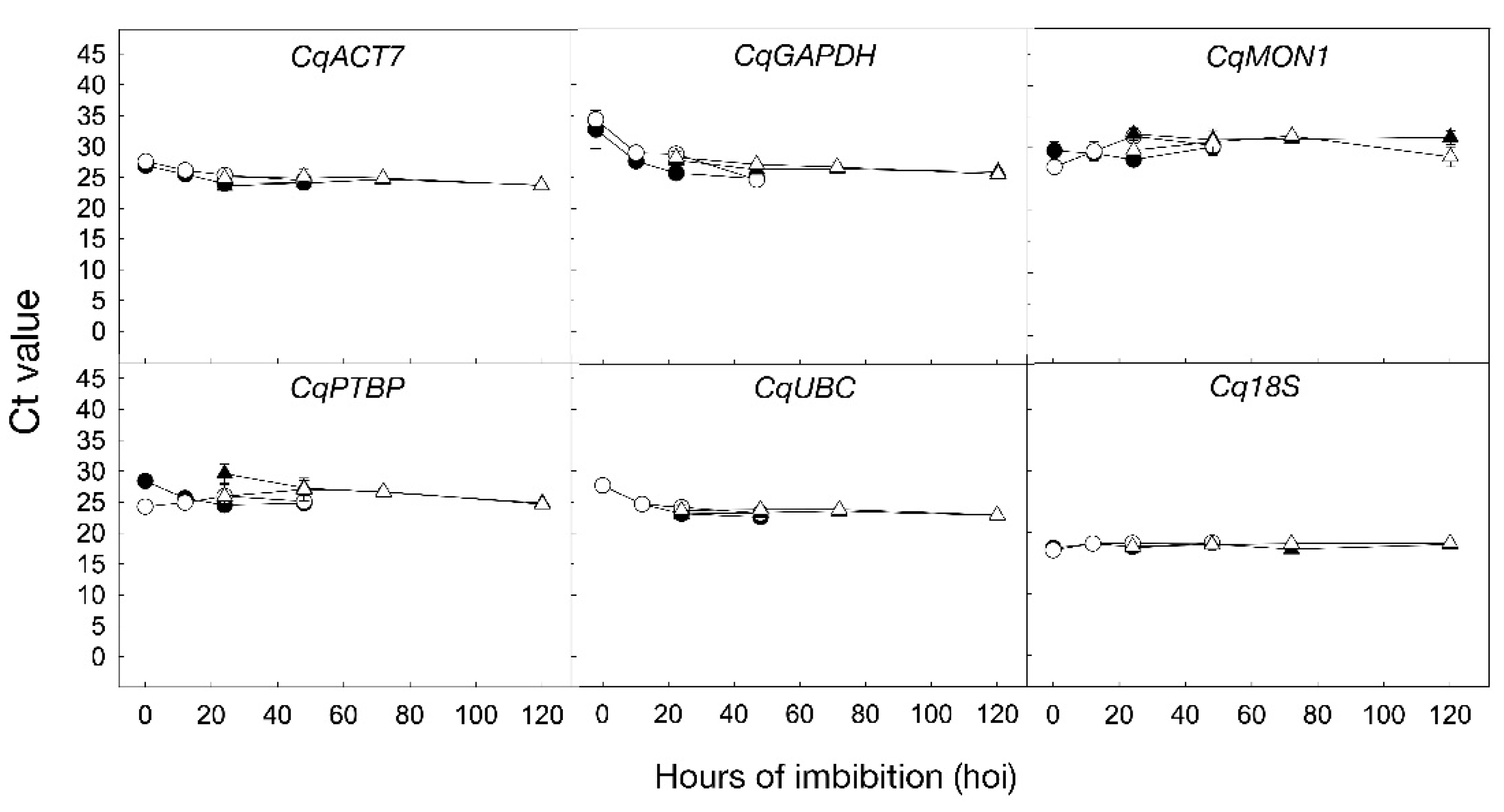
2.3. Stability of Candidate Reference Genes during Seed Germination under 250 mM NaCl in UDEC2 and UDEC4 Accessions
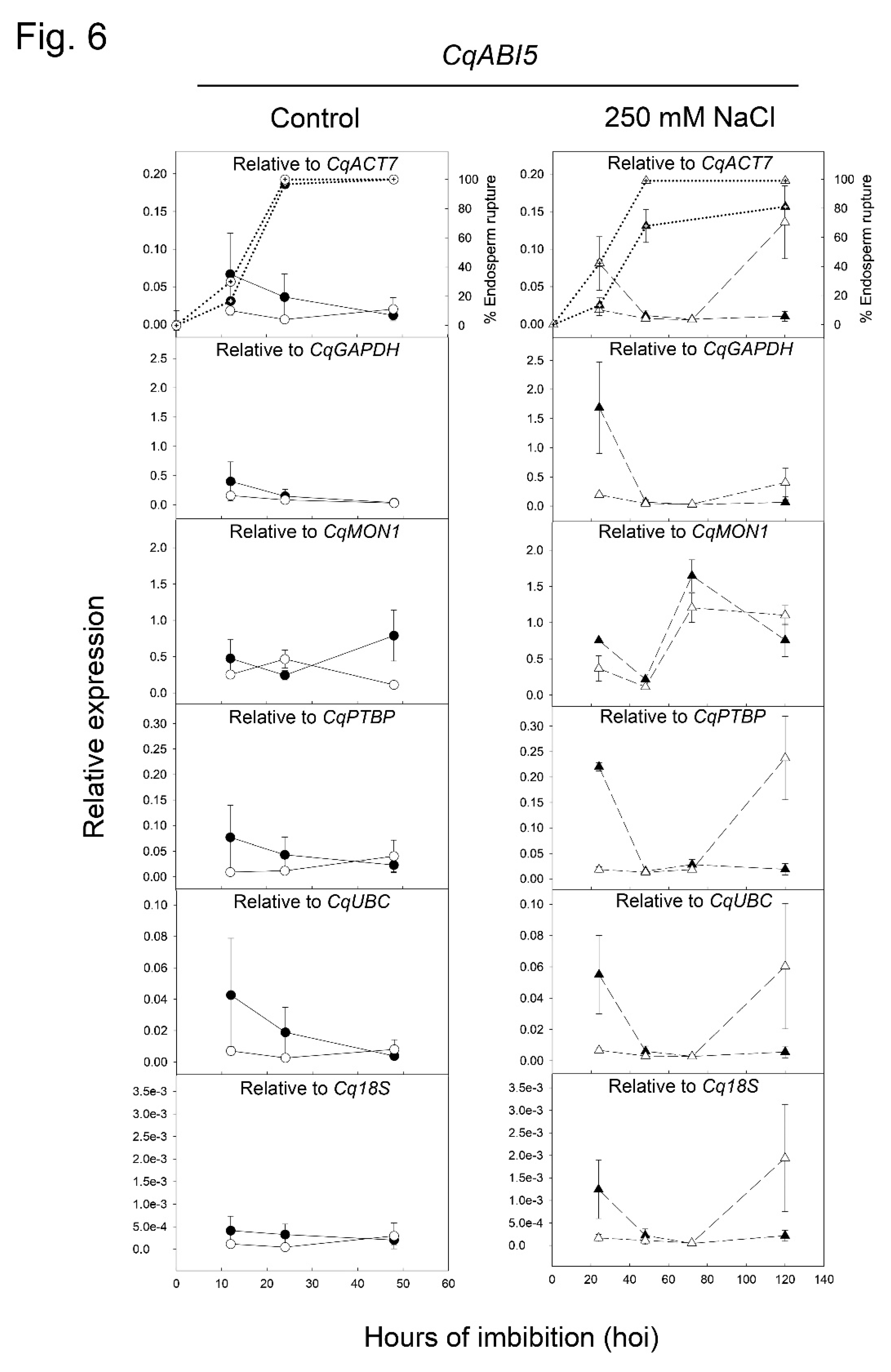
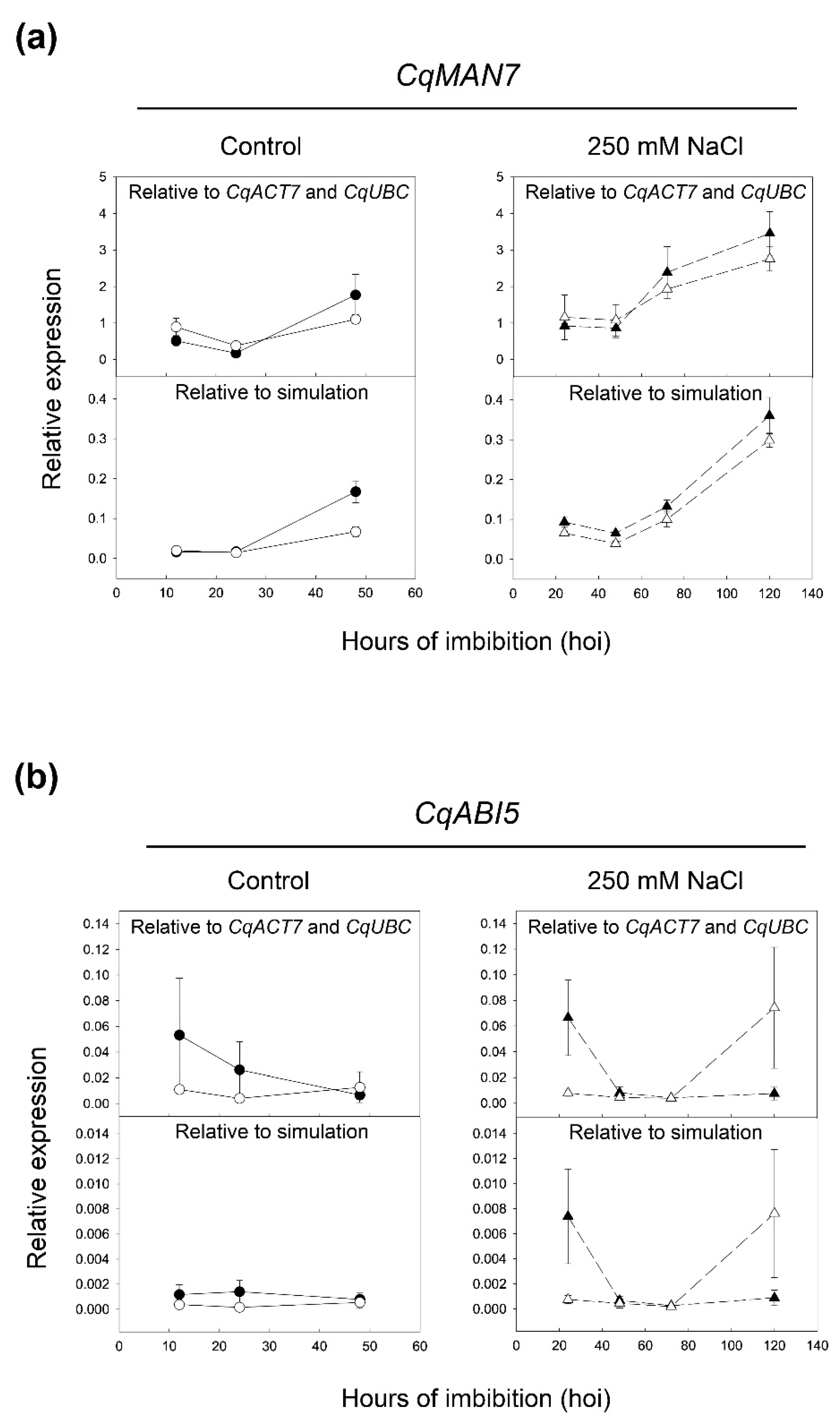
2.4. Validation of Selected Reference Genes during Seed Germination by Evaluation of CqMAN7 and CqABI5 Expression
3. Materials and Methods
3.1. Plant Materials and Growth Conditions
3.2. Light Microscopy: Fixation, Embedding, Sectioning and Polysaccharide and Protein Histological Staining
3.3. Germination Assays
3.4. Total RNA Isolation from Chenopodium quinoa Seeds and cDNA Synthesis
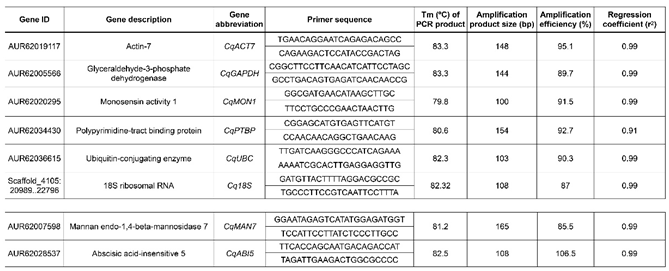
3.5. Selection of Candidate Reference and Target Genes
3.6. Primer Design
3.7. Quantitative PCR (qPCR)
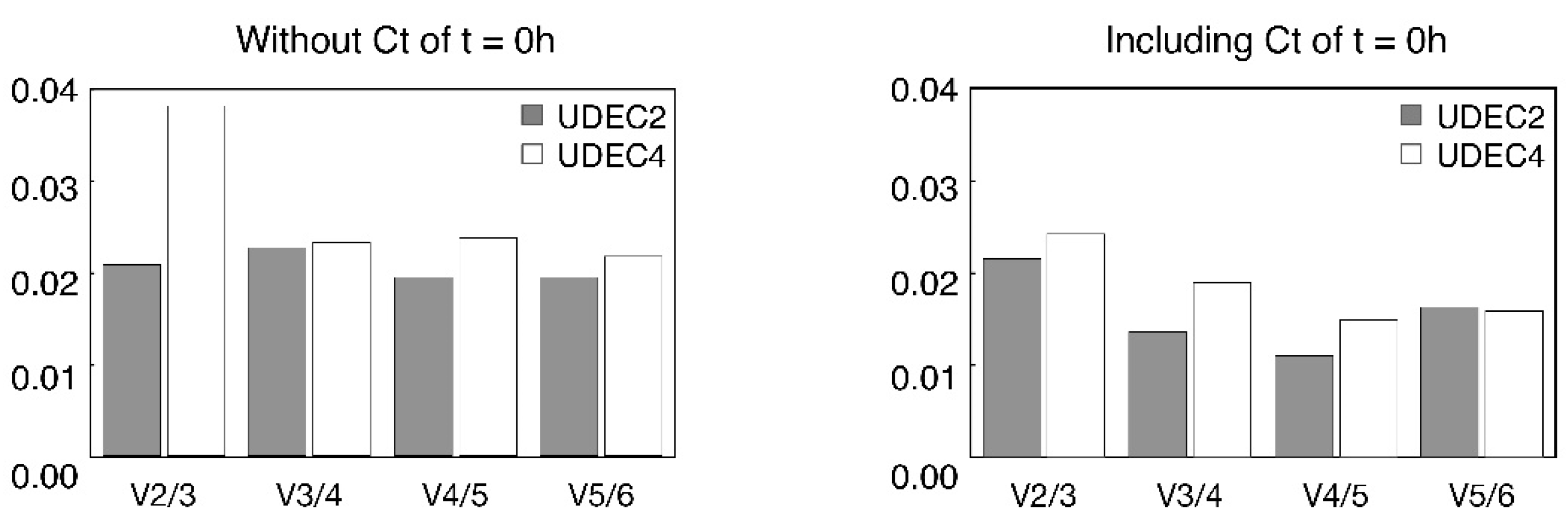
3.8. Gene stability analysis
- NFn = for each sample, geometric mean of the Ct obtained for n reference genes.
- An/n+1 = for each sample, log2 of NFn/NFn+1 ratio.
- Vn/n+1 = standard deviation of An/n+1 dataset.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.A.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; Kharbatia, N.M.; Rupper, R.R.; Sharp, A.R.; Dally, N.; Boughton, B.A.; Woo, Y.H.; Gao, G.; Schijlen, E.G.W.M.; Guo, X.; Momin, A.A.; Negrão, S.; Al-Babili, S.; Gehring, C.; Roessner, U.; Jung, C.; Murphy, K.; Arold, S.T.; Gojobori, T.; Linden, C.G.; van Loo, E.N.; Jellen, E.N.; Maughan, P.J.; Tester, M. The genome of Chenopodium quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Repo-Carrasco, R.; Espinoza, C.; Jacobsen, S.E. Nutritional value and use of the Andean crops quinoa (Chenopodium quinoa) and kañiwa (Chenopodium pallidicaule). Food Rev Int 2003, 19, 179–189. [Google Scholar] [CrossRef]
- Angeli, V.; Silva, P.M.; Massuela, D.C.; Khan, M.W.; Hamar, A.; Khajehei, F.; Grae, S.; Piatti, C. Quinoa (Chenopodium Quinoa Willd.): An overview of the potentials of the “golden grain” and socio-economic and environmental aspects of its cultivation and marketization. Foods 2020, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Craine, E.B.; Murphy, K.M. Seed composition and amino acid profiles for quinoa grown in Washington state. Front Nutr. 2020, 7, 126. [Google Scholar] [CrossRef] [PubMed]
- Stavi, I.; Thevs, N.; Simone, P. Soil salinity and sodicity in drylands: A review of causes, effects, monitoring, and restoration measures. Front Environ Sci 2021, 9, 712831. [Google Scholar] [CrossRef]
- Zurita-Silva, A.; Fuentes, F.; Zamora, P.; Jacobsen, S.E.; Schwembe, A.R. Breeding quinoa (Chenopodium quinoa Willd.): Potential and perspectives. Mol Breeding 2014, 34, 13–30. [Google Scholar] [CrossRef]
- Gómez Pando, L.; Aguilar Castellanos, E. Guía de cultivo de la quinua, 2nd ed.; FAO and Universidad Nacional Agraria La Molina: Lima, Perú, 2016. [Google Scholar]
- Otterbach, S.L.; Khoury, H.; Rupasinghe, T.; Mendis, H.; Kwan, K.H.; Lui, V.; Natera, S.H.A.; Klaiber, I.; Allen, N.M.; Jarvis, D.E.; Tester, M.; Roessner, U.; Schmöckel, S.M. Characterization of epidermal bladder cells in Chenopodium quinoa. Plant Cell Environ 2021, 44, 3606–3622. [Google Scholar] [CrossRef]
- Moog, M.W.; Trinh, M.D.L.; Nørrevang, A.F.; Bendtsen, A.K.; Wang, C.; Østerberg, J.T.; Shabala, S.; Hedrich, R.; Wendt, T.; Palmgren, M. The epidermal bladder cell-free mutant of the salt-tolerant quinoa challenges our understanding of halophyte crop salinity tolerance. New Phytol 2022, 236, 1409–1421. [Google Scholar] [CrossRef]
- Schmöckel, S.M.; Lightfoot, D.J.; Razali, R.; Tester, M.; Jarvis, D.E. Identification of putative transmembrane proteins involved in salinity tolerance in Chenopodium quinoa by integrating physiological data, RNAseq, and SNP analyses. Front Plant Sci. 2017, 8, 1023. [Google Scholar] [CrossRef]
- Poza-Viejo, L.; Redondo-Nieto, M.; Matías, J.; Granado-Rodríguez, S.; Maestro-Gaitán, I.; Cruz, V.; Olmos, E.; Bolaños, L.; Reguera, M. Shotgun proteomics of quinoa seeds reveals chitinases enrichment under rainfed conditions. Sci Rep 2023, 13, 4951. [Google Scholar] [CrossRef]
- Alandia, G.; Rodriguez, J.P.; Jacobsen, S.E.; Bazile, D.; Condori, B. Global expansion of quinoa and challenges for the Andean region. Glob Food Sec 2020, 26, 100429. [Google Scholar] [CrossRef]
- Matilla, A.J. The orthodox dry seeds are alive: A clear example of desiccation tolerance. Plants (Basel) 2021, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Burrieza, H.P.; López-Fernández, M.P.; Maldonado, S. Analogous reserve distribution and tissue characteristics in quinoa and grass seeds suggest convergent evolution. Front Plant Sci. 2014, 5, 546. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Bassel, G.W. Seed vigour and crop establishment: Extending performance beyond adaptation. J Exp Bot 2016, 67, 567–591. [Google Scholar] [CrossRef]
- Iglesias-Fernández, R.; Rodríguez-Gacio, M.C.; Barrero-Sicilia, C.; Carbonero, P.; Matilla, A. Three endo-β-mannanase genes expressed in the micropylar endosperm and in the radicle influence germination of Arabidopsis thaliana seeds. Planta 2011, 233, 25–36. [Google Scholar] [CrossRef] [PubMed]
- González-Calle, V.; Barrero-Sicilia, C.; Carbonero, P.; Iglesias-Fernández, R. Mannans and endo-β-mannanases (MAN) in Brachypodium distachyon: Expression profiling and possible role of the BdMAN genes during coleorhiza-limited seed germination. J Exp Bot 2015, 66, 3753–3764. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Mauriat, M.; Pelloux, J.; Bellini, C.; Van Wuytswinkel, O. Towards a systematic validation of references in real-time RT-PCR. Plant Cell 2008, 20, 1734–1735. [Google Scholar] [CrossRef]
- Wan, Q.; Chen, S.; Shan, Z.; Yang, Z.; Chen, L.; Zhang, C.; Yuan, S.; Hao, Q.; Zhang, X.; Qiu, D.; Chen, H.; Zhou, X. Stability evaluation of reference genes for gene expression analysis by RT-qPCR in soybean under different conditions. PLoS ONE 2017, 12, e0189405. [Google Scholar] [CrossRef]
- Nakabayashi, K.; Okamoto, M.; Koshiba, T.; Kamiya, Y.; Nambara, E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 2005, 41, 697–709. [Google Scholar] [CrossRef]
- Graeber, K.; Linkies, A.; Wood, A.T.A.; Leubner-Metzger, G.A. Guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a Brassicaceae cross-species seed germination case study. Plant Cell 2011, 23, 2045–2063. [Google Scholar] [CrossRef]
- Dekkers, B.J.; Willems, L.; Bassel, G.W.; van Bolderen-Veldkamp, R.P.; Ligterink, W.; Hilhorst, H.W.; Bentsink, L. Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol. 2012, 53, 28–37. [Google Scholar] [CrossRef]
- Maldonado-Taipe, N.; Patirange, D.S.R.; Schmockel, S.M.; Jung, C.; Emrani, N. Validation of suitable genes for normalization of seed germination gene expression studies in Chenopodium quinoa. PLoS ONE 2021, 16, 1–20. [Google Scholar] [CrossRef] [PubMed]
- De Santis, G.; Ronga, D.; Cardonia, F.; D’Ambrosio, T.; Jacopo, T.; Rascio, A.; Fragasso, M.; Pecchioni, N.; Rinaldi, M. Evaluation of two groups of quinoa (Chenopodium quinoa Willd.) accessions with different seed colours for adaptation to the Mediterranean environment. Crop Pasture Sci 2018, 69, 1264–1275. [Google Scholar] [CrossRef]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front Plant Sci 2016, 7, 1884. [Google Scholar] [CrossRef] [PubMed]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol 2006, 171, 501–523. [Google Scholar] [CrossRef]
- Ninfali, P.; Panato, A.; Bortolotti, F.; Valentini, L.; Gobbi, F. 2020. Morphological analysis of the seeds of three pseudocereals by using light microscopy and ESEM-EDS. Eur J Histochem 2020, 64, 1–10. [Google Scholar] [CrossRef]
- Hermann, K.; Meinhard, J.; Dobrev, P.; Linkies, A.; Pesek, B.; Heß, B.; Macháčková, I.; Fischer, U.; Leubner-Metzger, G. 1-Aminocyclopropane-1-carboxylic acid and abscisic acid during the germination of sugar beet (Beta vulgaris L.): A comparative study of fruits and seeds. J Exp Bot 2007, 58, 3047–3060. [Google Scholar] [CrossRef]
- Iglesias-Fernández, R.; Wozny, D.; Iriondo-de Hond, M.; Oñate-Sánchez, L.; Carbonero, P.; Barrero-Sicilia, C. The AtCathB3 gene, encoding a cathepsin B-like protease, is expressed during germination of Arabidopsis thaliana and transcriptionally repressed by the basic leucine zipper protein GBF1. J Exp Bot 2014, 65, 2009–2021. [Google Scholar] [CrossRef]
- Bolte, S.; Lanquar, V.; Soler, M.-N.; Beebo, A.; Satiat-Jeunemaitre, B.; Bouhidel, K.; Thomine, S. Distinct lytic vacuolar compartments are embedded inside the protein storage vacuole of dry and germinating Arabidopsis thaliana seeds. Plant Cell Physiol 2011, 52, 1142–1152. [Google Scholar] [CrossRef]
- Müller, K.; Tintelnot, S.; Leubner-Metzger, G. Endosperm-limited Brassicaceae seed germination: Abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 2006, 47, 864–877. [Google Scholar] [CrossRef]
- Carrillo-Barral, N.; Matilla, A.J.; Rodríguez-Gacio, M.D.C.; Iglesias-Fernández, R. Mannans and endo-β-mannanase transcripts are located in different seed compartments during Brassicaceae germination. Planta 2018, 247, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Nonogaki, H.; Gee, O.H.; Bradford, K.J. A germination-specific endo-beta-mannanase gene is expressed in the micropylar endosperm cap of tomato seeds. Plant Physiol 2000, 123, 1235–1246. [Google Scholar] [CrossRef] [PubMed]
- Loades, E.; Pérez, M.; Turečková, V.; Tarkowská, D.; Strnad, M.; Seville, A.; Nakabayashi, K.; Leubner-Metzger, G. Distinct hormonal and morphological control of dormancy and germination in Chenopodium album dimorphic seeds. Front Plant Sci 2023, 14, 1156794. [Google Scholar] [CrossRef]
- Barrero, J.M.; Talbot, M.J.; White, R.G.; Jacobsen, J.V.; Gubler, F. Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiol 2009, 150, 1006–1021. [Google Scholar] [CrossRef] [PubMed]
- Holloway, T.; Steinbrecher, T.; Pérez, M.; Seville, A.; Stock, D.; Nakabayashi, K.; Leubner-Metzger, G. Coleorhiza-enforced seed dormancy: A novel mechanism to control germination in grasses. New Phytol 2020, 229, 2179–2191. [Google Scholar] [CrossRef] [PubMed]
- Tarchoun, K.; Yousef, M.; Bánóczi, Z. Azapeptides as an Efficient Tool to Improve the Activity of Biologically Effective Peptides. Future Pharmacol. 2022, 2, 293–305. [Google Scholar] [CrossRef]
- Ruffino, A.M.C.; Rosa, M.; Hilal, M.; González, J.A.; Prado, F.E. The role of cotyledon metabolism in the establishment of quinoa (Chenopodium quinoa) seedlings growing under salinity. Plant Soil 2010, 326, 213–224. [Google Scholar] [CrossRef]
- Orsini, F.; Accorsi, M.; Gianquinto, G.; Dinelli, G.; Antognoni, F.; Ruiz Carrasco, K.B.; Martinez, E.A.; Alnayef, M.; Marotti, I.; Bosi, S.; Biondi, S. Beyond the ionic and osmotic response to salinity in Chenopodium quinoa: Functional elements of successful halophytism. Funct Plant Biol 2011, 38, 818–31. [Google Scholar] [CrossRef]
- Ruiz-Carrasco, K.; Antognoni, F.; Coulibaly, A.K.; Lizardi, S.; Covarrubias, A.; Martínez, E.A.; Molina-Montenegro, M.A.; Biondi, S.; Zurita-Silva, A. Variation in salinity tolerance of four lowland genotypes of quinoa (Chenopodium quinoa Willd.) as assessed by growth, physiological traits, and sodium transporter gene expression. Plant Physiol Biochem 2011, 49, 1333–1341. [Google Scholar] [CrossRef]
- Cai, Z.-Q.; Gao, Q. Comparative physiological and biochemical mechanisms of salt tolerance in five contrasting highland quinoa cultivars. BMC Plant Biol 2020, 20, 1–15. [Google Scholar] [CrossRef]
- Panuccio, M.R.; Jacobsen, S.E.; Akhtar, S.S.; Muscolo, A. Effect of saline water on seed germination and early seedling growth of the halophyte quinoa. AoB Plants 2014, 6, plu047. [Google Scholar] [CrossRef] [PubMed]
- Polle, A.; Chen, S. On the salty side of life: Molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant Cell Environ 2015, 38, 1794–816. [Google Scholar] [CrossRef] [PubMed]
- Derbali, I.; Manaa, A.; Derbali, W.; Slama, I. Effects of osmotic stress on Chenopodium quinoa seed germination. J Oasis Agric Sustain Dev 2021, 3, 30–35. [Google Scholar] [CrossRef]
- Nadarajan, J.; Walters, C.; Pritchard, H.W.; Ballesteros, D.; Colville, L. Seed longevity-the evolution of knowledge and a conceptual framework. Plants (Basel) 2023, 12, 471. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Ma, J.; Wang, J.; Wu, X.; Li, P.; Yao, Y. Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front Plant Sci 2014, 5, 788. [Google Scholar] [CrossRef]
- Thellin, O.; Zorzi, W.; Lakaye, B.; De Borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping genes as internal standards: Use and limits. J Biotechnol 1999, 75, 291–295. [Google Scholar] [CrossRef]
- Bustin, S.; Huggett, J. qPCR primer design revisited. Biomol Detect Quantif 2017, 14, 19–28. [Google Scholar] [CrossRef]
- Okamoto, M.; Tatematsu, K.; Matsui, A.; Morosawa, T.; Ishida, J.; Tanaka, M.; Endo, T.A.; Mochizuki, Y.; Toyoda, T.; Kamiya, Y.; Shinozaki, K.; Nambara, E.; Seki, M. Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. Plant J 2010, 62, 39–51. [Google Scholar] [CrossRef]
- Fiallos-Jurado, J.; Pollier, J.; Moses, T.; Arendt, P.; Barriga-Medina, N.; Morillo, E.; Arahana, V.; Torres, M.L.; Goossens, A.; Leon-Reyes, A. Saponin determination, expression analysis and functional characterization of saponin biosynthetic genes in Chenopodium quinoa leaves. Plant Sci 2016, 250, 188–197. [Google Scholar] [CrossRef]
- Shi, P.; Gu, M. Transcriptome analysis and differential gene expression profiling of two contrasting quinoa genotypes in response to salt stress. BMC Plant Biol 2020, 568. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; Rokhsar, D.S. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res 2012, 40(D1), 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in quantitative RT-PCR. Nucleic Acids Res 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of quantitative quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of quantitative quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using quantitative PCR. BMC Molecular Biol 2006, 7, 33. [Google Scholar] [CrossRef]
- Wang, M.; Ren, T.; Marowa, P.; Du, H.; Xu, Z. Identification and selection of reference genes for gene expression analysis by quantitative real-time PCR in Suaeda glauca's response to salinity. Sci Rep 2021, 11, 8569. [Google Scholar] [CrossRef]
- Auler, P.A.; Benitez, L.C.; do Amaral, M.N.; Vighi, I.L.; Dos Santos Rodrigues, G.; da Maia, L.C.; Braga, E.J. Evaluation of stability and validation of reference genes for RT-qPCR expression studies in rice plants under water deficit. J Appl Genet. 2017, 58, 163–177. [Google Scholar] [CrossRef]
- Niñoles, R.; Planes, D.; Arjona, P.; Ruiz-Pastor, C.; Chazarra, R.; Renard, J.; Bueso, E.; Forment, J.; Serrano, R.; Kranner, I.; Roach, T.; Gadea, J. Comparative analysis of wild-type accessions reveals novel determinants of Arabidopsis seed longevity. Plant Cell Environ. 2022, 45, 2708–2728. [Google Scholar] [CrossRef]
- Wu, Q.; Meng, Y.T.; Feng, Z.H.; Shen, R.F.; Zhu, X.F. The endo-beta mannase MAN7 contributes to cadmium tolerance by modulating root cell wall binding capacity in Arabidopsis thaliana. J Integr Plant Biol. 2023, 65, 1670–1686. [Google Scholar] [CrossRef]
- Iglesias-Fernández, R.; Pastor-Mora, E.; Vicente-Carbajosa, J.; Carbonero, P. A possible role of the aleurone expressed gene HvMAN1 in the hydrolysis of the cell wall mannans of the starchy endosperm in germinating Hordeum vulgare L. seeds. Front Plant Sci 2020, 10, 1706. [Google Scholar] [CrossRef]
- Collin, A.; Daszkowska-Golec, A.; Szarejko, I. Updates on the role of abscisic acid insensitive 5 (ABI5) and abscisic Acid-responsive element binding factors (ABFs) in ABA signaling in different developmental stages in plants. Cells. 2021, 10, 1996. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Molina, L.; Mongrand, S.; Chua, N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci U S A. 2001, 98, 4782–4787. [Google Scholar] [CrossRef]
- Joosen, R.V.L.; Kodde, J.; Willems, L.A.J.; Ligterink, W.; Van Der Plas, L.H.V.; Hilhorst, H.W.M. Germinator: A software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. Plant J 2010, 62, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Oñate-Sánchez, L.; Vicente-Carbajosa, J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 2008, 2008 1, 93. [Google Scholar] [CrossRef]
- Waese, J.; Fan, J.; Pasha, A.; Yu, H.; Fucile, G.; Shi, R.; Cumming, M.; Kelley, L.A.; Sternberg, M.J.; Krishnakumar, V.; Ferlanti, E.; Miller, J.; Town, C.; Stuerzlinger, W.; Provart, N.J. ePlant: Visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 2017, 29, 1806–1821. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res 2012, 40, e115. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol Lett 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct Integr Genomics 2023, 23, 125. [Google Scholar] [CrossRef]

 |
 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





