Submitted:
19 September 2023
Posted:
21 September 2023
You are already at the latest version
Abstract
Keywords:
Introduction
Clinical Applications of Liquid Biopsy in Brain Tumors
i. Pre-operative setting: Cancer Screening, Early Detection, and Diagnostic Differentiation
ii. Identification of Post-Operative Tumor Residual and Progression Surveillance
iii. Selection of Precision Therapies and Understanding Mechanisms of Resistance
| Histopathology | Biopsy Source | Tumoral content | Molecular alterations studied | Isolation Technique | Application/Findings |
|---|---|---|---|---|---|
| GBM | |||||
| [38] | Serum | cfDNA | MGMT, p16, DAPK, RASSF1A methylation | MS-PCR | Correlation with time to progression and response to 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU) and temozolomide |
| [39] | Plasma | ctDNA | P16, MGMT, p73, and RARβ methylation |
MS-PCR | Identification of tumor-specific promoter methylation |
| [40] | Urine | Panel of 23 miRNAs | - | Nanowire | Screening method for early detection of tumor |
| [41] | Neurosurgical aspirate fluid | EVs, miR-486 | - | NGS | Distinguishing GBM from Lower-Grade Astrocytoma |
| LGG | |||||
| [42] | Serum | miR-21, miR-20e, miR-223 | - | ddPCR | Post-operative monitoring |
| [5] | CSF | ctDNA | DH1, 1P19Q, CIC, ATRX, TP53 mutation |
NGS | Monitor evolution of the glioma genome through disease course Correlation with disease burden |
| Meningioma | |||||
| [43] | Serum | ctDNA | MGMT, RASSF1A, p15INK4B, and p14ARF methylation | MS-PCR | RASSF1A hypermethylation differentiates between metastatic and primary CNS cancers two groups. |
| [44] | Plasma, CSF | cfDNA | NF2, AKT1 mutation | ddPCR | Higher cfDNA concentrations in CSF than in plasma; CSF may be used for disease detection despite low plasma cfDNA concentrations. |
| [34] | Plasma | EVs | 22q and 1p deletion, NF2 and TRAF7 mutation | Nanoparticle tracking analysis | Tumor detection and classification, pre-operative tumor assessment and residual tumor monitoring, correlation with tumor size, grade and peritumoral edema. |
| [45] | Serum | miR-15a, miR16_1, miR−15b, miR-497, miR-195 | - | qPCR | Differentiating low-grade from high-grade meningioma |
| [46] | Serum | miRNA 200a, miRNAs 34a, miRNA 409 | Aberrations of parts of chromosomes 1, 14, 18, and 22 | qPCR | Predicting recurrent meningiomas |
The Role of Imaging in Liquid Biopsy of Brain Tumors
i. Identifying Factors that Affect Plasma cfDNA and ctDNA Detection
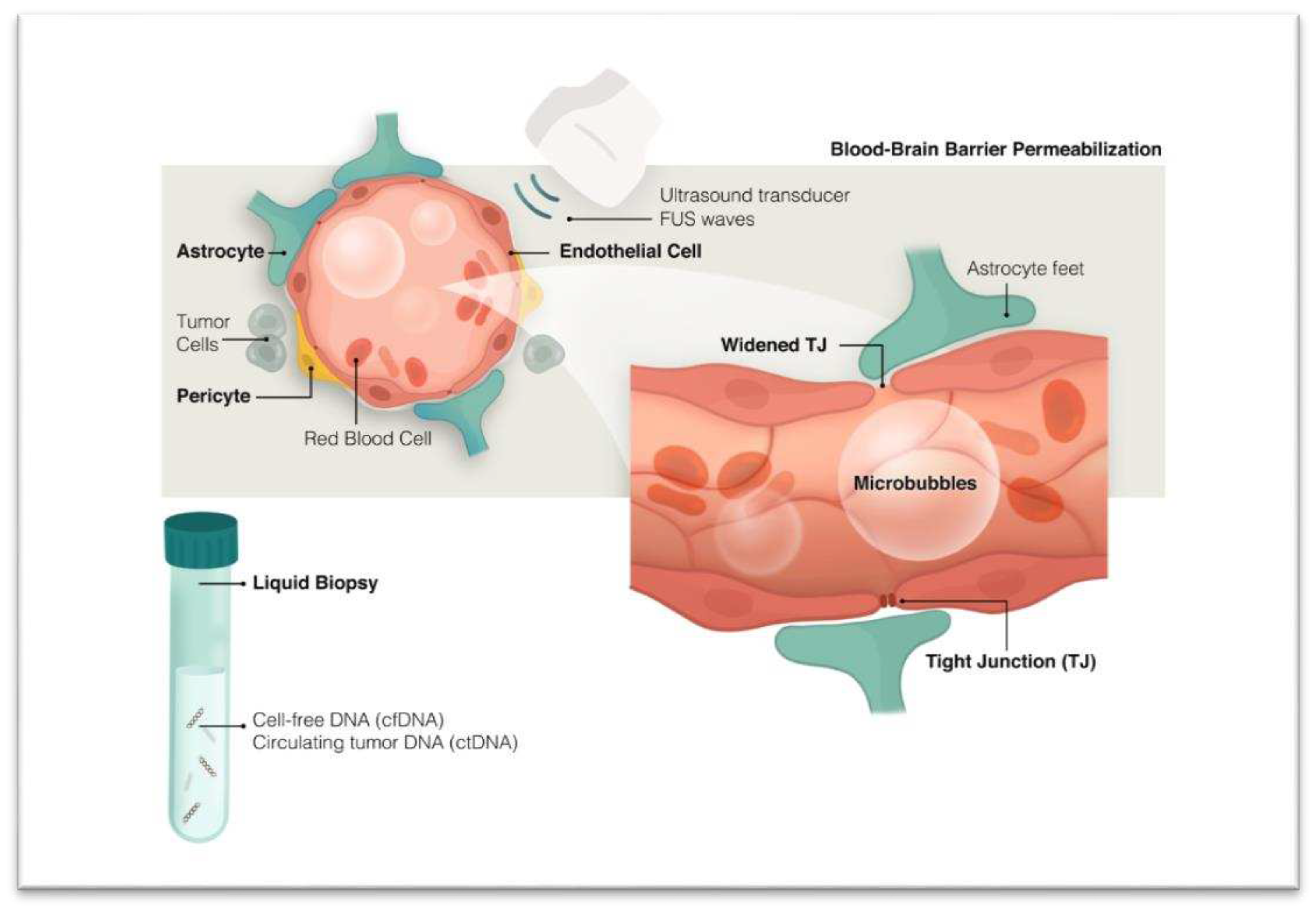
ii. Enhancing Presence of Tumor Biomarkers Through Blood-Brain Barrier (BBB) Disruption
The Role of Advanced Imaging Techniques in the Clinical Setting
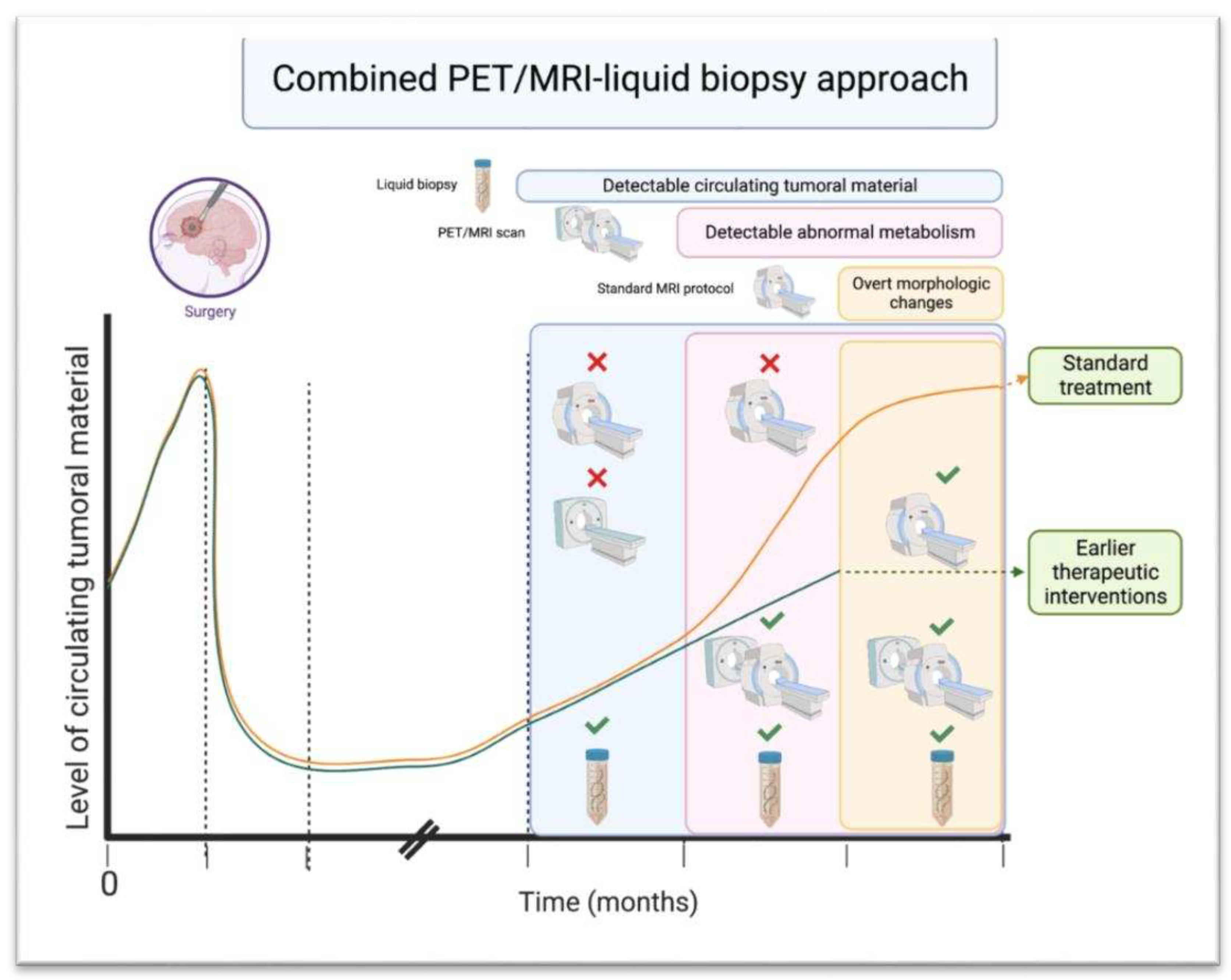
i. PET/MRI in Combination with Liquid Biopsy
ii. Integrating Radiomics with Liquid Biopsy
Challenges and Future Directions
Summary
Author Contributions
Acknowledgment
Conflicts of Interest
References
- Le Rhun E, Seoane J, Salzet M, Soffietti R, Weller MJCl. Liquid biopsies for diagnosing and monitoring primary tumors of the central nervous system. 2020;480:24-8. [CrossRef]
- Corcoran RB, Chabner BAJNEJoM. Application of cell-free DNA analysis to cancer treatment. 2018;379(18):1754-65.
- Alix-Panabières C, Pantel K. Liquid biopsy: from discovery to clinical implementation. Mol Oncol. 2021;15(6):1617-21. [CrossRef]
- Saenz-Antoñanzas A, Auzmendi-Iriarte J, Carrasco-Garcia E, Moreno-Cugnon L, Ruiz I, Villanua J, et al. Liquid Biopsy in Glioblastoma: Opportunities, Applications and Challenges. Cancers (Basel). 2019;11(7).
- Miller AM, Shah RH, Pentsova EI, Pourmaleki M, Briggs S, Distefano N, et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565(7741):654-8.
- Khristov V, Lin A, Freedman Z, Staub J, Shenoy G, Mrowczynski O, et al. Tumor-Derived Biomarkers in Liquid Biopsy of Glioblastoma. World Neurosurgery. 2023;170:182-94. [CrossRef]
- McEwen AE, Leary SES, Lockwood CM. Beyond the Blood: CSF-Derived cfDNA for Diagnosis and Characterization of CNS Tumors. Frontiers in Cell and Developmental Biology. 2020;8. [CrossRef]
- Raphael BJ, Dobson JR, Oesper L, Vandin F. Identifying driver mutations in sequenced cancer genomes: computational approaches to enable precision medicine. Genome Medicine. 2014;6(1):5.
- Lun M, Lok E, Gautam S, Wu E, Wong ET. The natural history of extracranial metastasis from glioblastoma multiforme. J Neurooncol. 2011;105(2):261-73. [CrossRef]
- Lavon I, Refael M, Zelikovitch B, Shalom E, Siegal T. Serum DNA can define tumor-specific genetic and epigenetic markers in gliomas of various grades. Neuro-Oncology. 2010;12(2):173-80. [CrossRef]
- Barciszewska A-M. Total DNA methylation as a biomarker of DNA damage and tumor malignancy in intracranial meningiomas. BMC Cancer. 2020;20(1):509. [CrossRef]
- Liu APY, Smith KS, Kumar R, Paul L, Bihannic L, Lin T, et al. Serial assessment of measurable residual disease in medulloblastoma liquid biopsies. Cancer Cell. 2021;39(11):1519-30.e4. [CrossRef]
- Yang C, Wang C, Chen X, Chen S, Zhang Y, Zhi F, et al. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer. 2013;132(1):116-27. [CrossRef]
- Morad G, Moses MA. Brainwashed by extracellular vesicles: the role of extracellular vesicles in primary and metastatic brain tumour microenvironment. J Extracell Vesicles. 2019;8(1):1627164. [CrossRef]
- Lyu Y, Guo Y, Okeoma CM, Yan Z, Hu N, Li Z, et al. Engineered extracellular vesicles (EVs): Promising diagnostic/therapeutic tools for pediatric high-grade glioma. Biomedicine & Pharmacotherapy. 2023;163:114630. [CrossRef]
- Nilsson RJ, Balaj L, Hulleman E, van Rijn S, Pegtel DM, Walraven M, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood. 2011;118(13):3680-3. [CrossRef]
- Chen WW, Balaj L, Liau LM, Samuels ML, Kotsopoulos SK, Maguire CA, et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol Ther Nucleic Acids. 2013;2(7):e109. [CrossRef]
- Carr C, O'Neill BE, Hochhalter CB, Strong MJ, Ware ML. Biomarkers of Pineal Region Tumors: A Review. Ochsner J. 2019;19(1):26-31. [CrossRef]
- Nabavizadeh A, Bagley SJ, Doot RK, Ware JB, Young AJ, Ghodasara S, et al. Distinguishing Progression from Pseudoprogression in Glioblastoma Using (18)F-Fluciclovine PET. J Nucl Med. 2023;64(6):852-8. [CrossRef]
- . [CrossRef]
- Bagley SJ, Nabavizadeh SA, Mays JJ, Till JE, Ware JB, Levy S, et al. Clinical Utility of Plasma Cell-Free DNA in Adult Patients with Newly Diagnosed Glioblastoma: A Pilot Prospective Study. Clinical cancer research : an official journal of the American Association for Cancer Research. 2020;26(2):397-407. [CrossRef]
- Ma C, Nguyen HPT, Luwor RB, Stylli SS, Gogos A, Paradiso L, et al. A comprehensive meta-analysis of circulation miRNAs in glioma as potential diagnostic biomarker. PLoS One. 2018;13(2):e0189452. [CrossRef]
- Ravegnini G, Cargnin S, Sammarini G, Zanotti F, Bermejo JL, Hrelia P, et al. Prognostic Role of miR-221 and miR-222 Expression in Cancer Patients: A Systematic Review and Meta-Analysis. Cancers (Basel). 2019;11(7). [CrossRef]
- An Y, Fan F, Jiang X, Sun K. Recent Advances in Liquid Biopsy of Brain Cancers. Frontiers in genetics. 2021;12:720270. [CrossRef]
- Kumar DM, Thota B, Shinde SV, Prasanna KV, Hegde AS, Arivazhagan A, et al. Proteomic identification of haptoglobin α2 as a glioblastoma serum biomarker: implications in cancer cell migration and tumor growth. J Proteome Res. 2010;9(11):5557-67. [CrossRef]
- Björkblom B, Wibom C, Jonsson P, Mörén L, Andersson U, Johannesen TB, et al. Metabolomic screening of pre-diagnostic serum samples identifies association between α- and γ-tocopherols and glioblastoma risk. Oncotarget. 2016;7(24):37043-53. [CrossRef]
- Shen J, Song R, Hodges TR, Heimberger AB, Zhao H. Identification of metabolites in plasma for predicting survival in glioblastoma. Mol Carcinog. 2018;57(8):1078-84. [CrossRef]
- Provencio M, Torrente M, Calvo V, Pérez-Callejo D, Gutiérrez L, Franco F, et al. Prognostic value of quantitative ctDNA levels in non small cell lung cancer patients. Oncotarget. 2018;9(1):488-94. [CrossRef]
- Pan RJ, Hong HJ, Sun J, Yu CR, Liu HS, Li PY, Zheng MH. Detection and Clinical Value of Circulating Tumor Cells as an Assisted Prognostic Marker in Colorectal Cancer Patients. Cancer Manag Res. 2021;13:4567-78. [CrossRef]
- Coombes RC, Page K, Salari R, Hastings RK, Armstrong A, Ahmed S, et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clinical Cancer Research. 2019;25(14):4255-63. [CrossRef]
- Pentsova EI, Shah RH, Tang J, Boire A, You D, Briggs S, et al. Evaluating Cancer of the Central Nervous System Through Next-Generation Sequencing of Cerebrospinal Fluid. J Clin Oncol. 2016;34(20):2404-15. [CrossRef]
- Nieland L, Morsett LM, Broekman MLD, Breakefield XO, Abels ER. Extracellular Vesicle-Mediated Bilateral Communication between Glioblastoma and Astrocytes. Trends Neurosci. 2021;44(3):215-26. [CrossRef]
- Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016;30(6):836-48. [CrossRef]
- Ricklefs FL, Maire CL, Wollmann K, Dührsen L, Fita KD, Sahm F, et al. Diagnostic potential of extracellular vesicles in meningioma patients. Neuro-Oncology. 2022;24(12):2078-90.
- Garnier D, Meehan B, Kislinger T, Daniel P, Sinha A, Abdulkarim B, et al. Divergent evolution of temozolomide resistance in glioblastoma stem cells is reflected in extracellular vesicles and coupled with radiosensitization. Neuro Oncol. 2018;20(2):236-48. [CrossRef]
- Liu T, Xu H, Huang M, Ma W, Saxena D, Lustig RA, et al. Circulating Glioma Cells Exhibit Stem Cell-like Properties. Cancer Res. 2018;78(23):6632-42. [CrossRef]
- Weller M, Butowski N, Tran DD, Recht LD, Lim M, Hirte H, et al. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017;18(10):1373-85. [CrossRef]
- Balaña C, Ramirez JL, Taron M, Roussos Y, Ariza A, Ballester R, et al. O6-methyl-guanine-DNA methyltransferase methylation in serum and tumor DNA predicts response to 1,3-bis(2-chloroethyl)-1-nitrosourea but not to temozolamide plus cisplatin in glioblastoma multiforme. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9(4):1461-8.
- Weaver KD, Grossman SA, Herman JG. Methylated tumor-specific DNA as a plasma biomarker in patients with glioma. Cancer Invest. 2006;24(1):35-40. [CrossRef]
- Kitano Y, Aoki K, Ohka F, Yamazaki S, Motomura K, Tanahashi K, et al. Urinary MicroRNA-Based Diagnostic Model for Central Nervous System Tumors Using Nanowire Scaffolds. ACS Applied Materials & Interfaces. 2021;13(15):17316-29.
- Hallal S, Ebrahim Khani S, Wei H, Lee MYT, Sim HW, Sy J, et al. Deep Sequencing of Small RNAs from Neurosurgical Extracellular Vesicles Substantiates miR-486-3p as a Circulating Biomarker that Distinguishes Glioblastoma from Lower-Grade Astrocytoma Patients. Int J Mol Sci. 2020;21(14). [CrossRef]
- Morokoff A, Jones J, Nguyen H, Ma C, Lasocki A, Gaillard F, et al. Serum microRNA is a biomarker for post-operative monitoring in glioma. J Neurooncol. 2020;149(3):391-400. [CrossRef]
- Majchrzak-Celińska A, Paluszczak J, Kleszcz R, Magiera M, Barciszewska AM, Nowak S, Baer-Dubowska W. Detection of MGMT, RASSF1A, p15INK4B, and p14ARF promoter methylation in circulating tumor-derived DNA of central nervous system cancer patients. J Appl Genet. 2013;54(3):335-44. [CrossRef]
- Pan W, Gu W, Nagpal S, Gephart MH, Quake SR. Brain Tumor Mutations Detected in Cerebral Spinal Fluid. Clinical Chemistry. 2015;61(3):514-22. [CrossRef]
- Negroni C, Hilton DA, Ercolano E, Adams CL, Kurian KM, Baiz D, Hanemann CO. GATA-4, a potential novel therapeutic target for high-grade meningioma, regulates miR-497, a potential novel circulating biomarker for high-grade meningioma. EBioMedicine. 2020;59:102941. [CrossRef]
- Urbschat S, Landau B, Bewersdorf N-C, Schuster C, Wagenpfeil G, Schulz-Schaeffer WJ, et al. MicroRNA 200a as a histologically independent marker for meningioma recurrence: Results of a four microRNA panel analysis in meningiomas. Cancer Medicine. 2023;12(7):8433-44. [CrossRef]
- Izquierdo E, Proszek P, Pericoli G, Temelso S, Clarke M, Carvalho DM, et al. Droplet digital PCR-based detection of circulating tumor DNA from pediatric high grade and diffuse midline glioma patients. Neuro-oncology advances. 2021;3(1). [CrossRef]
- Panditharatna E, Kilburn LB, Aboian MS, Kambhampati M, Gordish-Dressman H, Magge SN, et al. Clinically Relevant and Minimally Invasive Tumor Surveillance of Pediatric Diffuse Midline Gliomas Using Patient-Derived Liquid Biopsy. Clinical Cancer Research. 2018;24(23):5850-9.
- Huang TY, Piunti A, Lulla RR, Qi J, Horbinski CM, Tomita T, et al. Detection of Histone H3 mutations in cerebrospinal fluid-derived tumor DNA from children with diffuse midline glioma. Acta Neuropathologica Communications. 2017;5(1):28. [CrossRef]
- Cantor E, Wierzbicki K, Tarapore RS, Ravi K, Thomas C, Cartaxo R, et al. Serial H3K27M cell-free tumor DNA (cf-tDNA) tracking predicts ONC201 treatment response and progression in diffuse midline glioma. Neuro-Oncology. 2022;24(8):1366-74.
- Sun Y, Li M, Ren S, Liu Y, Zhang J, Li S, et al. Exploring genetic alterations in circulating tumor DNA from cerebrospinal fluid of pediatric medulloblastoma. Scientific reports. 2021;11(1):5638. [CrossRef]
- Escudero L, Llort A, Arias A, Diaz-Navarro A, Martínez-Ricarte F, Rubio-Perez C, et al. Circulating tumour DNA from the cerebrospinal fluid allows the characterisation and monitoring of medulloblastoma. Nature Communications. 2020;11(1):5376.
- Li J, Zhao S, Lee M, Yin Y, Li J, Zhou Y, et al. Reliable tumor detection by whole-genome methylation sequencing of cell-free DNA in cerebrospinal fluid of pediatric medulloblastoma. Science Advances. 2020;6(42):eabb5427. [CrossRef]
- Nabavizadeh SA, Ware JB, Guiry S, Nasrallah MP, Mays JJ, Till JE, et al. Imaging and histopathologic correlates of plasma cell-free DNA concentration and circulating tumor DNA in adult patients with newly diagnosed glioblastoma. Neuro-oncology advances. 2020;2(1):vdaa016. [CrossRef]
- Siegal T, Charbit H, Paldor I, Zelikovitch B, Canello T, Benis A, et al. Dynamics of circulating hypoxia-mediated miRNAs and tumor response in patients with high-grade glioma treated with bevacizumab. J Neurosurg. 2016;125(4):1008-15. [CrossRef]
- Wang Y, Springer S, Zhang M, McMahon KW, Kinde I, Dobbyn L, et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(31):9704-9. [CrossRef]
- Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. International Journal of Cancer. 2009;125(6):1407-13. [CrossRef]
- Ram Z, Cohen ZR, Harnof S, Tal S, Faibel M, Nass D, et al. MAGNETIC RESONANCE IMAGING-GUIDED, HIGH-INTENSITY FOCUSED ULTRASOUND FOR BRAIN TUMOR THERAPY. 2006;59(5):949-56. [CrossRef]
- MacDonell J, Patel N, Rubino S, Ghoshal G, Fischer G, Burdette EC, et al. Magnetic resonance–guided interstitial high-intensity focused ultrasound for brain tumor ablation %J Neurosurgical Focus FOC. 2018;44(2):E11.
- McDannold N, Vykhodtseva N, Hynynen K. Targeted disruption of the blood-brain barrier with focused ultrasound: association with cavitation activity. Physics in medicine and biology. 2006;51(4):793-807. [CrossRef]
- Wasielewska JM, White AR. "Focused Ultrasound-mediated Drug Delivery in Humans - a Path Towards Translation in Neurodegenerative Diseases". Pharmaceutical research. 2022;39(3):427-39. [CrossRef]
- Gandhi K, Barzegar-Fallah A, Banstola A, Rizwan SB, Reynolds JNJ. Ultrasound-Mediated Blood-Brain Barrier Disruption for Drug Delivery: A Systematic Review of Protocols, Efficacy, and Safety Outcomes from Preclinical and Clinical Studies. Pharmaceutics. 2022;14(4).
- Mainprize T, Lipsman N, Huang Y, Meng Y, Bethune A, Ironside S, et al. Blood-Brain Barrier Opening in Primary Brain Tumors with Non-invasive MR-Guided Focused Ultrasound: A Clinical Safety and Feasibility Study. Scientific reports. 2019;9(1):321. [CrossRef]
- Zhang DY, Gould A, Happ HC, Youngblood MW, Dmello C, Kang SJ, et al. Ultrasound-mediated blood–brain barrier opening increases cell-free DNA in a time-dependent manner. 2021;3(1):vdab165. [CrossRef]
- Zhu L, Nazeri A, Pacia CP, Yue Y, Chen H. Focused ultrasound for safe and effective release of brain tumor biomarkers into the peripheral circulation. PLOS ONE. 2020;15(6):e0234182. [CrossRef]
- Pacia CP, Zhu L, Yang Y, Yue Y, Nazeri A, Michael Gach H, et al. Feasibility and safety of focused ultrasound-enabled liquid biopsy in the brain of a porcine model. Scientific reports. 2020;10(1):7449. [CrossRef]
- Meng Y, Pople CB, Suppiah S, Llinas M, Huang Y, Sahgal A, et al. MR-guided focused ultrasound liquid biopsy enriches circulating biomarkers in patients with brain tumors. Neuro Oncol. 2021;23(10):1789-97. [CrossRef]
- Pacia CP, Yuan J, Yue Y, Xu L, Nazeri A, Desai R, et al. Sonobiopsy for minimally invasive, spatiotemporally-controlled, and sensitive detection of glioblastoma-derived circulating tumor DNA. Theranostics. 2022;12(1):362-78. [CrossRef]
- Santra A, Kumar R, Sharma P, Bal C, Kumar A, Julka PK, Malhotra A. F-18 FDG PET-CT in patients with recurrent glioma: comparison with contrast enhanced MRI. Eur J Radiol. 2012;81(3):508-13. [CrossRef]
- Shooli H, Assadi M, Nabavizadeh SA, Aboian M. Amino Acid PET/MRI in Neuro-oncology. In: Franceschi AM, Franceschi D, editors. Hybrid PET/MR Neuroimaging: A Comprehensive Approach. Cham: Springer International Publishing; 2022. p. 137-65.
- Shooli H, Assadi M, Aboian M. [18F]-FDG PET/MR Neuroimaging: Focus on Neuro-Oncology Applications. In: Franceschi AM, Franceschi D, editors. Hybrid PET/MR Neuroimaging: A Comprehensive Approach. Cham: Springer International Publishing; 2022. p. 89-98.
- Zheng W, Quan B, Gao G, Zhang P, Huang L. Combination of Circulating Cell-Free DNA and Positron Emission Tomography to Distinguish Non-Small Cell Lung Cancer from Tuberculosis. Lab Med. 2023;54(2):130-41. [CrossRef]
- González de Aledo-Castillo JM, Casanueva-Eliceiry S, Soler-Perromat A, Fuster D, Pastor V, Reguart N, et al. Cell-free DNA concentration and fragment size fraction correlate with FDG PET/CT-derived parameters in NSCLC patients. Eur J Nucl Med Mol Imaging. 2021;48(11):3631-42. [CrossRef]
- Jiménez-Ubieto A, Poza M, Martin-Muñoz A, Ruiz-Heredia Y, Dorado S, Figaredo G, et al. Real-life disease monitoring in follicular lymphoma patients using liquid biopsy ultra-deep sequencing and PET/CT. Leukemia. 2023;37(3):659-69. [CrossRef]
- Gill AB, Rundo L, Wan JCM, Lau D, Zawaideh JP, Woitek R, et al. Correlating Radiomic Features of Heterogeneity on CT with Circulating Tumor DNA in Metastatic Melanoma. Cancers (Basel). 2020;12(12). [CrossRef]
- Heye AK, Culling RD, Valdés Hernández Mdel C, Thrippleton MJ, Wardlaw JM. Assessment of blood-brain barrier disruption using dynamic contrast-enhanced MRI. A systematic review. Neuroimage Clin. 2014;6:262-74.
- Klostranec JM, Vucevic D, Bhatia KD, Kortman HGJ, Krings T, Murphy KP, et al. Current Concepts in Intracranial Interstitial Fluid Transport and the Glymphatic System: Part II—Imaging Techniques and Clinical Applications. Radiology. 2021;301(3):516-32. [CrossRef]
- Choi JJ, Reich CF, 3rd, Pisetsky DS. The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology. 2005;115(1):55-62.

| Histopathology | Biopsy source | Tumoral Content | Molecular alteration studied | Isolation Technique | Application/Findings |
|---|---|---|---|---|---|
| DMG/DIPG/HGG | |||||
| [47] | Plasma, CSF, cystic fluid | ctDNA | H3K27M, IDH1, BRAF, MYCN | ddPCR | Increased cfDNA concentrations was associated with shorter time to progression in DIPG and conversely, better survival in HGG patients, tumor-specific DNA alterations more readily identified in CSF than plasma |
| [48] | CSF, Plasma, cystic fluid | ctDNA | H3K27M | ddPCR | Assessing response to radiotherapy and recurrence |
| [49] | CSF | ctDNA | H3K27, H3.3G34 |
PCR | Detecting mutations |
| [50] |
CSF, blood | ctDNA, cfDNA | H3K27 | ddPCR | Predicting recurrence prior to imaging, predicting response to therapy, differentiating progression and pseudo-progression |
| Medulloblastoma | |||||
| [12] | CSF | ctDNA | CTNNB1, SUFU, KMT2D, CREBBP, KBTBD4, PT53, DDX3X, PTCH1 KDM6A |
qPCR | Detection of different methylation patterns, metastasis status, correlation with tumor burden and location, prediction of disease progression, evolution of the genome in response to therapy |
| [51] | CSF, blood | ctDNA | KMT2D, KMT2C, SMARCA4, BCOR, TP53, PTCH1, EP300, NF1, SETD2, MED12, SPEN |
qPCR |
ctDNA correlated with disease progression and metastasis; tumor-specific alterations detected more frequently in CSF than tumor tissue |
| [52] | CSF | ctDNA | TP53, PTEN, PTCH1, BCOR mutation, 17p deletion | qPCR | Assessing minimal residual disease and tumor evolution, Identifying intra- and intertumoral heterogeneity |
| [53] | CSF | cfDNA | CpG methylation | qPCR | Detecting tumor and its subtype, monitoring treatment response and recurrence |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





