Submitted:
19 September 2023
Posted:
19 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
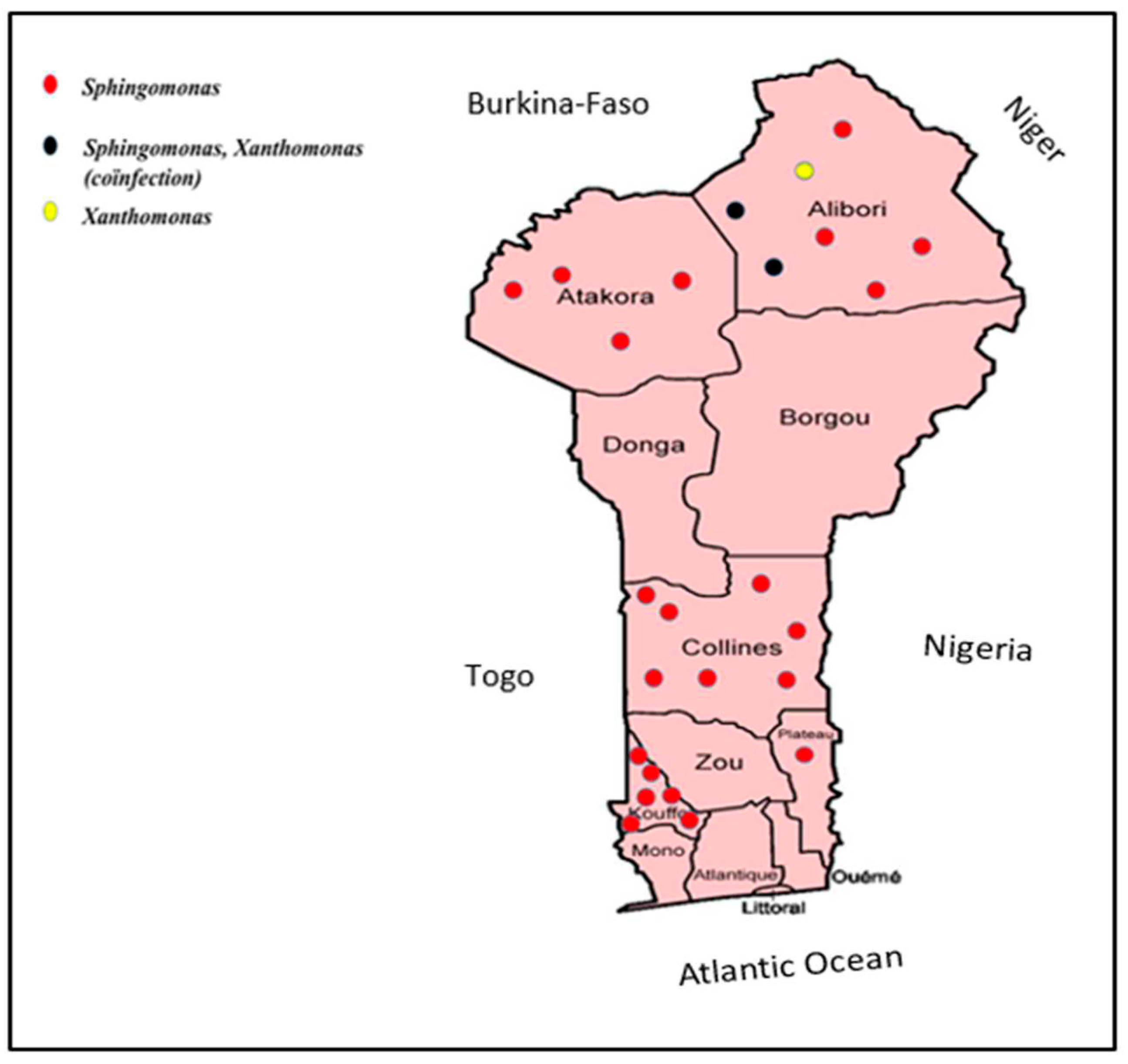
2.1. Study Area and collection of diseased leaves
| No | Accession code | Department/Town/Village | No | Accession code | Department/Town/Village |
|---|---|---|---|---|---|
| 1 | Bagou 18 | Alibori/Gogounou/Bagou | 46 | Nana30 | Atacora/Cobly/Nanagadé |
| 2 | Gou10 | Alibori/Karimama/Gouroubéri | 47 | Bagou 21 | Alibori/Gogounou/Bagou |
| 3 | Tchaka38 | Atacora/Touncoutouna/Tchakalakou | 48 | Koung65 | Atacora/Wassa Pehonco/Koungarou |
| 4 | Kotch70 | Atacora/Tanguiéta/Kotchessi | 49 | Doko122 | Atlantique/Abomey-Calavi/Dokomey |
| 5 | Tchaka41 | Atacora/Touncoutouna/Tchakalakou | 50 | Kik96 | Donga/Bassila/Kikélé-Lokpa |
| 6 | Kan58 | Atacora/Matéri/Kankini-Séri | 51 | Nippon bar | Alibori/Kandi/Angaradébou |
| 7 | Bagou 25 | Alibori/Gogounou/Bagou | 52 | Ang 6 | Alibori/Kandi/Angaradébou |
| 8 | Gami74 | Borgou/Bembèrèkè/Gamia | 53 | Bagou 23 | Alibori/Gogounou/Bagou |
| 9 | Nana32 | Atacora/Cobly/Nanagadé | 54 | Bagou 28 | Alibori/Gogounou/Bagou |
| 10 | Tchaka33 | Atacora/Touncoutouna/Tchakalakou | 55 | Ang 2 | Alibori/Kandi/Angaradébou |
| 11 | Bagou 19 | Alibori/Gogounou/Bagou | 56 | Tog 5307 | Alibori/Kandi/Angaradébou |
| 12 | Okouta98 | Collines/Bantè/Okouta-Ossè | 57 | Koum53 | Atacora/Boukoumbé/Koumadogou |
| 13 | Kan61 | Atacora/Matéri/Kankini-Séri | 58 | IR64 | Atlantique/Zè/Awokpa |
| 14 | Gou11 | Alibori/Karimama/Gouroubéri | 59 | Gami76 | Borgou/Bembèrèkè/Gamia |
| 15 | Bagou 17 | Alibori/Gogounou/Bagou | 60 | Koud43 | Atacora/Natitingou/Koudengou |
| 16 | Bori84 | Borgou/N’dali/Bori | 61 | Agbab 101 | Collines/Savè/Agbaboué |
| 17 | Koud45 | Atacora/Natitingou/Koudengou | 62 | Gou12 | Alibori/Karimama/Gouroubéri |
| 18 | Kpatab100 | Collines/Savalou/Kpataba | 63 | Ang 16 | Alibori/Kandi/Angaradébou |
| 19 | Koum54 | Atacora/Boukoumbé/Koumadogou | 64 | Tchaka39 | Atacora/Touncoutouna/Tchakalakou |
| 20 | Bagou 22 | Alibori/Gogounou/Bagou | 65 | Koud42 | Atacora/Natitingou/Koudengou |
| 21 | Kan60 | Atacora/Matéri/Kankini-Séri | 66 | Foun15 | Alibori/Banikoara/Founougo |
| 22 | Kan59 | Atacora/Matéri/Kankini-Séri | 67 | Tot82 | Borgou/Nikki/Totorou |
| 23 | Bagou 26 | Alibori/Gogounou/Bagou | 68 | Nana29 | Atacora/Cobly/Nanagadé |
| 24 | Kotch71 | Atacora/Tanguiéta/Kotchessi | 69 | Bori83 | Borgou/N’dali/Bori |
| 25 | Bagou 24 | Alibori/Gogounou/Bagou | 70 | Kotch73 | Atacora/Tanguiéta/Kotchessi |
| 26 | Koum47 | Atacora/Boukoumbé/Koumadogou | 71 | Moroberekan | Atacora/Tanguiéta/Kotchessi |
| 27 | Koud46 | Atacora/Natitingou/Koudengou | 72 | Koum55 | Atacora/Boukoumbé/Koumadogou |
| 28 | Man118 | Mono/Houéyogbé/Manonkpon | 73 | Koud 44 | Atacora/Natitingou/Koudengou |
| 29 | Tchaka36 | Atacora/Touncoutouna/Tchakalakou | 74 | Koum 51 | Atacora/Boukoumbé/Koumadogou |
| 30 | Bagou 27 | Alibori/Gogounou/Bagou | 75 | IR841 | INRAB |
| 31 | Koum49 | Atacora/Boukoumbé/Koumadogou |
Positif controls |
IRBB60 | |
| 32 | NERICA19 | Atacora/Matéri | IRBB5 | ||
| 33 | Okouta97 | Collines/Bantè/Okouta-Ossè | IRBB21 | ||
| 34 | Koung69 | Atacora/Wassa Pehonco/Koungarou | |||
| 35 | ONK93 | Donga/Djougou/Onklou | |||
| 36 | Kotch72 | Atacora/Tanguiéta/Kotchessi | |||
| 37 | Tchal89 | Donga/Ouaké/Tchalinga | |||
| 38 | Dev116 | Couffo/Dogbo/Dévé | |||
| 39 | Koung67 | Atacora/Wassa Pehonco/Koungarou | |||
| 40 | Ang1 | Alibori/Kandi/Angaradébou | |||
| 41 | Koum50 | Atacora/Boukoumbé/Koumadogou | |||
| 42 | ONK93b | Donga/Djougou/Onklou | |||
| 43 | Gami77 | Borgou/Bembèrèkè/Gamia | |||
| 44 | Bagou 20 | Alibori/Gogounou/Bagou | |||
| 45 | 6R2B9 | Alibori/Gogounou/Bagou | |||
2.2. Sowing and sampling of rice leaves for DNA extraction.
2.3. DNA Extraction from collected leaves showing BLB symptoms and from leaves of cultivated rice accessions of the laboratory of molecular biology and bioinformatics applied to genomic.
2.4. Control of total genomic DNA quality by electrophoresis
2.5. Polymerase chain reaction (PCR) design methodology for Xanthomonas oryzae pv Oryzae identification.
2.6. Molecular screening of rice accessions cultivated in Benin for the detection of resistance genes to bacterial leaf blight caused by Xanthomonas oryzae pv Oryzae
2.7. Electrophoresis of PCR products
2.8. Data analysis
3. Results and discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sere, Y.; Onasanya, A.; Verdier, V.; Akator, K.; Ouedraogo, L.S.; Segda, Z.; Mbare, M.M.; Sido, A.Y.; Baso., A. Rice bacterial leaf blight in West Africa: preliminary studies on disease in farmer’s field and screening. Asian J. Plant Sci. 2005, 4, 577–579. [Google Scholar] [CrossRef]
- Gonzalez, C.; Szurek, B.; Manceau, C.; Mathieu, T.; Séré, Y.; Verdier, V. “Molecular and Pathotypic Characterization of New Xanthomonas oryzae Strains from West Africa.”. Mol. Plant Microbe Interact. 2007, 20(5), 534–546. [Google Scholar] [CrossRef]
- FAO; 2020. Available: www.fao.org com. Accessed on 05/12/2023.
- Pandey, M. K.; Shobha R., N.; Sundaram, R.; Laha, G.; Madhav, M.; Srinivasa R., K.; Sudharshan, I.; Hari, Y.; Varaprasad, G.; Subba R., L. Improvement of two traditional Basmati rice varieties for bacterial blight resistance and plant stature through morphological and marker-assisted selection. Mol. Breed. 2013, 31, 239-246. [Google Scholar] [CrossRef]
- Afolabi, O.; Amoussa, R.; Bilé, M.; Oludare, A.; Gbogbo, V.; Poulin, L.; Koebnik, R.; Szurek, B.; Silué, D. First Report of Bacterial Leaf Blight of Rice Caused by Xanthomonas oryzae pv. oryzae in Benin. Plant Dis. 2016, 100, 515. [Google Scholar] [CrossRef]
- Islam, Md. R.; Alam, Md. S.; Khan, A. I.; Hossain, I.; Adam, L. R.; Daayf, F. “Analyses of Genetic Diversity of Bacterial Blight Pathogen, Xanthomonas oryzae pv. oryzae Using IS1112 in Bangladesh.” C.R. Biologies 2016, 399–407.
- Mundt, C.C. “Durable Resistance: A Key to Sustainable Management of Pathogens and Pests.”Infect. Genet. Evol. 2014, 27 446–55. [CrossRef]
- Verdier, V. Characterization of new races of Xanthomonas oryzae pv. Oryzae in Mali informs resistance gene deployment. Phytopathology 2020, 110(2), 267-277.
- Gu, K.; Yang, B.; Tian, D.; Wu, L.; Wang, D.; Sreekala, C.; Yang, F.; Chu, Z.; Wang, G. L.; White, F. F.; Yin, Z. R. gene expression induced by a type-III effector triggers disease resistance in rice. Nature 2005, 435, 1122–1125. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.; Fu, B.; Yang, H.; Xu, C.; Li, Z; Sanchez, A. Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor. Appl. Genet 2006, 12, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Mohiuddin, M.S.; Rao, Y.P.; Mohan, S.K.; Verma, J.P. Role of Leptocorisaacuta Thun in the spread of bacterial blight of rice. Curr. Sci. 1976, 45, 11, p 426 427. [Google Scholar]
- Furuya, N.; Taura, S.; Goto., T.; Thuy, B.T.; Ton, P.H.; Tsuchiya, K.; Yoshimura, A. Diversity in virulence of Xanthomonas oryzae pv. oryzae from Northern Vietnam. Jpn. Agric. Res. Q. 2012, 46(4), 329–338. [Google Scholar] [CrossRef]
- Petpisit, V.; Khush, G.S.; Kauffman, H.E. Inheritance of Resistance to Bacterial-Blight in Rice. Crop Sci. 1977, 17, 551–554. [Google Scholar] [CrossRef]
- Blair, M. W.; Garris, A. J.; Iyer, A. S.; Chapman, B.; Kresovich, S.; McCouch, S.R. 2003: High-resolution genetic mapping and candidate gene identification at the xa5 locus for 44 bacterial blight resistance in rice (Oryza sativa L.). Theor. Appl. Genet. 2003, 107, 62–73. [Google Scholar] [CrossRef]
- Jiang, G.H.; Xia, Z.H.; Zhou, Y.L.; Wan, J.; Li, D.Y.; Chen, R.S.; Zhai, W.X.; Zhu, L.H. Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAγ1. Mol. Genet. Genom 2006, 275(4), 354–366. [Google Scholar] [CrossRef]
- Chen, X.; Liu, P.; Mei, L.; He, X.; Chen, L.; Liu, H.; Shen, S.; Ji, Z.; Zheng, X.; Zhang, Y. (2021. Xa7, a new executor R gene that confers durable and broad-spectrum resistance to bacterial blight disease in rice. Plant Comm. 2021, 2(3), 100143. [Google Scholar] [CrossRef]
- Dossa, G. S.; Quibod, I.; Atienza-Grande, G.; Oliva, R.; Maiss, E.; Vera Cruz, C.; Wydra, K. Rice pyramided line IRBB67 (Xa4/Xa7) homeostasis under combined stress of high temperature and bacterial blight. Sci. Rep 2020, 10(1), 683. [Google Scholar] [CrossRef] [PubMed]
- Khan, M. A.; Naeem, M.; Iqbal, M. Breeding approaches for bacterial leaf blight resistance in rice (Oryza sativa L.), current status and future directions. Eur. J. Plant Pathol. 2014, 139, 27-37. [Google Scholar] [CrossRef]
- Vikal, Y.; Bhatia, D. Genetics and genomics of bacterial blight resistance in rice. Advances in international rice research, 2017,175-213.
- Moumouni, B. Le flétrissement bactérien du riz au Niger : Diversité pathologique d’isolats collectés sur les périmètres irrigués. J. appl. biosci, 2007, 38, 2551-2563. [Google Scholar]
- Priya, L.B.; Ujjal, K.N.; Sharmistha, G.; Gayatri, G.; Shalim, U.; Omar, M.A.; Arafat, A.H.; Alison, M.L.; Yong-Ming, G.; Akbar, H. Introgression of bacterial blight resistance genes in the rice cultivar ciherang: Response against Xanthomonas oryzae pv. oryzae in the F6 generation. Plants 2021, 10, 2048. [Google Scholar]
- Djedatin, G.; Nanoukon, C.; Missihoun, A.; Lomou, M.; Sedah, P.; Agbangla, C. Molecular Identification of Xa4 Resistance Gene to Xanthomonas oryzae pv. Oryzae in Cultivated Rice in Northwest Benin. Asian J. Agric. Res 2022, 15(4), 11–22. [Google Scholar] [CrossRef]
- Lang, J M.; Hamilton, J. P.; Diaz, M. G.Q.; Van Sluys, M.A.M.; Burgos, R.G.; Vera Cruz, C.M.; Buell, Tisserat, C.R., Leach, N.A. “Genomics-Based Diagnostic Marker Development for Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola.” Plant Dis, 2010, 94 (3): 311–19.
- Kini, K.; Agnimonhan, R.; Dossa, R.; Soglonou, B.; Gbogbo, V.; Ouedraogo, I.; Kpemoua, K.; Traoré, M.; Silue, D. First report of Sphingomonas sp. causing bacterial leaf blight of rice in Benin, Burkina Faso, The Gambia, Ivory Coast, Mali, Nigeria, Tanzania and Togo. New Dis. Rep. 2027, 35, 32. [Google Scholar] [CrossRef]
- Fanou, A. A.; Missihoun,A.A.; Sovegnon,P.; Behoundja-kotoko,O.; Baimey, H.; Agbangla C. Molecular genetic identification of viruses affecting pepper crop (Capsicum spp.) in Western North of Benin. Int. J. Curr. Res. Biosci. Plant Biol. 2019, 6(3), 9-14. [CrossRef]
- Chen, X.; Temnykh, S.; Xu, Y.; Cho, Y.; McCouch, S. Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor. Appl. Genet, 2017, 95, 553-567. [Google Scholar] [CrossRef]
- Porter, B.; Chittoor, J.; Yano, M.; Sasaki, T.; White, F. Development and mapping of markers linked to the rice bacterial blight resistance gene Xa7. Crop Sci, 2003, 43(4), 1484-1492. [CrossRef]
- Hajira, S. K.; Sundaram, R. M.; Laha, G. S.; Yugander, A.; Balachandran, S. M.; Viraktamath, B. C.; Sujatha, K.; Balachiranjeevi, C. H.; Pranathi, K.; Anila, M. A single-tube, functional marker-based multiplex PCR assay for simultaneous detection of major bacterial blight resistance genes Xa21, xa13 and xa5 in rice. Rice Sci, 2016, 23(3), 144-151. [CrossRef]
- Yan-Chang, L.; Shou-hai, W.; Cheng-quan, L.; Shuang, W.; De-zheng, W.; Shi-yun, D. Improvement of resistance to bacterial blight by marker-assisted selection in a wide compatibility restorer line of hybrid rice. Rice Sci 2004, 11(5-6), 231. [Google Scholar]
- Ullah, I.; Jamil, S.; Iqbal, M.; Shaheen, H.; Hasni, S.; Jabeen, S.; Mehmood, A.; Akhter, M. Detection of bacterial blight resistance genes in basmati rice landraces. Genet Mol Res 2012, 11(3), 1960–1966. [Google Scholar] [CrossRef] [PubMed]
- Sabar, M.; Bibi, T.; Farooq, H. U.; Haider, Z.; Naseem, I.; Mahmood, A.; Akhter, M. Molecular screening of rice (Oryza sativa L.) germplasm for Xa4, xa5 and Xa21 bacterial leaf blight (BLB) resistant genes using linked marker approach. Afr. J. Biotechnol 2016, 15(41), 2317–2324. [Google Scholar]
- Kamhun, W.; Pheng-am, S.; Uppananchai, T.; Ratanasut, K.; Rungrat, T. Effects of nitrogen levels on sucrose content, disease severity of Xanthomonas oryzae pv. Oryzae and yield of hybrid rice (BC4F5). Agric. Nat. Resour. 2022, 56(5), 909–916. [Google Scholar]
- Dossa, G. S. , Oliva, R., Maiss, E., Vera Cruz, C., & Wydra, K. High temperature enhances the resistance of cultivated African rice, Oryza glaberrima, to bacterial blight. Plant Dis. 2016, 100(2), 380–387. [Google Scholar] [CrossRef]
- Ullah, I. , Jamil, S., Iqbal, M., Shaheen, H., Hasni, S., Jabeen, S., Mehmood, A., & Akhter, M. Detection of bacterial blight resistance genes in basmati rice landraces. Genet Mol Res. 2012, 11(3), 1960–1966. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.P.; Jeung, J.U.; Noh, T.H.; Cho, Y.C.; Park, S.H.; Park, H.S.; Shin, M.S.; Kim, C.K.; Jena, K. K;. Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice, 2013, 6, 1-11. [Google Scholar] [CrossRef] [PubMed]
- Ronald, P. C.; Albano, B.; Tabien, R.; Abenes, L.; Wu, K.; McCouch, S.; Tanksley, S. D. Genetic and physical analysis of the rice bacterial blight disease resistance locus, Xa21. Mol. Genet. Genom. 1992, 236, 113-120. [Google Scholar] [CrossRef]
- Gonzalez, C.; Szurek, B.; Manceau, C.; Mathieu, T.; Séré, Y.; Verdier, V. Molecular and pathotypic characterization of new Xanthomonas oryzae strains from West Africa. Mol. Plant Microbe Interact. 2007, 20(5), 534–546. [Google Scholar] [CrossRef]
- Ramalingam, J.; Raveendra, C.; Savitha, P.; Vidya, V.; Chaithra, T. L.; Velprabakaran, S.; Saraswathi, R.; Ramanathan, A.; Arumugam Pillai, M. P.; Arumugachamy, S. Gene pyramiding for achieving enhanced resistance to bacterial blight, blast, and sheath blight diseases in rice. Front. Plant Sci., 2020, 11, 591457. [Google Scholar] [CrossRef]
- Krishnakumar, R.; Kumaravadivel, N. Marker-assisted selection for biotic stress (Bacterial leaf blight and gall midge) tolerance in Bc4F4 generation of rice (Oryza sativa L.). Electron. J. Plant Breed. 2018, 9(1), 275–282. [Google Scholar] [CrossRef]
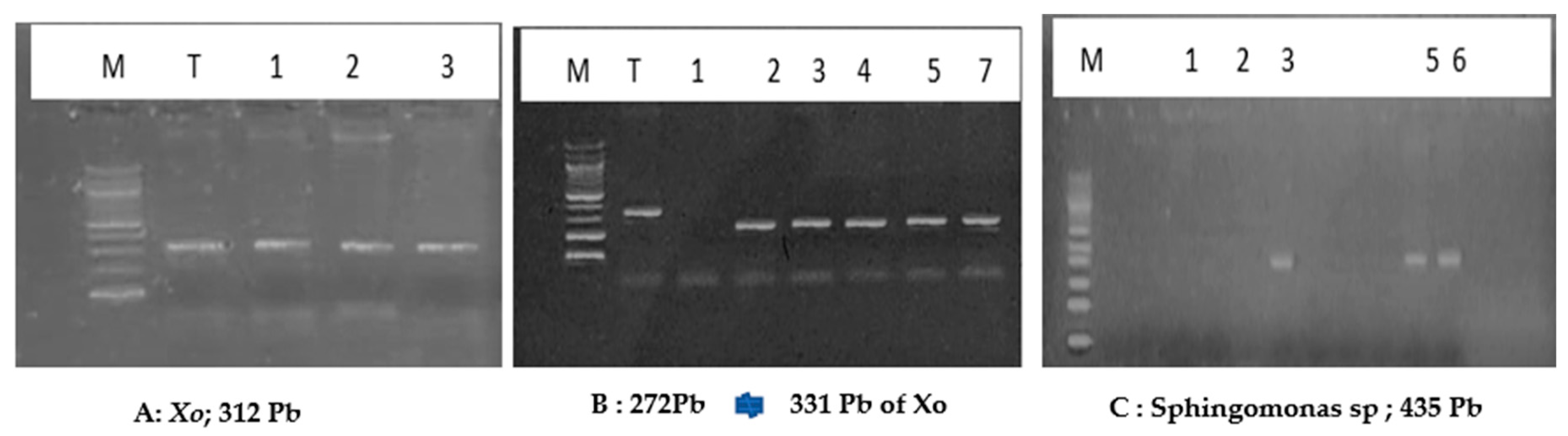
| Target genes | Primers sequence (5’-3’) | Size (bp) | References | |
|---|---|---|---|---|
| Hypothetical protein (X. orysae) | F | CATCGTTAGGACTGCCAGAAG | 331 | [23] |
| R | GTGAGAACCACCGCCATCT | |||
| Hypothetical protein (X. oryzae pv. Oryzae) | F | GCCGCTAGGAATGAGCAAT | 162 | [24] |
| R | GCGTCCTCGTCTAAGCGATA | |||
| Sphingomonas sp | F | CGGCTGCTAATACCGGATGAT | 435 | [24] |
| R | AGGCAGTTCTGGAGTTGAGC | |||
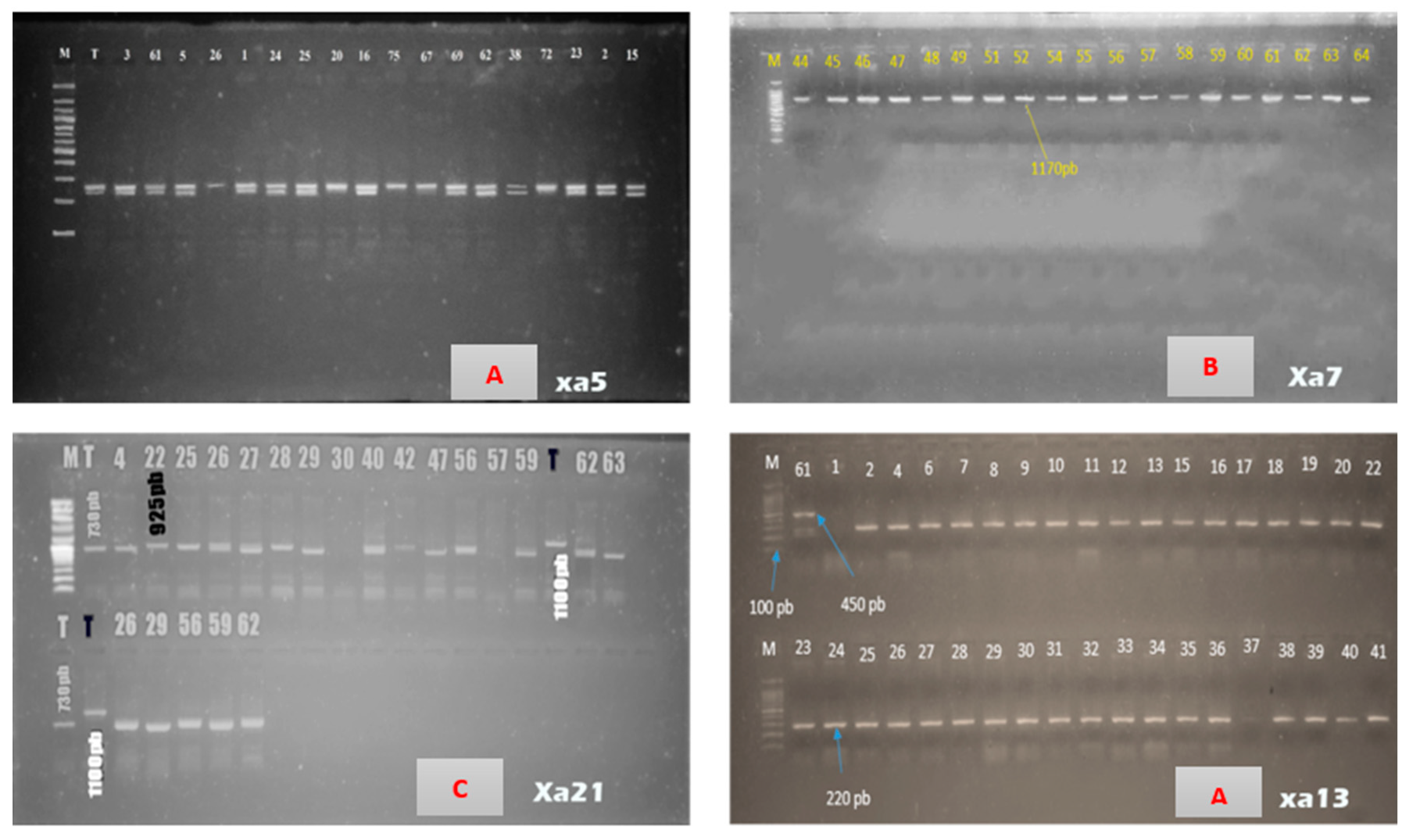
| Genes | chromosome | Marqueur types / Nom | Primers sequence (5’-3’) | Resistance allele (pb) |
Susceptibility allele (pb) | References | |
|---|---|---|---|---|---|---|---|
| xa5 | 5 |
STS/ RM 122 |
F | GAGTCGATGTAATGTCATCAGTGC | 240pb | 230pb | [26] |
| R | GAAGGAGGTATCGCTTTGTTGGAC | ||||||
| Xa7 | 6 | STS/M5 | F | CTGGATACGGAACCTTCTAAC | 294pb | 1170pb | [27] |
| xa13 | 8 | STS/xa13-prom | R | AGAGAACCTTCTCCTTCAGTG | |||
| F | GGCCATGGCTCAGTGTTTAT | 450pb | 220pb | [28] | |||
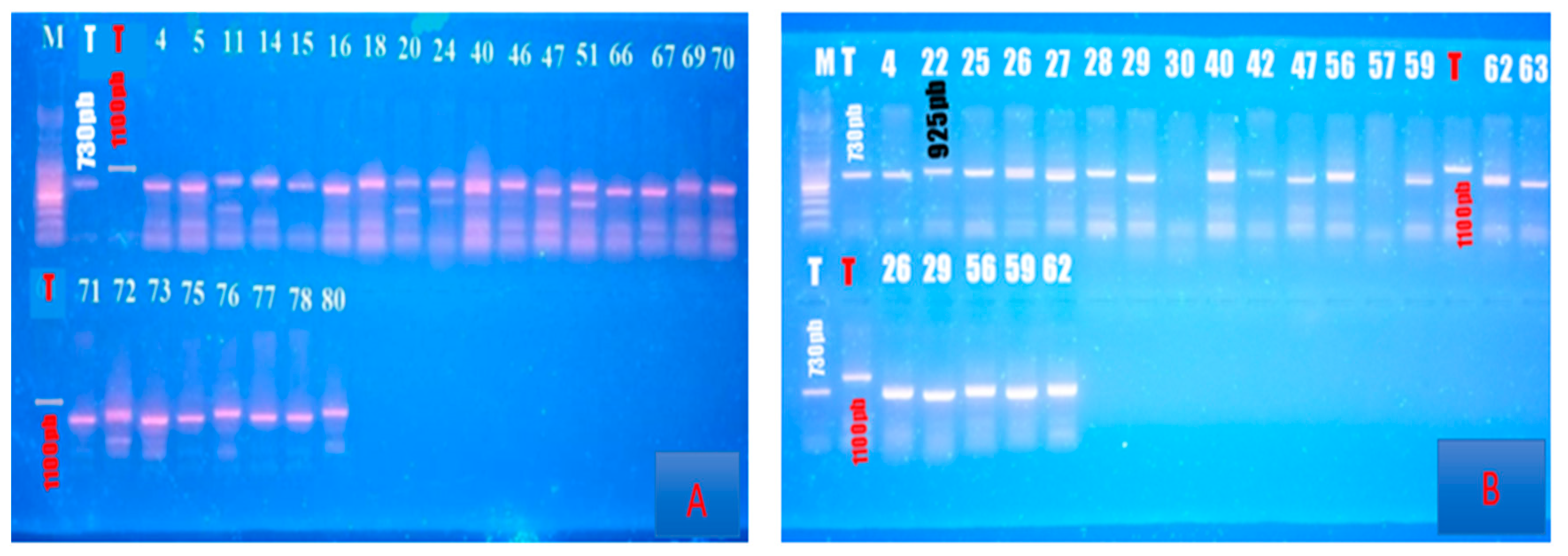
| Xa21 | 11 | STS/ pTA248 | R | GAGTCCAGCTCTCCAAATG | |||
| F | AGACGCGAAGGGTGGTTCCCGA | 925pb | 730pb | [29] | |||
| R | AGACGCGGTAATCGAAGATGAAA | ||||||
| xA5 | Xa21 | Xa7 | xa13 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Department | Samples | Resistanceallele (240pb) | Susceptibility allele (230pb) |
Genotypes | Resistance allele(1100pb) |
Resistance allele (925pb) |
Susceptibility allele (730pb) |
Genotypes | Resistance Allele (294pb) |
Susceptibility Allele (1170pb) |
Genotypes | Resistance allele (450pb) |
Susceptibility allele (220pb) |
Genotypes |
| Alibori | Bagou 18 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 |
| Gou 10 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Bagou 25 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Bagou 19 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Gou 11 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Bagou 17 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Bagou 22 | + | - | xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Bagou 26 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Bagou 24 | + | - | xa5/xa5 | - | + | + | Xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Bagou 27 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Ang 1 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Bagou 20 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Bagou 21 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Ang 6 | + | + | Xa5/xa5 | - | + | + | Xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Bagou 23 | + | + | Xa5/xa5 | - | + | + | Xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Bagou 28 | - | - | Ø | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Ang 2 | + | + | Xa5/xa5 | - | + | + | Xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koum 53 | - | + | Xa5/xa5 | - | + | + | Xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koud 43 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Gou 12 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Ang 16 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Foun 15 | + | - | xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Borgou | Gami 74 | + | - | xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 |
| Bori 84 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Gami 77 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Gami 76 | + | + | Xa5/Xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Tot 82 | + | - | xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Bori 83 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Atacora | Koum 51 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 |
| Tchaka 38 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Kotch 70 | + | - | xa5/xa5 | - | - | + | xa21/xa21 | + | xa7/xa7 | - | + | Xa13/Xa13 | ||
| Tchaka 41 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Kan 58 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Nana 32 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Tchaka 33 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Kan 61 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koud 45 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koum 54 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Kan 60 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Kan 59 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Kotch 71 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koum 47 | + | + | Xa5/xa5 | - | + | + | Xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koud 46 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | ||
| Tchaka 36 | + | + | Xa5/xa5 | - | - | - | ϕ | - | + | xa7/xa7 | - | + | ||
| Koung 69 | - | + | Xa5/Xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | ||
| Kotch 72 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koung 67 | + | + | Xa5/xa5 | - | + | + | Xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koum 50 | - | - | Ø | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Nana 30 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Nana 29 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koud 44 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Kotch 73 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koum 55 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koum 49 | + | + | Xa5/xa5 | - | + | + | Xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koung 65 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| NERICA 19 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Tchaka 39 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Koud 42 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Donga | ONK 93 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 |
| Tchal 89 | - | + | Xa5/Xa5 | - | + | + | Xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| ONK 93b | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Kik 96 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Colline | Okouta 98 | + | + | Xa5/xa5 | - | + | + | Xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 |
| Kpatab 100 | + | - | xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Okouta 97 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Agbab 101 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Mono | Man 118 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 |
| Couffo | Dev 116 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 |
| Atlantique | Doko 122 | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 |
| 6R2B9 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| IR841 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | ||||||||
| IR64 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | ||||||||
| Moroberekan | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Nippon bar | + | + | Xa5/xa5 | - | + | - | Xa21/Xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
| Tog 5307 | + | + | Xa5/xa5 | - | - | + | xa21/xa21 | - | + | xa7/xa7 | - | + | Xa13/Xa13 | |
|
Positif controls |
IRBB5 | + | - | xa5/xa5 | ||||||||||
| IRBB60 | + | + | Xa5/xa5 | + | - | - | Xa21/Xa21 | + | + | xa13/Xa13 | ||||
| IRBB21 | - | - | + | xa21/xa21 | ||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





