1. Introduction
Passive immunotherapy approach, based on antibodies introduction, started by Emil von Behring and Shibasaburo Kitasato in the late nineteenth century using serum to protect from infectious diseases such as diphtheria and tetanus [
1]. This approach was effective to several infectious diseases however, its use had been reduced by the antimicrobial drugs approvals, and it was restricted to treat venom intoxication and some viral infections [
2]. The technological revolution in the discovery strategies to obtain monoclonal antibodies (mAbs), followed by microbial resistance to certain drugs, opened opportunity to develop passive immunotherapy to a wide variety of infectious diseases without drug approved for the prophylaxis and therapeutics [
2], although few mAbs targeting infectious diseases were approved until now [
3]. There are, up to now, mAbs targeting only 3 viruses, respiratory syncytial virus (RSV), human immunodeficiency virus (HIV-1) and Ebola virus (EBOV) considering only viral infections [
3]. The development of mAb therapy was initially directed to cancer and immunological diseases such as autoimmune disorder due to increased incidences of these conditions and efficient drugs for treatment were unavailable [
2].
Recently, pandemic of severe acute respiratory syndrome coronavirus 2 infection and COVID-19 disease changed this scenario since the rapid spread of this infection and the urgent need to accelerate the development of drugs and vaccines to combat the pandemic. The efforts from the scientific community were intensive and vaccines and antiviral drugs, including neutralizing mAbs, have obtained emergency use authorization from US Food and Drug Administration (FDA) in a record time [
3].
Infection diseases caused by arthropod-borne viruses (arboviruses) are a challenging public health problem in tropical and subtropical developing countries that needs a dynamic state of emergence due to the potential outbreaks [
4]. It is considered as a global medical concern since emerging diseases do not impact only the population of these countries in function of the intensive circulation of the people due to the globalization and tourism [
5]. Furthermore, the dispersion of the (re)emerging diseases is rapid when happens and thus it is important having a continuous effort and support for studies developing drugs and vaccines to control the outbreaks.
It was reported several cases of the human emerging diseases caused by arboviruses in both hemispheres in recent centuries, such as yellow fever virus (YFV), dengue virus (DENV), West Nile virus (WNV), chikungunya virus (CHIKV) and Zika virus (ZIKV) [
5]. Highly effective vaccines for some arboviruses were approved such as 17D YFV [
6], Japanese encephalitis [
7] and tetravalent DENV [
8] and others are under development [
4]. However, antivirals or other therapeutics are not available for clinical use and the development of specific therapeutics such as mAbs are under development. Passive immunotherapy using neutralizing mAbs in animal model-based studies protected from the infection by genus
Flaviviruses [
9] and
Alphaviruses [
10] using specific mAbs. It was observed that mAbs with poor neutralizing activity can also provide protection in animals [
10]. The biggest challenge is to demonstrate the protection and the efficacy in the human therapy.
This review focus on the mAb-based therapeutics for infectious diseases caused by arboviruses, specifically on Aedes-borne RNA arboviruses DENV, ZIKV, CHIKV and WNV which is not Aedes-borne RNA arbovirus. The mAb discovery technologies and improved strategies for the mAb development are highlighted. Firstly, we present an overview of mAb isolation technologies, then neutralizing mAbs have been developed to the arboviruses and outstanding approaches to obtain mAbs with high efficacy and potency will be shown.
2. Technologies for mAb discovery
2.1. Hybridoma technology
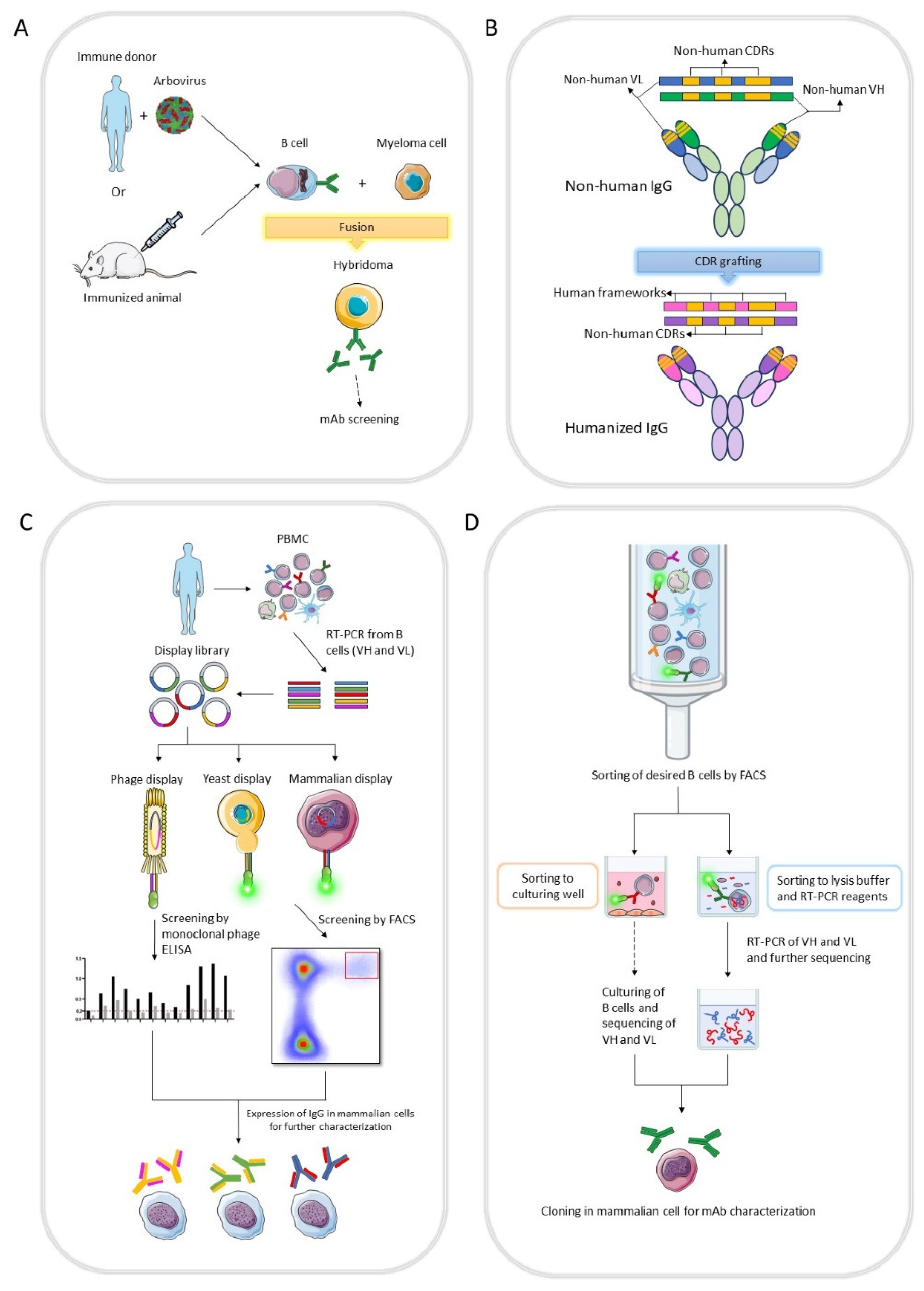
The first technology of mAb production was hybridoma developed by Köhler and Milstein in the 1975 [
11]. Hybridoma technology involves fusing antibody-producing B cells from an immunized animal, mainly mice, with murine myeloma cells resulting in hybrid cells that can produce unlimited quantities of a specific antibody (
Figure 1A). The first therapeutic mAb, Orthoclone OKT3, was approved in 1986 [
12]. However, clinical trials with hybridoma-derived mAbs were largely unsuccessful due to murine mAbs were recognized as heterologous proteins and were highly immunogenic, generating human anti-mouse antibodies (HAMA) that affected the safety and therapeutic efficacy of the antibody [
13,
14].
The mAbs obtained from hybridoma retain the natural gene pairing of variable and constant regions and ensure that they can function properly. Besides, this technology relies on B cells that were matured in secondary lymphatic organs in response to an antigen [
15].
Human hybridoma produces human mAbs by fusion of human B cells and fully human myeloma [
16]. However, the application for the therapeutic purposes has been limited since presented several technical disadvantages. For example, high efficacy human fusion partners are unavailable, the number of B cells is limited, the fusion efficiency is low and the production cost is high (6). Hetero-myelomas were used to generate mAbs against viral diseases such as HIV, CHKV and DENV [
15].
2.2. Antibody humanization
Administration of non-human mAbs in the therapy may function as antigens and therefore, antibodies against them could be elicited. To overcome this problem by reducing heterologous domains or residues in non-human mAbs, antibody humanization techniques were developed, replacing them with human domains or residues to minimize the immunogenicity. The first technique was to obtain chimeric mAbs: antibody variable domains (light chain and heavy chain) from heterologous origin was combined with human constant domains [
17]. The rational choice of constant domains is crucial to obtain chimeric mAbs. Although the variable domains are responsible for the antigen binding, the constant regions may also play a role in this function [
18]. Avidity is the strength of multiple interactions between antibody and antigen to form a stable complex and is another important factor that interferes in binding properties. Chimeric mAbs have altered avidity due to constant domain exchanges [
19] and binding decrease could happen in function of allosteric effects [
20]. The isotype selection could also have impact on the chimeric mAb specificity [
21]. Furthermore, these mAbs triggered immunogenicity responses generating human anti-chimeric antibodies (HACA) [
22].
Then techniques to obtain humanized antibodies were developed to lower immunogenicity response. One promising technique are based on the transplantation of the highly specific sequence of the antibodies called complementarity-determining regions (CDRs) of the non-human origin on human framework sequences using molecular biology approaches and was designated CDR grafting [
23]. The first methodology cloned heterologous CDRs into human frameworks, however, this lead to reduced affinity due to the alterations of critical residues present in heterologous frameworks related to the binding and grafting did not preserve these features [
24,
25]. These residues in the frameworks that affect the conformation of CDR loops and contribute for mAbs characteristics are called Vernier zones [
26]. The back mutation approach to revert these framework residues of humanized mAbs was used in the development of Zenapax® (daclizumab), the FDA approved mAb in 1997 for the therapeutic use in patients with kidney transplantation [
27]. All CDR grafting methodologies rely on the homology between the non-human mAb with human antibodies to decide the suitable human framework for the humanization. Most of humanized antibodies presented reduced affinity toward antigens [
28] and about 9% of them elicited human anti-humanized antibodies (HAHA) [
22].
2.3. In vitro display technology
In vitro display technology is the selection of DNA-binding protein or antibody based on the directed evolution approach [
29]. In the field of the antibody discovery, it mimics the
in vivo process of antibody generation and has four mainly characteristics: (1) the generation of the genotypic diversity by library construction, (2) the linkage between the genotype and the phenotype, (3) the recovery of the DNA sequence encoding the selected antibody and finally (4) the antibody amplification. The isolated clones can be expressed to be characterized and the best candidates are selected [
30] (
Figure 1B). Further characterizations can be performed by IgG production in mammalian cells. Antibody libraries can be derived from either non-immunized (
naïve and synthetic) or immunized donors.
Naïve libraries should have a high diversity, but the selected clones may not have high affinity. On other hand, libraries from immunized have a lower diversity with higher affinity candidates [
31,
32].
The main advantage of this technology is the capability to select antibodies against a wide range of targets and epitopes, since the selection from a
naïve or synthetic antibody repertoire does not rely on an
in vivo immune response. Thus, the isolation of antibodies targeting even self-antigens, toxic, unstable, and non-immunogenic antigens is possible using library from non-immunized donors [
30].
Phage, yeast and mammalian cell surface displays are examples of
in vitro display technologies to select antibody fragments such as the single-chain variable fragment (scFv), fragment antigen-binding (Fab) and single domain antibody (sdAb) [
31,
32,
33].
Phage display was the first technology developed to select exogenous peptides on the filamentous M13 bacteriophage (phage) surface with specific binding properties by Smith et al. in 1985 [
34]. Antibody phage display was developed by McCafferty and colleagues five years later [
35]. It is widely used in the discovery of therapeutic mAbs due to several advantages: low cost of the
E. coli expression system [
36], finding antibodies with desired properties by large
naïve libraries [
31], selection process is versatile with possibility of using diverse conditions [
30], affinity maturation approach can be done to improve binding properties [
36], antibody can be humanized by guided selection technique [
30], and a high level of customization and also full control of the experiment at each stage by direct and rational approach [
36].
2.4. B cell sorting technology
After the revolutionary hybridoma technology and
in vitro display technologies, single B cell sorting technology was developed for mAb discovery. Antibody secreting cells (ASC) and B cells have a very short lifespan
ex vivo, therefore there is a need for the immortalization of these cells e.i. by hybridoma technology, in order to gain time for antibodies screening derived from these cells. Novel advances allowing cell culture of ASC and B cells were developed enabling the screening of a large number of mAbs [
37,
38,
39]. Thus, to recover broadly neutralizing mAbs, an efficient screening approach of a larger number of mAbs is important, given that a minority of B cells have these features.
A well-known method to isolate single B cells is the fluorescence-activated cell sorting (FACS). Surface markers, such as membrane bound antibodies (B cell receptor, BCR) in B cells are a suitable candidate to isolate cells with specific antibodies for a given antigen. The antigen of interest must be attached to a fluorophore and used as a bait to sort the single B cells bearing antibodies. Cells can be sorted into wells for culture, further for mAb screening and accessing of the genetic material [
37,
39] (
Figure 1D) or even prolonging their lifespan through Epstein-Barr virus (EBV) immortalization [
40,
41]. Alternatively, cells can be sorted into wells with reagents for high-throughput sequencing [
42,
43].
While FACS is one of the most commonly used methods for the mAb discovery from single B cells, there are limitations. Plasma cells, a differentiated stage of B cells that are highly specialized in secreting antibodies, do not possess a BCR. However, using FACS for B cells has significant advantages, such as a relatively low cost, simplicity, and a high throughput platform [
44].
Modern technology for single B cell investigation use microfluidic platforms. Berkeley Lights Beacon
TM is an example of a highly efficient and automated microfluidic system that enables rapid interrogation of B cells in a high-throughput manner [
45]. In this technology, B cells are sorted into individual channels using OptoElectro Positioning (OEP), each having a very small volume (usually ranging from 740 pL to 1.7 nL), with culture chips customized for specific needs [
44,
46,
47]. The culture supernatant is further analyzed, and several parameters are assessed related to the mAb and cell clone screening, such as IgG productivity, cell expansion rate, and surface markers [
44,
46,
47]. It represents an example of an open system, where information about ASC can be readily accessed in real time.
Another example of an open microfluidic system is based on microengraved polydimethylsiloxane (PDMS) chips. These chips contain nanowells into which cultured B cells are added and sorted [
48]. After sorting, the ASCs with specific antibodies for a given antigen are analyzed in microarrays, and genetic material can be recovered for further analyses [
48]. Although this methodology enables high-throughput analyses, it does not provide the same level of information as the Berkeley Lights Beacon
TM, although it is a cheaper technology, making it more accessible.
Closed microfluidic systems, also known as closed systems, are based on encapsulating cells in droplets embedded in an oil emulsion. They are referred as closed systems due to the need to break the encapsulated droplet to access the information inside the reaction. The sequencing of individual B cells may be done with the cDNA synthesized inside the droplets [
49,
50]. Other closed microfluidic methodologies do not sort B cells prior to microfluidic encapsulation, instead, they rely on the B cell identification with specific antibodies for antigens of interest within the droplets, and material for sequencing is also obtained within the droplets [
51].
The greatest advantage of microfluidic systems is that they operate with very small volumes, allowing for minimal reagent use, rapid screening of mAbs, and quick achievement of the molar amount of antibody needed for identifying ligands and other characteristics [
44]. Additionally, they can also be employed for high-throughput studies [
44]. However, these systems do not currently support functional analyses, such as neutralization assays, an important characteristic to be assessed for therapeutic mAbs, therefore these assays should be performed after mAb isolation [
44].
The primary advantage of B cell-based technologies for clinical purposes is the preservation of the native pairing of the variable domains of the B cell. Antibodies undergo affinity maturation
in vivo, and those with auto-reactivity undergo clonal deletion [
52]. This mechanism helps minimize the selection of potential autoreactive and non-specific mAbs [
44]. Native paired mAbs may also exhibit lower immunogenicity and have greater developability, a common problem faced by mAbs obtained from libraries that do not retain the native pairing [
44].
3. Development of the therapeutic mAbs targeting arboviruses
3.1. Characterization of the arboviruses
Arboviruses are a diverse group of viruses whose vectors are arthropods, including different families such as
Flaviviridae and
Togaviridae among others. These two families have a worldwide medical importance due to representing human viral pathogens causing several emerging diseases [
53].
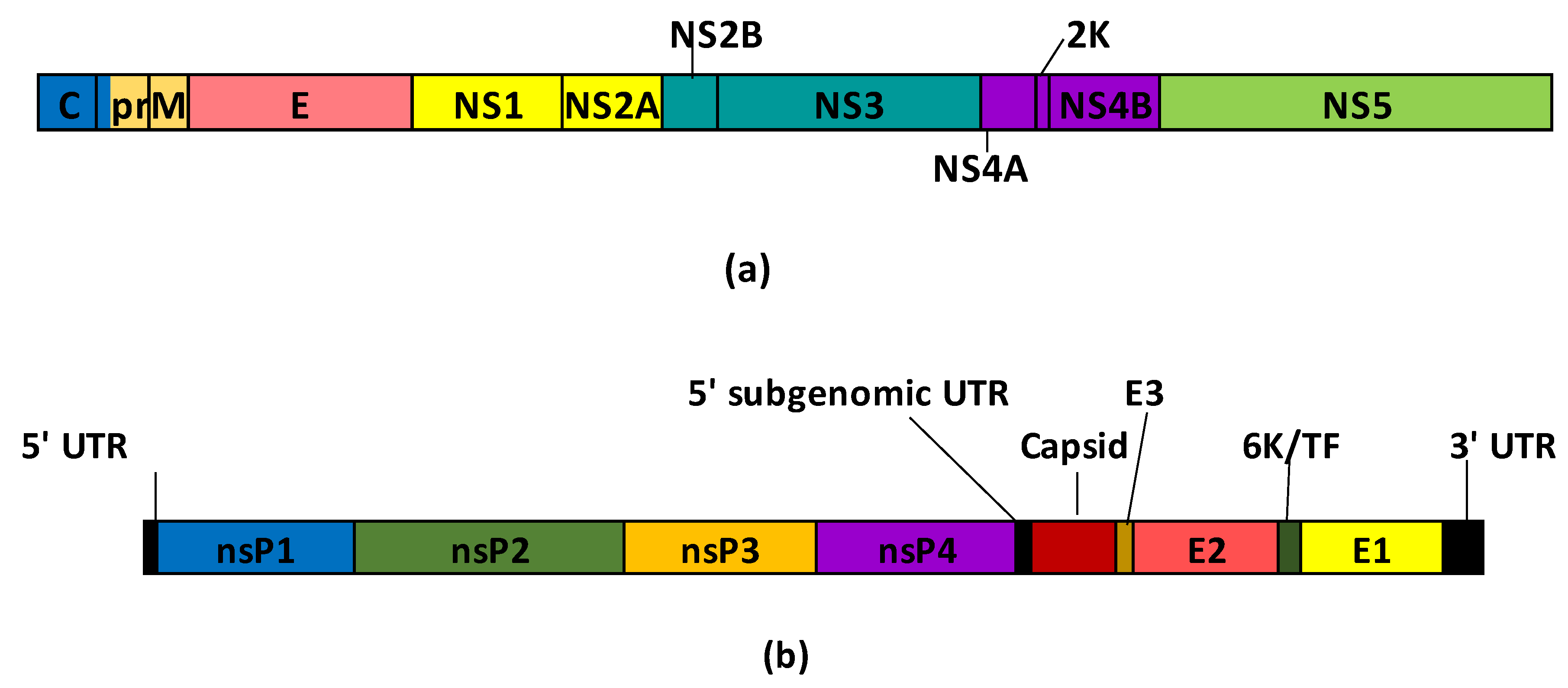
Flaviviruses are spherical particles containing positive-sense RNA that encodes three structural proteins, such as pre-membrane (prM), envelope (E) and capsid (C), and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) [
9,
54] (
Figure 2a). The E protein forms dimeric structure anchoring on the M protein, mediates virus entry and has three domains: DI, DII and DIII [
54]. Flavivirus infected individuals presented neutralizing antibodies to E, prM and NS1 proteins [
55,
56].
Alphaviruses are positive-sense RNA viruses of the
Togaviridae family with an icosahedral nucleocapsid encoding four nonstructural proteins (nsP1, nsP2, nsP3 and nsP4) and five structural proteins (capsid, E3, E2, 6K/TF and E1) [
10] (
Figure 2b). Host humoral antibodies have as target E1 and E2 proteins [
10,
57].
The vector species, such as mosquito or tick, responsible for flavivirus diseases primarily define the epidemiology [
9]. The clinical manifestation which differentiates the diseases caused by two types of the mosquitoes is that
Aedes-borne viruses are characterized mainly by fever, influenza-like symptoms and/or hemorrhagic illness, whereas
Culex-borne viruses such as WNV are encephalitic infections [
5].
Figure 2.
Schematic representation of the polypeptides encoded by flavivirus (a) and alphavirus (b) genomes. Flavivirus scheme was adapted from
Pierson and Diamond 2020 [
54] and alphavirus was adapted from Rupp
et al. [
58].
Figure 2.
Schematic representation of the polypeptides encoded by flavivirus (a) and alphavirus (b) genomes. Flavivirus scheme was adapted from
Pierson and Diamond 2020 [
54] and alphavirus was adapted from Rupp
et al. [
58].
3.2. Dengue (DENV)
Dengue is one of the most common mosquito-borne disease in tropical countries [
59] with four serotypes: DENV1, DENV2 DENV3 and DENV4, and all of them can cause asymptomatic to severe cases, including hemorrhagic fever [
60]. DENV1 is the most prevalent DENV serotype. The infection can elicit long-lasting memory B cells however, serotype cross-reactive antibodies can bind or cause weak DENV neutralization, leading to the virus entrance into cells via FcɣR and causing antibody-dependent enhancement (ADE) [
61,
62]. Besides, non-specific T memory cells lead to cytokine storm and poor outcomes in the second infection, a phenomenon known as original antigenic sin [
63]. These two immunological aspects are an impairment in the development of vaccines for DENV. Currently, Butantan-DV, a tetravalent live-attenuated dengue vaccine [
64], is in phase III clinical trial and registered with ID
NCT02406729 [
65]
. There are also two mAbs for dengue treatment in the clinical trials, both in phase I [
66,
67]
.
The mechanism underling the DENV entry into host cells is not fully understood yet, it is known that the virus concentration gradually increases over the cell surface, leading its entry. The adhesion molecule of dendritic cells (DC-SIGN) such as glycosaminoglycan and the mannose receptor (MR) of macrophages are cell targets for the virus, suggesting a non-specific entry and indicating that the virus has evolved by using as target various cells [
68]. Nevertheless, there is evidence that the E protein mediates virus entry. It is proposed that the DIII of the E protein is involved in the entry into host cells, since mutated DIII was not able to enter cells [
69,
70]. Since the diversity among DENV serotypes, some mAbs may not be neutralized by others [
9].
MAb-based therapies have certain advantages in dengue treatment since most of the antibodies elicited by the infection are non-neutralizing with ADE risk that comes with polyclonal antibodies. Development of the recombinant mAbs are promising drugs without presenting adverse events. As ADE phenomenon is more likely to occurs with serotype specific antibodies that just cross-react with other serotypes [
71], we focused on the DENV antibodies that cross-neutralize different serotypes (
Table 1).
3.2.1. MAbs for DENV
Development of the mAbs targeting specifically DENV1 serotype is urgent since it causes predominant infection. Mice were immunized with UV-inactivated DENV1 particle and recombinant DENV1 E protein, and hybridomas producing DV1-E1 and DV1-E2 mAbs were isolated. The epitope was characterized as the DIII of the E protein, in the region ranging from Gln347 and Asp360 residues, that has important role in the neutralization and it is not a conserved region among other serotypes [
83].
The 4E11 Fab was obtained by the hybridoma technology from mice immunized with DENV4 and has as target the DIII of the E protein. It neutralized all four DENV serotypes with different potency [
72]. The IgG2a format of this mAb neutralized DENV with greater potency than the Fab, indicating that the
in vivo properties of the mAb are desirable for therapeutic purposes [
72].
MAbs were generated by the fusion of the PBMC from DENV2 acute-phase or DENV-infected convalescent donors with SPYMEG cells and reveled that mAbs derived from acute-phase patients had cross-reactivity and neutralization capacity to all four serotypes [
84].
In other approach, mAbs to DENV E protein were identified from two patients during acute-phase secondary infection by single plasmablast sorting [
80]. These mAbs displayed strong neutralization activity against multiple serotypes
in vitro and showed higher protection to the previous serotype infection due to antigenic sin phenomenon [
80].The same group also that used plasmablast sorting and selected SIgN-3C mAb, which binds to a quaternary epitope in inter-dimer interface of the E protein, specifically to the DII fusion loop and DIII [
79]. The epitope of this antibody is extensive, giving the antibody the capacity to cover the inter-dimer region due to the long CDR3 sequence [
79]. SIgN-3C has neutralizing capacity across all four serotypes of DENV. A prophylactic and therapeutic approach was applied to AG129 mice model which can develop high viremia, and showed efficacy for all serotypes reducing significantly the viremia [
79].
Neutralizing mAbs were also isolated by DENV2 E envelope specific memory B cell from individuals recovered from natural infections, and some mAbs presented neutralization potency to more than one serotype [
85]. Memory B cells represents a very small part of PBMC, needing to choose the right period for mAb selection.
Discovery of mAbs targeting diverse epitopes helps the design of effective therapeutic drugs. The d488 is a broadly DENV neutralizing mAb, obtained by the B cell sorting of Rhesus macaques immunized with dengue vaccine, binds to DII at the interface of E and M proteins and it was the first report of this epitope [
82]. This mAb has cross-reactivity and cross-neutralization potency to four DENV serotypes [
82].
The fusion loop of the E protein is the epitope for weak neutralizing antibodies [
75], but potent antibodies targeting this epitope have been selected. The 3G9 mAb is one example obtained by a hybridoma generated from a patient in acute phase of a secondary DENV infection and the variable region presented high somatic hypermutation rates [
75]. This mAb neutralized
in vitro all four serotypes and also JEV, WNV and ZIKV. Besides, 3G9 displayed
in vivo therapeutic potential, increasing survival and reducing the viremia in mice [
75].
Other mAb targeting the fusion loop is 2A10G6, that binds to a new epitope, 98DRXW101 motif, highly conserved among different flavivirus [
73]. This antibody was obtained by a hybridoma using B cells of a mice immunized with DENV2 [
73]. The 2A10G6 showed protection against lethal infection caused by DENV1-4 when administered at the same time of the viral challenge in a suckling model. The effects of 2A10G6 were dose-dependent and gave full protection against DENV2 and partial protection against DENV1, 3, and 4 [
73].
A panel of mAbs targeting a new epitope called envelope dimer epitope (EDE) that connects two E protein subunits were identified by plasmablast sorting from a DENV secondary infected patients with hemorrhagic fever symptoms. These mAbs are broadly reactive capable of neutralizing multiple DENV serotypes [
86].
Epitopes in other DENV proteins can also trigger neutralizing antibodies, as 2E8 and 33D2 mAbs targeting NS1 obtained by murine hybridoma, and presented neutralizing activity against all DENV serotypes [
78]. NS1 is an antigen that can raise protection but it has molecular mimicry with proteins of the mammalian host, and the group synthesized as antigen NS1 wing domain region that it is not recognized by host [
78]. These mAbs were humanized by CDR grafting and
in vivo therapeutic potential was evaluated in a mice model, and both mAbs provided a reduction on DENV-induced prolonged bleeding time and skin hemorrhage and the humanized mAb, h33D2, also decreased the viremia [
76].
3.3. Zika (ZIKV)
Zika is a disease caused by ZIKV, transmitted by
Aedes spp. mosquitoes [
87] and also sexually [
88]. While asymptomatic infections account for around of 80% of cases, the zika symptoms include rash, fever, headaches, and Guillain-Barré syndrome that can be a sequalae [
89]. Importantly, ZIKV shows tropism to immune-privileged sites [
90], such as the brain [
91] and the placenta [
92]. This tropism to the placenta and the ZIKV vertical transmission can lead to abnormalities in brain development [
93,
94] and even spontaneous abortion [
94]. Microcephaly is a common morphologic abnormality, as well as, the brain calcification [
93,
94], but even without apparent morphologic disorders in the brain development, the congenital zika may lead to neurodevelopment impairments in children [
95]. Although there is no FDA approved vaccines for ZIKV, there are currently one mAb for zika treatment in phase I clinical trial [
96].
ZIKV and DENV E protein show similarity around of 56%, as consequence, antibodies triggered by ZIKV infection are widely cross-reactive with DENV [
97]. Similarly to DENV, the depletion of B cells reactive to the soluble ZIKV E protein does not significantly affect the neutralization of ZIKV immune sera, suggesting that the quaternary epitopes, i.e. those present in structural virion arrangement may be responsible for the neutralizing response [
98].
Plasmablasts from ZIKV-infected patients with previous dengue experience showed the clonal expansion and high rates of the somatic hypermutation due to a secondary infection, differing from DENV-naïve ZIKV-infected patients derived plasmablasts. ZIKV neutralizing antibody from dengue experienced-donors developed DENV cross-reactivity response [
99]. In other study, higher levels of ZIKV neutralizing antibodies after ZIKV exposure was associated with their reactivity to DENV1 DIII of the E protein [
100].
Antibodies for DENV, presenting cross-reactivity with ZIKV in sub-optimal concentrations or with poor neutralization potency conducted to ADE in
in vitro models [
101] and also in mouse models [
102]. Since ZIKV circulates in DENV endemic areas, ADE phenomenom happens during infections among these viruses, being of fundamental importance to investigate this characteristic in therapeutic antibodies. The effects of the ZIKV in fetal development is also relevant when therapeutic effects are evaluated and gestational models should be considered.
3.3.1. Therapeutic mAbs for zika
Therapeutic mAbs developed for zika are summarized in
Table 2.
A panel of human DENV mAbs that binds to E protein was identified and presented in DENV section [
86]. Some potent and neutralizing DENV mAbs targeting envelope dimer epitope (EDE) [
86] presented cross-reactivity with ZIKV and inhibited ADE of ZIKV infection [
101]. EDE1-B10 has therapeutic potential to DENV and ZIKV, since inhibited virus strains and protected ZIKV-infected mice against lethality. Besides this mAb reduced viral persistence in immune-privileged tissues, such as brain or testicles, it protected animals against tissue injury and also the transmission to the fetus [
109]. In rhesus monkey study, EDE1-B10 presented efficacy for therapeutic and prophylaxis against ZIKV [
110].
SIgN-3C is another DENV mAb binding to a quaternary epitope, mentioned on the DENV section, that showed
in vivo efficacy to zika [
111]. In non-pregnant mice, mAb administration exhibited therapeutic effects against lethal infection, viremia and weight loss [
111]. In pregnant mice infected on embryonic day 10.5, mAb administration resulted in normal fetal weight and reduced viral load in amniotic fluid and organs, showing potential therapeutic and prophylactic effects [
111].
ZIKV-117 is a representative potent mAb against ZIKV that bind to a quaternary epitope in the interdimer interface of the protein E. It was obtained by hybridoma fusing B cells from a convalescent patient that was selected after deeper serum response investigation [
103]. ZIKV-117 neutralized ZIKV strains and failed to neutralize DENV all serotypes. When administrated for mice treatment, ZIKV-117 reduced zika lethality and in the prophylactic approach, the post-exposure therapy in mice dams it was observed significant viral burden decrease in placenta, maternal tissues, serum and fetus tissues and also the reduction of the fetal demise [
103]. Besides, no mutant escapes were detected after six passages in the presence of ZIKV-117 [
103]. ZIKV-117 was formulated as lipid nanostructures with the mRNA encoding the mAb and a robust protection was shown in unpregnant mice [
105].
Z23 and Z3L1 mAbs were isolated from memory B cells sorted from a convalescent zika patient and recognizes different tertiary epitopes in the E protein [
108]. Z23 binding site is at the top of DIII, while Z3L1 binds to an epitope ranging from DI and DII [
108]. These mAbs strongly neutralize ZIKV with no cross-reactivity with all four DENV serotypes and mice treated with each mAb were completely protected against ZIKV infection without the weight loss [
108].
Anti-NS1 antibodies represent an alternative approach to protect against ZIKV. A panel of ZIKV murine and human mAbs targeting NS1 were identified through hybridomas secreting murine mAbs [
104] or B cell sorting of ZIKV-infected donors presented previously [
103]. 749-A4 mAb was isolated by plasmablast sorting from a dengue patient, showing cross-reactivity with ZIKV NS1 and was included in the study [
104]. Murine Z17, human ZIKV-292 and human 794-A4 mAbs recognize NS1 residues in C-terminal region of the β-platform and gave protection in immunocompromised mice receiving lethal ZIKV challenge and the viral RNA level was reduced in the fetuses of the pregnant mice [
104]. Z15 and Z17 are murine mAbs with different epitopes and the cocktail approach showed reduced viral RNA in fetal brain however, it was not statistically significant [
104].
An approach to avoid escape mutants in zika is the administration of a combination of two or more mAbs targeting distinct epitopes. Z004 mAb was selected from memory B cell sorting of ZIKV-infected patients, presenting reactivity to ZIKV and DENV-1, and targets the lateral ridge of DIII of the E protein with potent neutralizing activity
in vitro. It also protected mice and decreased symptoms and the lethality when given pre- and post-infection [
100]. High dose ZIKV challenge in rhesus macaques was tested and showed prolonged viremia and conducted to escape virus mutants. Z021 mAb was isolated from the same B cell sorting strategy as Z004, with similar characteristics of this mAb related to reactivity, neutralizing potency
in vitro and
in vivo, however, Z021 recognizes EDIII nearby or overlapping epitope by Z004 [
107]. When Z004 and Z021 were administrated as a cocktail in monkeys before receiving high dose ZIKV challenge, the infection was delayed and the viremia was reduced, preventing ZIKV escape mutants [
107].
Therapeutical antibodies are usually IgG, but memory B cell sorting of an pregnant patient with secondary ZIKV infection has yielded the ultrapotent DH1017.IgM targeting a quaternary epitope in the E dimer whitin DII [
40]. All variable domains of the DH1017.IgM could bind to the same virion, but neutralization seem to occur due to the viral aggregation [
40]. The neutralization of this mAb is dependent on isotype, since IgG was expressed and did not neutralize ZIKV and induced ADE, besides, protected more efficiently mice from the infection and the viremia was reduced [
40].
Development of new strategies to accelerate therapeutic mAb discovery should be relevant since the selection of potential candidates is a long process. An integrated workflow that combined the discovery and the validation of the protective efficacy in animals was designated to identify promising ZIKV mAbs in shorter time. High-throughput investigation of B cells from immune donors can rapidly provide a wide amount of mAbs and this strategy could be applied for a rapid response in case new outbreak disease may emerge in the future [
49].
3.4. Chikungunya (CHIKV)
CHIKV is an alphavirus associated with musculoskeletal disease (arthritogenic alphaviruses), including Mayaro virus (MAYV), Ross River virus (RRV), O’nyong-nyong virus (ONNV) and Semliki Forest virus (SFV). These viruses are globally distributed [
112] and the infection causes a severe and debilitating illness. The most common symptom is fever, but can also provoke joint pain, rash, and headache. In some cases, the joint pain can be so severe that can last for months or even years [
113,
114,
115].
The genotypes Asian, East/Central/South African (ECSA) and West African (WA) are known and CHIKV has limited genetic diversity among strains with 95.2 to 99.8% amino acid identity [
116]. Antibodies raised against one strain can react with all others, leading to the broad consensus that CHIKV lineages constitute a single serotype [
114,
117,
118,
119].
Antivirals for the prophylaxis or therapeutic treatment were not approved yet, and current treatment options are purely symptomatic. Development of mAbs against CHIKV has been developed (
Table 3) and could be an alternative for clinical treatment.
3.4.1. MAbs for CHIKV
CHK-152 and CHK-166 are murine mAbs that target CHIKV E2 and E1 proteins, respectively, and were selected from a panel of CHIKV-neutralizing MAbs produced by hybridomas using mice immunized with CHIKV virus-like particles (VLPs) [
114]. A single dose of the combination of two mAbs was effective in limiting the resistance development to the antibodies and protected immunocompromised mice from the disease when given 24 to 36 h before CHIKV-induced death in post-exposure therapeutic trials [
114]. Rhesus macaques received the mAb combination and it was effective in reducing viral spread and infection at distant tissue sites, however, residual viral RNA was present in tissues and organs and additional treatments may be needed to fully eliminate CHIKV [
120].
The E2 protein is a major target for mAbs and some mAbs were developed: 4N12 [
115], SVIR023 [
121] and DC2.271B [
116].
4N12 is a fully human IgG2 kappa mAb that neutralizes
in vitro three genotypes of CHIKV and was selected from a single individual who had CHIKV infection in Sri Lanka in 2006. A stable hybridoma was generated and 4N12 neutralized the wild-type virus, as well as mutant forms, and was able to protect mice against CHIKV-induced death, even when administered after infection [
115]. Then 4N12 mAb was improved by moving CDR sequences to another framework and SVIR001 mAb was generated, presenting a similar antigen binding and neutralizing activity as the parental mAb. This modification was made to address some of the challenges that 4N12 mAb faced with, such as the limited ability to control acute infection, the inability to reduce viral persistence, and the potential to cause long-term joint disease. SVIR001 was administered in CHIKV-infected Rhesus macaques and showed reduced viremia and potential to decrease CHIKV-associated inflammatory diseases [
122].
SVIR023 is a human IgG1 mAb derived from a human hybridoma that showed neutralizing activity against three clades of the CHIKV. When administered until three days post-infection, it reduced virus number in various tissues. It was effective in preventing disease in mice when administered up to 4 weeks prior to the virus challenge. CHIKV-infected mice treated with mAb were resistant to the secondary challenge, and no evidence of the ADE was detected [
121].
DC2.271B is a human IgG1 mAb that binds CHIKV E2 protein between the β-connector region and the B-domain. Single B cell sorting was used to isolate human mAbs from CHIKV-infected convalescent donors. The lethal viral dose challenge in mice showed that this mAb was highly protective after prophylactic and therapeutic administration [
116].
Immune library was constructed with CHIKV-infected individuals and IM-CKV063 was selected by phage display technology using structurally intact E1/E2 on VLPs as target. This mAb was highly neutralizing, and therapeutic and prophylactic protection in multiple animal models up to 24 h post-exposure was observed [
123].
Single-domain antibodies (sdAbs) are alternatives to conventional antibodies for diagnostic and therapeutic applications. CC3 and CA6 are nanobodies, obtained by llama immunized with CHIKV VLPs, they bind to both the VLPs and the recombinant E1 protein and have neutralizing activity [
124].
Broadly neutralizing antibodies that protect from several arthritogenic alphaviruses should be an interesting therapeutic approach. DC2.M16 and DC2.M357 are examples of human mAbs isolated from a donor previously exposed to CHIKV by a single B cell sorting using MAYV E3-E2-E1 recombinant protein as target. They have neutralizing potency to CHIKV and MAYV and DC2.M357 also neutralized RRV, ONNV and SFV. Both mAbs protected mice from CHIKV- and MAYV-induced musculoskeletal disease [
1].
Pan-protective mAb refers to mAbs that can protect against more than one disease [
113,
114,
115] and it is another relevant therapeutic approach. DC2.112 and DC2.315 are pan-protective and weak neutralizing human mAbs that bind to a conserved epitope in the DII of the E1 protein, located near the fusion peptide, from alphaviruses. They were selected by B cell sorting from CHIKV seropositive individuals, bind to a variety of alphaviruses, those causing arthritis (CHIKV and MAYV) and encephalitis (Venezuelan, Eastern and Western equine encephalitis). Passive transfer approach of each mAb was tested in mice to evaluate the protection. Mice were protected from musculoskeletal disease induced by CHIKV and MAYV and the lethal neurological infectious disease provoked by encephalitis viruses [
125].
3.5. West Nile virus (WNV)
WNV emerged as an important cause of viral encephalitis and is maintained between mosquito vectors and birds in an enzootic cycle. However, it can cause infection disease in humans, horses and other vertebrate animals [
126,
127]. In humans, the infection is characterized by febrile illness that could advance to meningitis, encephalitis, and even fatal disease, especially for elderly and immunocompromised individuals [
128].
There are two lineages of the WNV, the lineage I is distributed globally and associated with severe disease in human and in avian, while the lineage II was isolated in Central and Southern Africa and Asia and it is not associated with severe disease in human [
126,
129]. WNV disease outbreak was in Middle East, Europe and Africa and then spread in North America and other Americas [
126].
3.5.1. MAbs for WNV
Table 4 presents mAbs have been developed for WNV.
E16 mAb, derived from hybridoma, recognized WNV DIII of the E protein and had
in vitro and
in vivo inhibitory potency. Hm-E16 or hE16 is the humanized mAb version, obtained by the CDR grafting technique, that presented similar affinity and efficacy when mice were administrated post-exposure [
130,
131]. Studies in WNV-infected hamsters showed that, when hE16 was administered after virus reached the neurons in the brain, the treatment improved the survival [
132] and ameliorated neurological disease after viral replication [
133]. Then the humanized mAb was designated as MGAWN1 to enter phase 1 clinical study [
135]. The phase 1 clinical trial was registered with NCT00515385 and was completed in 2009, confirming the safety of a single intravenous infusion up to 30 mg/kg of MGAWN1 in healthy adults [
135,
140]. The phase 2 trial (NCT00927953) was designed to study the WNV treatment with MGAWN1 and was terminated early due to the low enrollment [
141].
Phage display scFv immune library was constructed with peripheral blood donated by three WNV-infected individuals and neutralizing mAbs targeting DIII of the E protein were selected [
129]. CR4354 is a fully human IgG1 mAb derived from this immune library showing strong neutralizing activity against WNV I and protected mice against lethal infection [
137]. It neutralizes WNV infection at a post attachment stage in the viral life cycle by blocking the pH-induced rearrangement of the E protein that prevents the virus fusion to the endosomal membrane [
137,
138]. Structural determination of WNV-antibody complex was performed to determine the epitope and the neutralization mechanism was suggested as blocking virus fusion with the endosomal membrane [
138].
Non-immune human phage display library was screened with WNV E protein and identified some scFvs. The scFv epitope was within DI and DII sites of the E protein. The scFv-Fc were developed and five mAbs protected 100% of the mice from death when given prior to the virus infection [
142].
MIT89 is a fully human IgG1 neutralizing mAb specific for WNV E protein, identified from WNV convalescent subjects, by combining the single B cell sorting and the next-generation sequencing (NGS). Immune response study of the infection disease, using the integration of single cell data, serum analysis and repertoire sequencing data, is a relevant approach that can be applied to other diseases [
48].
WNV-86 is a neutralizing mAb to the WNV DII E protein that recognizes mature virions inhibiting the virus infection and dissemination. This contrasts with other flavivirus-specific antibodies, whose tend to recognize immature virions and may rely on Fc-mediated functions to provide protection. Wild-type and LALA version of WNV-86 significantly reduced viral burden in the spinal cord and brain of infected animals [
136].
WN_83 is a human mAb that binds to WNV DIII of the E protein and was isolated from B cell from individuals vaccinated with inactivated Japanese encephalitis virus (JEV) presenting WNV- and JEV-neutralizing antibodies in the sera. Recombinant WNV E protein was used to isolate WN_83 and it neutralized WNV both
in vitro and in a WNV-inoculated mouse model [
139].
4. Outstanding strategies for therapeutic mAbs
Earlier studies of the mAbs discovery targeting arboviruses selected non-human mAbs. However, advances in technological platforms, such as engineering techniques, have been conducting to isolate human mAbs with characteristics for therapeutic purposes.
Table 5 summarizes mAbs obtained by outstanding strategies to obtain improved mAbs.
One point that should be taken in consideration when developing therapeutic mAbs for infection diseases, specially to RNA virus such as arbovirus, is high mutation rates generating variants. Therapeutic use of the neutralizing antibodies may also pose a selective pressure, obtaining escape mutants and leading to an ineffective treatment [
53]. One approach to avoid this is the combination of various mAbs targeting different neutralization epitopes as mentioned in previous sections that was applied to some therapeutic mAbs developed for arboviruses.
ADE phenomenon is another point of attention when therapeutic mAbs have been developed for flaviruses. ADE is well known to DENV antibodies in the secondary infection, leading to the hemorrhagic fever [
61,
62], but not only antibodies for the same virus may cause this phenomenon. It occurred for DENV antibodies used for zika infection treatment in
in vitro model [
101] and also in murine model [
102]. The main mechanism proposed for ADE is the more efficient infection of Fcɣ receptor-expressing myeloid cells
in vivo [
61]. This inconvenience can be addressed by antibody engineering approach, focusing on the Fc portion. The L234A/L235A (LALA) and N297A Fc mutations are commonly used mutations that dramatically reduce the affinity of Fc to the Fcɣ receptor [
149]. Many antibodies for arboviruses have the LALA mutation in order to abrogate ADE phenomenon (
Table 5). However, in some cases this engineering approach abolished completely Fc-dependent responses, i.e. effector functions, therefore careful evaluation is needed when this strategy is applied [
150].
The application of both approaches, the mAb cocktail and LALA mutation, could generate improved mAbs for therapeutic purpose. SMZAb1, SMZAb2 and SMZAb5 are mAbs to ZIKV derived by plasmablast sorting from ZIKV-infected subjects and presented therapeutic potential when used as a cocktail since each mAb binds an epitope overlapping the fusion loop of the E protein. SMZAb1 and SMZAb5 have as target DIII, while SMZAb2 bind to DII. Some cross-reactivity with DENV were observed in these three mAbs and Fc LALA mutations were introduced to prevent potential ADE. The cocktail of engineered mAbs were administered in non-pregnant Rhesus monkeys one day before ZIKV challenge and viral replication was completely prevented in a prophylaxis regimen [
147]. The same cocktail was administered in pregnant Rhesus macaques ZIKV-infected at peak viremia and the treatment was effective to clear the virus however, viral RNA was present in amniotic fluid and failed to prevent fetal demise [
151]. Thus, the treatment was not functional to stop the vertical transmission.
Antibody engineering is a promising approach to obtain improved therapeutic mAbs. VIS513 is a humanized IgG1 antibody for DENV obtained by antibody engineering of the murine 4E11 mAb that neutralized all four DENV serotypes [
72]. Initially, the combination of predicted mutations obtained by
in silico approach, without any crystal structure, promoted 450-fold increase in affinity to DENV4, while the affinity to DENV1-3 was maintained [
143]. Then structure-guided approach was applied introducing other mutations to improve the affinity and lead to broadly neutralization mAb [
144]. Cynomolgus macaques infected with DENV2 were treated with VIS513 24 h post-onset of viremia and 5 days after infection, at the peak of viremia, and the mAb abrogated the infection, while viremia was detectable during post-treatment in lower level compared to control animals [
145]. VIS513 was evaluated in two DENV-infected mice model, AG129 for primary infection and A129 for secondary infection of maternal antibody mediated enhanced infection. Both groups showed viral load reduction and no mortality [
146]. All studies of the VIS513 mAb showed that this engineered mAb is a promising DENV therapeutic antiviral drug.
Development of bispecific mAbs is an alternative approach for the combination of two or more therapeutic mAbs to minimize viral escape and ADE. Bispecfic mAb for ZIKV was developed. Initially, a panel of anti-ZIKV mAbs were identified from four ZIKV-infected patients with two of them DENV-naïve, then grouped by the affinity of B cells to the ZIKV NS1 and E protein and also by neutralizing mAbs to ZIKV and DENV DIII of the E protein or quaternary epitope displayed on the infectious virions [
41]. The epitope of the ZKA190 mAb isolated from this panel was further investigated, and it was located to the E protein in DI-DIII linker and the lateral ridge region of DIII, a conserved epitope [
148]. In a mice model, ZIK190 was capable of delaying the morbidity and mortality in a prophylactic approach and its LALA variant did not show ADE. In the therapeutic approach, >80% survival rates were achieved, as well as reduction in morbidity [
148]. Despite that, an ZIKV escape mutant was detected
in vivo that completely abrogated ZIK190 neutralization activity [
148]. Bispecific mAb was developed to bypass this problem, and ZIK185 mAb, targeting DII of the E protein, was chosen to combine with ZIK190 [
148]. Escape mutants were also emerged from ZIK185 mAb, indicating that this phenomenon occurs in mAbs binding to distinct epitopes [
148]. A tetravalent symmetric format Fabs-in-tandem-Ig (FIT-Ig) was constructed with ZIK190 and ZIK185 Fabs, combined with the engineered Fc backbone with the LALA mutation, and FIT-1 bispecific antibody was produced [
148]. In a mice model, FIT-1 provided protection against the lethal infection in all cases and no viral load was detected, nonetheless no escape mutant was detected
in vitro neither
in vivo [
148].
5. Conclusions
The lessons learned with COVID-19 pandemic showed that it is important a continuous support to develop mAbs for emergent diseases to be prepared to answer faster when new threads may appear. Passive immunotherapy should be an effective alternative for the treatment of the infectious diseases, especially for immunocompromised patients and individuals whom the vaccination is not indicated.
Neutralizing mAbs with therapeutic potential have been developed to arboviruses using advanced mAb discovery technologies. Many B cell extensive studies allowed to deeply analyze and understand the immune response elicited by infected donors during the acute-phase, post-infection period, secondary infection phase and others.
Studies related to the discovery of the protective arboviruses mAbs are also important for the rational vaccine design giving complement information in order to obtain potent vaccines with better protective features.
There are many challenges to obtain mAbs for arbovirus diseases with desired features, as highly potent and broadly neutralizing agents. Engineering approaches could improve mAb characteristics and lead to this objective. The technology is always advancing and novel techniques will be introduced as soon as the proof of concept is shown to them, that will allow to select mAbs with improved and refined features.
Author Contributions
Conceptualization, L.F.O., C.T.B. and L.R.T.; writing—original draft preparation, L.F.O., C.T.B. and L.R.T.; writing—review and editing, L.R.T.; visualization, L.F.O., C.T.B. and L.R.T.; supervision, L.R.T.; project administration, L.R.T.; funding acquisition, L.R.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by São Paulo Research Foundation (FAPESP), grant number 2020/15833-1.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Graham, B.S.; Ambrosino, D.M. History of Passive Antibody Administration for Prevention and Treatment of Infectious Diseases: Curr. Opin. HIV AIDS 2015, 10, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Dadachova, E.; Pirofski, L. Passive Antibody Therapy for Infectious Diseases. Nat. Rev. Microbiol. 2004, 2, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Pantaleo, G.; Correia, B.; Fenwick, C.; Joo, V.S.; Perez, L. Antibodies to Combat Viral Infections: Development Strategies and Progress. Nat. Rev. Drug Discov. 2022, 21, 676–696. [Google Scholar] [CrossRef] [PubMed]
- Ogunlade, S.T.; Meehan, M.T.; Adekunle, A.I.; Rojas, D.P.; Adegboye, O.A.; McBryde, E.S. A Review: Aedes-Borne Arboviral Infections, Controls and Wolbachia-Based Strategies. Vaccines 2021, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Gould, E.; Pettersson, J.; Higgs, S.; Charrel, R.; De Lamballerie, X. Emerging Arboviruses: Why Today? One Health 2017, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wieten, R.W.; Goorhuis, A.; Jonker, E.F.F.; De Bree, G.J.; De Visser, A.W.; Van Genderen, P.J.J.; Remmerswaal, E.B.M.; Ten Berge, I.J.M.; Visser, L.G.; Grobusch, M.P.; et al. 17D Yellow Fever Vaccine Elicits Comparable Long-Term Immune Responses in Healthy Individuals and Immune-Compromised Patients. J. Infect. 2016, 72, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-L.; Chang, J.-K.; Tang, R.-B. Current Recommendations for the Japanese Encephalitis Vaccine. J. Chin. Med. Assoc. 2015, 78, 271–275. [Google Scholar] [CrossRef]
- Pintado Silva, J.; Fernandez-Sesma, A. Challenges on the Development of a Dengue Vaccine: A Comprehensive Review of the State of the Art. J. Gen. Virol. 2023, 104. [Google Scholar] [CrossRef]
- Sevvana, M.; Kuhn, R.J. Mapping the Diverse Structural Landscape of the Flavivirus Antibody Repertoire. Curr. Opin. Virol. 2020, 45, 51–64. [Google Scholar] [CrossRef]
- Kim, A.S.; Diamond, M.S. A Molecular Understanding of Alphavirus Entry and Antibody Protection. Nat. Rev. Microbiol. 2023, 21, 396–407. [Google Scholar] [CrossRef]
- Köhler, G.; Milstein, C. Continuous Cultures of Fused Cells Secreting Antibody of Predefined Specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef] [PubMed]
- Norman, D.J.; Shield, C.F., 3rd; Barry, J.; Henell, K.; Funnell, M.B.; Lemon, J. A U.S. Clinical Study of Orthoclone OKT3 in Renal Transplantation. Transplant. Proc. 1987, 19 (Suppl. 1), 21–27. [Google Scholar] [PubMed]
- Tsuruta, L.R.; Dos, M.L.; Moro, A.M. Display Technologies for the Selection of Monoclonal Antibodies for Clinical Use. In Antibody Engineering; Böldicke, T., Ed.; InTech, 2018. ISBN 978-953-51-3825-9.
- Nagano, K.; Tsutsumi, Y. Phage Display Technology as a Powerful Platform for Antibody Drug Discovery. Viruses 2021, 13, 178. [Google Scholar] [CrossRef] [PubMed]
- Parray, H.A.; Shukla, S.; Samal, S.; Shrivastava, T.; Ahmed, S.; Sharma, C.; Kumar, R. Hybridoma Technology a Versatile Method for Isolation of Monoclonal Antibodies, Its Applicability across Species, Limitations, Advancement and Future Perspectives. Int. Immunopharmacol. 2020, 85, 106639. [Google Scholar] [CrossRef]
- Smith, S.A.; Crowe, J.E., Jr. Use of Human Hybridoma Technology To Isolate Human Monoclonal Antibodies. Microbiol. Spectr. 2015, 3, 3–1. [Google Scholar] [CrossRef] [PubMed]
- Morrison, S.L.; Johnson, M.J.; Herzenberg, L.A.; Oi, V.T. Chimeric Human Antibody Molecules: Mouse Antigen-Binding Domains with Human Constant Region Domains. Proc. Natl. Acad. Sci. 1984, 81, 6851–6855. [Google Scholar] [CrossRef]
- Janda, A.; Bowen, A.; Greenspan, N.S.; Casadevall, A. Ig Constant Region Effects on Variable Region Structure and Function. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Morelock, M.M.; Rothlein, R.; Bright, S.M.; Robinson, M.K.; Graham, E.T.; Sabo, J.P.; Owens, R.; King, D.J.; Norris, S.H.; Scher, D.S. Isotype Choice for Chimeric Antibodies Affects Binding Properties. J. Biol. Chem. 1994, 269, 13048–13055. [Google Scholar] [CrossRef]
- Torres, M.; Fernandez-Fuentes, N.; Fiser, A.; Casadevall, A. Exchanging Murine and Human Immunoglobulin Constant Chains Affects the Kinetics and Thermodynamics of Antigen Binding and Chimeric Antibody Autoreactivity. PLoS ONE 2007, 2, e1310. [Google Scholar] [CrossRef]
- McLean, G.R.; Torres, M.; Elguezabal, N.; Nakouzi, A.; Casadevall, A. Isotype Can Affect the Fine Specificity of an Antibody for a Polysaccharide Antigen. J. Immunol. 2002, 169, 1379–1386. [Google Scholar] [CrossRef]
- Hwang, W.Y.K.; Foote, J. Immunogenicity of Engineered Antibodies. Methods 2005, 36, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.T.; Dear, P.H.; Foote, J.; Neuberger, M.S.; Winter, G. Replacing the Complementarity-Determining Regions in a Human Antibody with Those from a Mouse. Nature 1986, 321, 522–525. [Google Scholar] [CrossRef] [PubMed]
- Caldas, C.; Coelho, V.; Kalil, J.; Moro, A.M.; Maranhão, A.Q.; Brı́gido, M.M. Humanization of the Anti-CD18 Antibody 6.7: An Unexpected Effect of a Framework Residue in Binding to Antigen. Mol. Immunol. 2003, 39, 941–952. [Google Scholar] [CrossRef] [PubMed]
- Queen, C.; Schneider, W.P.; Selick, H.E.; Payne, P.W.; Landolfi, N.F.; Duncan, J.F.; Avdalovic, N.M.; Levitt, M.; Junghans, R.P.; Waldmann, T.A. A Humanized Antibody That Binds to the Interleukin 2 Receptor. Proc. Natl. Acad. Sci. 1989, 86, 10029–10033. [Google Scholar] [CrossRef] [PubMed]
- Makabe, K.; Nakanishi, T.; Tsumoto, K.; Tanaka, Y.; Kondo, H.; Umetsu, M.; Sone, Y.; Asano, R.; Kumagai, I. Thermodynamic Consequences of Mutations in Vernier Zone Residues of a Humanized Anti-Human Epidermal Growth Factor Receptor Murine Antibody, 528. J. Biol. Chem. 2008, 283, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Tsurushita, N.; Hinton, P.R.; Kumar, S. Design of Humanized Antibodies: From Anti-Tac to Zenapax. Methods 2005, 36, 69–83. [Google Scholar] [CrossRef]
- Safdari, Y.; Farajnia, S.; Asgharzadeh, M.; Khalili, M. Antibody Humanization Methods – a Review and Update. Biotechnol. Genet. Eng. Rev. 2013, 29, 175–186. [Google Scholar] [CrossRef]
- O’Neil, K.T.; Hoess, R.H. Phage Display: Protein Engineering by Directed Evolution. Curr. Opin. Struct. Biol. 1995, 5, 443–449. [Google Scholar] [CrossRef]
- Tsuruta, L.R.; Dos, M.L.; Moro, A.M. Display Technologies for the Selection of Monoclonal Antibodies for Clinical Use. In Antibody Engineering; Böldicke, T., Ed.; InTech, 2018. ISBN 978-953-51-3825-9.
- Valldorf, B.; Hinz, S.C.; Russo, G.; Pekar, L.; Mohr, L.; Klemm, J.; Doerner, A.; Krah, S.; Hust, M.; Zielonka, S. Antibody Display Technologies: Selecting the Cream of the Crop. Biol. Chem. 2022, 403, 455–477. [Google Scholar] [CrossRef]
- Ledsgaard, L.; Kilstrup, M.; Karatt-Vellatt, A.; McCafferty, J.; Laustsen, A. Basics of Antibody Phage Display Technology. Toxins 2018, 10, 236. [Google Scholar] [CrossRef]
- Bradbury, A.R.M.; Sidhu, S.; Dübel, S.; McCafferty, J. Beyond Natural Antibodies: The Power of in Vitro Display Technologies. Nat. Biotechnol. 2011, 29, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Filamentous Fusion Phage: Novel Expression Vectors That Display Cloned Antigens on the Virion Surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- McCafferty, J.; Griffiths, A.D.; Winter, G.; Chiswell, D.J. Phage Antibodies: Filamentous Phage Displaying Antibody Variable Domains. Nature 1990, 348, 552–554. [Google Scholar] [CrossRef] [PubMed]
- Jaroszewicz, W.; Morcinek-Orłowska, J.; Pierzynowska, K.; Gaffke, L.; Węgrzyn, G. Phage Display and Other Peptide Display Technologies. FEMS Microbiol. Rev. 2022, 46, fuab052. [Google Scholar] [CrossRef] [PubMed]
- Whaley, R.E.; Ameny, S.; Arkatkar, T.; Seese, A.; Wall, A.; Khan, I.; Carter, J.J.; Scherer, E.M.; Rawlings, D.J.; Galloway, D.A.; et al. Generation of a Cost-Effective Cell Line for Support of High-Throughput Isolation of Primary Human B Cells and Monoclonal Neutralizing Antibodies. J. Immunol. Methods 2021, 488, 112901. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.M.; Phogat, S.K.; Chan-Hui, P.-Y.; Wagner, D.; Phung, P.; Goss, J.L.; Wrin, T.; Simek, M.D.; Fling, S.; Mitcham, J.L.; et al. Broad and Potent Neutralizing Antibodies from an African Donor Reveal a New HIV-1 Vaccine Target. Science 2009, 326, 285–289. [Google Scholar] [CrossRef]
- Cox, K.S.; Tang, A.; Chen, Z.; Horton, M.S.; Yan, H.; Wang, X.-M.; Dubey, S.A.; DiStefano, D.J.; Ettenger, A.; Fong, R.H.; et al. Rapid Isolation of Dengue-Neutralizing Antibodies from Single Cell-Sorted Human Antigen-Specific Memory B-Cell Cultures. mAbs 2016, 8, 129–140. [Google Scholar] [CrossRef]
- Singh, T.; Hwang, K.-K.; Miller, A.S.; Jones, R.L.; Lopez, C.A.; Dulson, S.J.; Giuberti, C.; Gladden, M.A.; Miller, I.; Webster, H.S.; et al. A Zika Virus-Specific IgM Elicited in Pregnancy Exhibits Ultrapotent Neutralization. Cell 2022, 185, 4826–4840. [Google Scholar] [CrossRef]
- Stettler, K.; Beltramello, M.; Espinosa, D.A.; Graham, V.; Cassotta, A.; Bianchi, S.; Vanzetta, F.; Minola, A.; Jaconi, S.; Mele, F.; et al. Specificity, Cross-Reactivity, and Function of Antibodies Elicited by Zika Virus Infection. Science 2016, 353, 823–826. [Google Scholar] [CrossRef]
- Busse, C.E.; Czogiel, I.; Braun, P.; Arndt, P.F.; Wardemann, H. Single-cell Based High-throughput Sequencing of Full-length Immunoglobulin Heavy and Light Chain Genes. Eur. J. Immunol. 2014, 44, 597–603. [Google Scholar] [CrossRef]
- Liao, H.-X.; Levesque, M.C.; Nagel, A.; Dixon, A.; Zhang, R.; Walter, E.; Parks, R.; Whitesides, J.; Marshall, D.J.; Hwang, K.-K.; et al. High-Throughput Isolation of Immunoglobulin Genes from Single Human B Cells and Expression as Monoclonal Antibodies. J. Virol. Methods 2009, 158, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Pedrioli, A.; Oxenius, A. Single B Cell Technologies for Monoclonal Antibody Discovery. Trends Immunol. 2021, 42, 1143–1158. [Google Scholar] [CrossRef] [PubMed]
- Zost, S.J.; Gilchuk, P.; Chen, R.E.; Case, J.B.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; Sutton, R.E.; Suryadevara, N.; Chen, E.C.; et al. Rapid Isolation and Profiling of a Diverse Panel of Human Monoclonal Antibodies Targeting the SARS-CoV-2 Spike Protein. Nat. Med. 2020, 26, 1422–1427. [Google Scholar] [CrossRef] [PubMed]
- Winters, A.; McFadden, K.; Bergen, J.; Landas, J.; Berry, K.A.; Gonzalez, A.; Salimi-Moosavi, H.; Murawsky, C.M.; Tagari, P.; King, C.T. Rapid Single B Cell Antibody Discovery Using Nanopens and Structured Light. mAbs 2019, 11, 1025–1035. [Google Scholar] [CrossRef]
- Broketa, M.; Bruhns, P. Single-Cell Technologies for the Study of Antibody-Secreting Cells. Front. Immunol. 2022, 12, 821729. [Google Scholar] [CrossRef]
- Tsioris, K.; Gupta, N.T.; Ogunniyi, A.O.; Zimnisky, R.M.; Qian, F.; Yao, Y.; Wang, X.; Stern, J.N.H.; Chari, R.; Briggs, A.W.; et al. Neutralizing Antibodies against West Nile Virus Identified Directly from Human B Cells by Single-Cell Analysis and next Generation Sequencing. Integr. Biol. 2015, 7, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Gilchuk, P.; Bombardi, R.G.; Erasmus, J.H.; Tan, Q.; Nargi, R.; Soto, C.; Abbink, P.; Suscovich, T.J.; Durnell, L.A.; Khandhar, A.; et al. Integrated Pipeline for the Accelerated Discovery of Antiviral Antibody Therapeutics. Nat. Biomed. Eng. 2020, 4, 1030–1043. [Google Scholar] [CrossRef]
- Setliff, I.; Shiakolas, A.R.; Pilewski, K.A.; Murji, A.A.; Mapengo, R.E.; Janowska, K.; Richardson, S.; Oosthuysen, C.; Raju, N.; Ronsard, L.; et al. High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell 2019, 179, 1636–1646. [Google Scholar] [CrossRef]
- Gérard, A.; Woolfe, A.; Mottet, G.; Reichen, M.; Castrillon, C.; Menrath, V.; Ellouze, S.; Poitou, A.; Doineau, R.; Briseno-Roa, L.; et al. High-Throughput Single-Cell Activity-Based Screening and Sequencing of Antibodies Using Droplet Microfluidics. Nat. Biotechnol. 2020, 38, 715–721. [Google Scholar] [CrossRef]
- Rees, A.R. Understanding the Human Antibody Repertoire. mAbs 2020, 12, 1729683. [Google Scholar] [CrossRef]
- Crowe, J.E. Human Antibodies for Viral Infections. Annu. Rev. Immunol. 2022, 40, 349–386. [Google Scholar] [CrossRef] [PubMed]
- Pierson, T.C.; Diamond, M.S. The Continued Threat of Emerging Flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef] [PubMed]
- Slon Campos, J.L.; Mongkolsapaya, J.; Screaton, G.R. The Immune Response against Flaviviruses. Nat. Immunol. 2018, 19, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-Monzón, A.M.; Cordero-Rivera, C.D.; Farfan-Morales, C.N.; Osuna-Ramos, J.F.; De Jesús-González, L.A.; Reyes-Ruiz, J.M.; Ángel, R.M. The Role of Anti-flavivirus Humoral Immune Response in Protection and Pathogenesis. Rev. Med. Virol. 2020, 30. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, I.S.B.; Tanabe, E.L.L.; Santos, E.C.; Martins, W.V.; Araújo, I.M.T.C.; Cavalcante, M.C.A.; Lima, A.R.V.; Câmara, N.O.S.; Anderson, L.; Yunusov, D.; et al. Cellular and Molecular Immune Response to Chikungunya Virus Infection. Front. Cell. Infect. Microbiol. 2018, 8, 345. [Google Scholar] [CrossRef]
- Rupp, J.C.; Sokoloski, K.J.; Gebhart, N.N.; Hardy, R.W. Alphavirus RNA Synthesis and Non-Structural Protein Functions. J. Gen. Virol. 2015, 96, 2483–2500. [Google Scholar] [CrossRef]
- Dengue and Severe Dengue. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 20 July 2023).
- Halstead, S.B. Dengue. The Lancet 2007, 370, 1644–1652. [Google Scholar] [CrossRef]
- Halstead, S.; O’Rourke, E. Dengue Viruses and Mononuclear Phagocytes. I. Infection Enhancement by Non-Neutralizing Antibody. J. Exp. Med. 1977, 146, 201–217. [Google Scholar] [CrossRef]
- Smith, S.A.; Zhou, Y.; Olivarez, N.P.; Broadwater, A.H.; De Silva, A.M.; Crowe, J.E. Persistence of Circulating Memory B Cell Clones with Potential for Dengue Virus Disease Enhancement for Decades Following Infection. J. Virol. 2012, 86, 2665–2675. [Google Scholar] [CrossRef]
- Rothman, A.L. Immunity to Dengue Virus: A Tale of Original Antigenic Sin and Tropical Cytokine Storms. Nat. Rev. Immunol. 2011, 11, 532–543. [Google Scholar] [CrossRef]
- Kallas, E.G.; Precioso, A.R.; Palacios, R.; Thomé, B.; Braga, P.E.; Vanni, T.; Campos, L.M.A.; Ferrari, L.; Mondini, G.; Da Graça Salomão, M.; et al. Safety and Immunogenicity of the Tetravalent, Live-Attenuated Dengue Vaccine Butantan-DV in Adults in Brazil: A Two-Step, Double-Blind, Randomised Placebo-Controlled Phase 2 Trial. Lancet Infect. Dis. 2020, 20, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Butantan Institute Phase III, Double-Blind, Randomized, Placebo-Controlled Trial to Evaluate the Efficacy, Safety, and Immunogenicity of the Dengue 1, 2, 3, 4 (Attenuated) Vaccine From Instituto Butantan; clinicaltrials.gov, 2023.
- AbViro LLC A Phase 1a, Double-Blind, Placebo-Controlled, Single Ascending Dose Study to Determine the Safety and Pharmacokinetics of AV-1 in Healthy Male and Female Adult Subjects; clinicaltrials.gov, 2022.
- Serum Institute of India Pvt. Ltd. A Phase I, Partially Blind (Observer-Blind), Randomized, Single Dose Ascending Study of Dengue Monoclonal Antibody (Dengushield) in Healthy Adults; clinicaltrials.gov, 2020.
- Cruz-Oliveira, C.; Freire, J.M.; Conceição, T.M.; Higa, L.M.; Castanho, M.A.R.B.; Da Poian, A.T. Receptors and Routes of Dengue Virus Entry into the Host Cells. FEMS Microbiol. Rev. 2015, 39, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Chin, J.F.L.; Chu, J.J.H.; Ng, M.L. The Envelope Glycoprotein Domain III of Dengue Virus Serotypes 1 and 2 Inhibit Virus Entry. Microbes Infect. 2007, 9, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hung, J.-J.; Hsieh, M.-T.; Young, M.-J.; Kao, C.-L.; King, C.-C.; Chang, W. An External Loop Region of Domain III of Dengue Virus Type 2 Envelope Protein Is Involved in Serotype-Specific Binding to Mosquito but Not Mammalian Cells. J. Virol. 2004, 78, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Morens, D.M.; Halstead, S.B.; Marchette, N.J. Profiles of Antibody-Dependent Enhancement of Dengue Virus Type 2 Infection. Microb. Pathog. 1987, 3, 231–237. [Google Scholar] [CrossRef]
- Thullier, P.; Lafaye, P.; Mégret, F.; Deubel, V.; Jouan, A.; Mazié, J.C. A Recombinant Fab Neutralizes Dengue Virus in Vitro. J. Biotechnol. 1999, 69, 183–190. [Google Scholar] [CrossRef]
- Deng, Y.-Q.; Dai, J.-X.; Ji, G.-H.; Jiang, T.; Wang, H.-J.; Yang, H.; Tan, W.-L.; Liu, R.; Yu, M.; Ge, B.-X.; et al. A Broadly Flavivirus Cross-Neutralizing Monoclonal Antibody That Recognizes a Novel Epitope within the Fusion Loop of E Protein. PLoS ONE 2011, 6, e16059. [Google Scholar] [CrossRef]
- Dai, L.; Song, J.; Lu, X.; Deng, Y.-Q.; Musyoki, A.M.; Cheng, H.; Zhang, Y.; Yuan, Y.; Song, H.; Haywood, J.; et al. Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody. Cell Host Microbe 2016, 19, 696–704. [Google Scholar] [CrossRef]
- Kotaki, T.; Kurosu, T.; Grinyo-Escuer, A.; Davidson, E.; Churrotin, S.; Okabayashi, T.; Puiprom, O.; Mulyatno, K.C.; Sucipto, T.H.; Doranz, B.J.; et al. An Affinity-Matured Human Monoclonal Antibody Targeting Fusion Loop Epitope of Dengue Virus with in Vivo Therapeutic Potency. Sci. Rep. 2021, 11, 12987. [Google Scholar] [CrossRef]
- Tien, S.-M.; Chang, P.-C.; Lai, Y.-C.; Chuang, Y.-C.; Tseng, C.-K.; Kao, Y.-S.; Huang, H.-J.; Hsiao, Y.-P.; Liu, Y.-L.; Lin, H.-H.; et al. Therapeutic Efficacy of Humanized Monoclonal Antibodies Targeting Dengue Virus Nonstructural Protein 1 in the Mouse Model. PLOS Pathog. 2022, 18, e1010469. [Google Scholar] [CrossRef]
- Wan, S.-W.; Chen, P.-W.; Chen, C.-Y.; Lai, Y.-C.; Chu, Y.-T.; Hung, C.-Y.; Lee, H.; Wu, H.F.; Chuang, Y.-C.; Lin, J.; et al. Therapeutic Effects of Monoclonal Antibody against Dengue Virus NS1 in a STAT1 Knockout Mouse Model of Dengue Infection. J. Immunol. 2017, 199, 2834–2844. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.-C.; Chuang, Y.-C.; Liu, C.-C.; Ho, T.-S.; Lin, Y.-S.; Anderson, R.; Yeh, T.-M. Antibodies Against Modified NS1 Wing Domain Peptide Protect Against Dengue Virus Infection. Sci. Rep. 2017, 7, 6975. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zuest, R.; Velumani, S.; Tukijan, F.; Toh, Y.X.; Appanna, R.; Tan, E.Y.; Cerny, D.; MacAry, P.; Wang, C.-I.; et al. A Potent Neutralizing Antibody with Therapeutic Potential against All Four Serotypes of Dengue Virus. Npj Vaccines 2017, 2, 2. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hadinoto, V.; Appanna, R.; Joensson, K.; Toh, Y.X.; Balakrishnan, T.; Ong, S.H.; Warter, L.; Leo, Y.S.; Wang, C.-I.; et al. Plasmablasts Generated during Repeated Dengue Infection Are Virus Glycoprotein–Specific and Bind to Multiple Virus Serotypes. J. Immunol. 2012, 189, 5877–5885. [Google Scholar] [CrossRef]
- Zhang, S.; Loy, T.; Ng, T.-S.; Lim, X.-N.; Chew, S.-Y.V.; Tan, T.Y.; Xu, M.; Kostyuchenko, V.A.; Tukijan, F.; Shi, J.; et al. A Human Antibody Neutralizes Different Flaviviruses by Using Different Mechanisms. Cell Rep. 2020, 31, 107584. [Google Scholar] [CrossRef]
- Li, L.; Meng, W.; Horton, M.; DiStefano, D.R.; Thoryk, E.A.; Pfaff, J.M.; Wang, Q.; Salazar, G.T.; Barnes, T.; Doranz, B.J.; et al. Potent Neutralizing Antibodies Elicited by Dengue Vaccine in Rhesus Macaque Target Diverse Epitopes. PLOS Pathog. 2019, 15, e1007716. [Google Scholar] [CrossRef]
- Chen, W.-H.; Chou, F.-P.; Wang, Y.-K.; Huang, S.-C.; Cheng, C.-H.; Wu, T.-K. Characterization and Epitope Mapping of Dengue Virus Type 1 Specific Monoclonal Antibodies. Virol. J. 2017, 14, 189. [Google Scholar] [CrossRef]
- Setthapramote, C.; Sasaki, T.; Puiprom, O.; Limkittikul, K.; Pitaksajjakul, P.; Pipattanaboon, C.; Sasayama, M.; Leuangwutiwong, P.; Phumratanaprapin, W.; Chamnachanan, S.; et al. Human Monoclonal Antibodies to Neutralize All Dengue Virus Serotypes Using Lymphocytes from Patients at Acute Phase of the Secondary Infection. Biochem. Biophys. Res. Commun. 2012, 423, 867–872. [Google Scholar] [CrossRef]
- Cox, K.S.; Tang, A.; Chen, Z.; Horton, M.S.; Yan, H.; Wang, X.-M.; Dubey, S.A.; DiStefano, D.J.; Ettenger, A.; Fong, R.H.; et al. Rapid Isolation of Dengue-Neutralizing Antibodies from Single Cell-Sorted Human Antigen-Specific Memory B-Cell Cultures. mAbs 2016, 8, 129–140. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Wongwiwat, W.; Supasa, S.; Zhang, X.; Dai, X.; Rouvinski, A.; Jumnainsong, A.; Edwards, C.; Quyen, N.T.H.; Duangchinda, T.; et al. A New Class of Highly Potent, Broadly Neutralizing Antibodies Isolated from Viremic Patients Infected with Dengue Virus. Nat. Immunol. 2015, 16, 170–177. [Google Scholar] [CrossRef]
- Chouin-Carneiro, T.; Vega-Rua, A.; Vazeille, M.; Yebakima, A.; Girod, R.; Goindin, D.; Dupont-Rouzeyrol, M.; Lourenço-de-Oliveira, R.; Failloux, A.-B. Differential Susceptibilities of Aedes Aegypti and Aedes Albopictus from the Americas to Zika Virus. PLoS Negl. Trop. Dis. 2016, 10, e0004543. [Google Scholar] [CrossRef] [PubMed]
- Counotte, M.J.; Kim, C.R.; Wang, J.; Bernstein, K.; Deal, C.D.; Broutet, N.J.N.; Low, N. Sexual Transmission of Zika Virus and Other Flaviviruses: A Living Systematic Review. PLOS Med. 2018, 15, e1002611. [Google Scholar] [CrossRef] [PubMed]
- Brasil, P.; Sequeira, P.C.; Freitas, A.D.; Zogbi, H.E.; Calvet, G.A.; De Souza, R.V.; Siqueira, A.M.; De Mendonca, M.C.L.; Nogueira, R.M.R.; De Filippis, A.M.B.; et al. Guillain-Barré Syndrome Associated with Zika Virus Infection. The Lancet 2016, 387, 1482. [Google Scholar] [CrossRef] [PubMed]
- Miner, J.J.; Diamond, M.S. Zika Virus Pathogenesis and Tissue Tropism. Cell Host Microbe 2017, 21, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, C.P.; Barros-Aragão, F.G.Q.; Neris, R.L.S.; Frost, P.S.; Soares, C.; Souza, I.N.O.; Zeidler, J.D.; Zamberlan, D.C.; De Sousa, V.L.; Souza, A.S.; et al. Zika Virus Replicates in Adult Human Brain Tissue and Impairs Synapses and Memory in Mice. Nat. Commun. 2019, 10, 3890. [Google Scholar] [CrossRef] [PubMed]
- Tabata, T.; Petitt, M.; Puerta-Guardo, H.; Michlmayr, D.; Wang, C.; Fang-Hoover, J.; Harris, E.; Pereira, L. Zika Virus Targets Different Primary Human Placental Cells, Suggesting Two Routes for Vertical Transmission. Cell Host Microbe 2016, 20, 155–166. [Google Scholar] [CrossRef]
- De Araújo, T.V.B.; Ximenes, R.A.D.A.; Miranda-Filho, D.D.B.; Souza, W.V.; Montarroyos, U.R.; De Melo, A.P.L.; Valongueiro, S.; De Albuquerque, M.D.F.P.M.; Braga, C.; Filho, S.P.B.; et al. Association between Microcephaly, Zika Virus Infection, and Other Risk Factors in Brazil: Final Report of a Case-Control Study. Lancet Infect. Dis. 2018, 18, 328–336. [Google Scholar] [CrossRef]
- Martines, R.B.; Bhatnagar, J.; De Oliveira Ramos, A.M.; Davi, H.P.F.; Iglezias, S.D.; Kanamura, C.T.; Keating, M.K.; Hale, G.; Silva-Flannery, L.; Muehlenbachs, A.; et al. Pathology of Congenital Zika Syndrome in Brazil: A Case Series. The Lancet 2016, 388, 898–904. [Google Scholar] [CrossRef]
- Kapogiannis, B.G.; Chakhtoura, N.; Hazra, R.; Spong, C.Y. Bridging Knowledge Gaps to Understand How Zika Virus Exposure and Infection Affect Child Development. JAMA Pediatr. 2017, 171, 478. [Google Scholar] [CrossRef]
- Tychan Pte Ltd. Phase 1 Time Lagged, Parallel-Group, Randomized, Placebo-Controlled, Single-Blind, Single Ascending Dose Study of Tyzivumab in ZIKV Infected Patients; clinicaltrials.gov, 2022.
- Priyamvada, L.; Quicke, K.M.; Hudson, W.H.; Onlamoon, N.; Sewatanon, J.; Edupuganti, S.; Pattanapanyasat, K.; Chokephaibulkit, K.; Mulligan, M.J.; Wilson, P.C.; et al. Human Antibody Responses after Dengue Virus Infection Are Highly Cross-Reactive to Zika Virus. Proc. Natl. Acad. Sci. 2016, 113, 7852–7857. [Google Scholar] [CrossRef]
- Collins, M.H.; Tu, H.A.; Gimblet-Ochieng, C.; Liou, G.-J.A.; Jadi, R.S.; Metz, S.W.; Thomas, A.; McElvany, B.D.; Davidson, E.; Doranz, B.J.; et al. Human Antibody Response to Zika Targets Type-Specific Quaternary Structure Epitopes. JCI Insight 2019, 4, e124588. [Google Scholar] [CrossRef] [PubMed]
- Rogers, T.F.; Goodwin, E.C.; Briney, B.; Sok, D.; Beutler, N.; Strubel, A.; Nedellec, R.; Le, K.; Brown, M.E.; Burton, D.R.; et al. Zika Virus Activates de Novo and Cross-Reactive Memory B Cell Responses in Dengue-Experienced Donors. Sci. Immunol. 2017, 2, eaan6809. [Google Scholar] [CrossRef] [PubMed]
- Robbiani, D.F.; Bozzacco, L.; Keeffe, J.R.; Khouri, R.; Olsen, P.C.; Gazumyan, A.; Schaefer-Babajew, D.; Avila-Rios, S.; Nogueira, L.; Patel, R.; et al. Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico. Cell 2017, 169, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Dejnirattisai, W.; Supasa, P.; Wongwiwat, W.; Rouvinski, A.; Barba-Spaeth, G.; Duangchinda, T.; Sakuntabhai, A.; Cao-Lormeau, V.-M.; Malasit, P.; Rey, F.A.; et al. Dengue Virus Sero-Cross-Reactivity Drives Antibody-Dependent Enhancement of Infection with Zika Virus. Nat. Immunol. 2016, 17, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Bardina, S.V.; Bunduc, P.; Tripathi, S.; Duehr, J.; Frere, J.J.; Brown, J.A.; Nachbagauer, R.; Foster, G.A.; Krysztof, D.; Tortorella, D.; et al. Enhancement of Zika Virus Pathogenesis by Preexisting Antiflavivirus Immunity. Science 2017, 356, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Sapparapu, G.; Fernandez, E.; Kose, N.; Bin, C.; Fox, J.M.; Bombardi, R.G.; Zhao, H.; Nelson, C.A.; Bryan, A.L.; Barnes, T.; et al. Neutralizing Human Antibodies Prevent Zika Virus Replication and Fetal Disease in Mice. Nature 2016, 540, 443–447. [Google Scholar] [CrossRef]
- Wessel, A.W.; Kose, N.; Bombardi, R.G.; Roy, V.; Chantima, W.; Mongkolsapaya, J.; Edeling, M.A.; Nelson, C.A.; Bosch, I.; Alter, G.; et al. Antibodies Targeting Epitopes on the Cell-Surface Form of NS1 Protect against Zika Virus Infection during Pregnancy. Nat. Commun. 2020, 11, 5278. [Google Scholar] [CrossRef]
- Erasmus, J.H.; Archer, J.; Fuerte-Stone, J.; Khandhar, A.P.; Voigt, E.; Granger, B.; Bombardi, R.G.; Govero, J.; Tan, Q.; Durnell, L.A.; et al. Intramuscular Delivery of Replicon RNA Encoding ZIKV-117 Human Monoclonal Antibody Protects against Zika Virus Infection. Mol. Ther. - Methods Clin. Dev. 2020, 18, 402–414. [Google Scholar] [CrossRef]
- Singh, T.; Lopez, C.A.; Giuberti, C.; Dennis, M.L.; Itell, H.L.; Heimsath, H.J.; Webster, H.S.; Roark, H.K.; Merçon De Vargas, P.R.; Hall, A.; et al. Efficient Transplacental IgG Transfer in Women Infected with Zika Virus during Pregnancy. PLoS Negl. Trop. Dis. 2019, 13, e0007648. [Google Scholar] [CrossRef]
- Keeffe, J.R.; Van Rompay, K.K.A.; Olsen, P.C.; Wang, Q.; Gazumyan, A.; Azzopardi, S.A.; Schaefer-Babajew, D.; Lee, Y.E.; Stuart, J.B.; Singapuri, A.; et al. A Combination of Two Human Monoclonal Antibodies Prevents Zika Virus Escape Mutations in Non-Human Primates. Cell Rep. 2018, 25, 1385–1394. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, H.; Liu, X.; Dai, L.; Ma, T.; Qi, J.; Wong, G.; Peng, R.; Liu, S.; Li, J.; et al. Molecular Determinants of Human Neutralizing Antibodies Isolated from a Patient Infected with Zika Virus. Sci. Transl. Med. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, E.; Dejnirattisai, W.; Cao, B.; Scheaffer, S.M.; Supasa, P.; Wongwiwat, W.; Esakky, P.; Drury, A.; Mongkolsapaya, J.; Moley, K.H.; et al. Human Antibodies to the Dengue Virus E-Dimer Epitope Have Therapeutic Activity against Zika Virus Infection. Nat. Immunol. 2017, 18, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Abbink, P.; Larocca, R.A.; Dejnirattisai, W.; Peterson, R.; Nkolola, J.P.; Borducchi, E.N.; Supasa, P.; Mongkolsapaya, J.; Screaton, G.R.; Barouch, D.H. Therapeutic and Protective Efficacy of a Dengue Antibody against Zika Infection in Rhesus Monkeys. Nat. Med. 2018, 24, 721–723. [Google Scholar] [CrossRef]
- Kam, Y.-W.; Lee, C.Y.-P.; Teo, T.-H.; Howland, S.W.; Amrun, S.N.; Lum, F.-M.; See, P.; Kng, N.Q.-R.; Huber, R.G.; Xu, M.-H.; et al. Cross-Reactive Dengue Human Monoclonal Antibody Prevents Severe Pathologies and Death from Zika Virus Infections. JCI Insight 2017, 2, e92428. [Google Scholar] [CrossRef] [PubMed]
- Malonis, R.J.; Earnest, J.T.; Kim, A.S.; Angeliadis, M.; Holtsberg, F.W.; Aman, M.J.; Jangra, R.K.; Chandran, K.; Daily, J.P.; Diamond, M.S.; et al. Near-Germline Human Monoclonal Antibodies Neutralize and Protect against Multiple Arthritogenic Alphaviruses. Proc. Natl. Acad. Sci. 2021, 118, e2100104118. [Google Scholar] [CrossRef] [PubMed]
- Fric, J.; Bertin-Maghit, S.; Wang, C.-I.; Nardin, A.; Warter, L. Use of Human Monoclonal Antibodies to Treat Chikungunya Virus Infection. J. Infect. Dis. 2013, 207, 319–322. [Google Scholar] [CrossRef]
- Pal, P.; Dowd, K.A.; Brien, J.D.; Edeling, M.A.; Gorlatov, S.; Johnson, S.; Lee, I.; Akahata, W.; Nabel, G.J.; Richter, M.K.S.; et al. Development of a Highly Protective Combination Monoclonal Antibody Therapy against Chikungunya Virus. PLoS Pathog. 2013, 9, e1003312. [Google Scholar] [CrossRef]
- Smith, S.A.; Silva, L.A.; Fox, J.M.; Flyak, A.I.; Kose, N.; Sapparapu, G.; Khomandiak, S.; Ashbrook, A.W.; Kahle, K.M.; Fong, R.H.; et al. Isolation and Characterization of Broad and Ultrapotent Human Monoclonal Antibodies with Therapeutic Activity against Chikungunya Virus. Cell Host Microbe 2015, 18, 86–95. [Google Scholar] [CrossRef]
- Quiroz, J.A.; Malonis, R.J.; Thackray, L.B.; Cohen, C.A.; Pallesen, J.; Jangra, R.K.; Brown, R.S.; Hofmann, D.; Holtsberg, F.W.; Shulenin, S.; et al. Human Monoclonal Antibodies against Chikungunya Virus Target Multiple Distinct Epitopes in the E1 and E2 Glycoproteins. PLOS Pathog. 2019, 15, e1008061. [Google Scholar] [CrossRef]
- Nitatpattana, N.; Kanjanopas, K.; Yoksan, S.; Satimai, W.; Vongba, N.; Langdatsuwan, S.; Nakgoi, K.; Ratchakum, S.; Wauquier, N.; Souris, M.; et al. Long-Term Persistence of Chikungunya Virus Neutralizing Antibodies in Human Populations of North Eastern Thailand. Virol. J. 2014, 11, 183. [Google Scholar] [CrossRef]
- Langsjoen, R.M.; Haller, S.L.; Roy, C.J.; Vinet-Oliphant, H.; Bergren, N.A.; Erasmus, J.H.; Livengood, J.A.; Powell, T.D.; Weaver, S.C.; Rossi, S.L. Chikungunya Virus Strains Show Lineage-Specific Variations in Virulence and Cross-Protective Ability in Murine and Nonhuman Primate Models. mBio 2018, 9, e02449–17. [Google Scholar] [CrossRef] [PubMed]
- Hucke, F.I.L.; Bestehorn-Willmann, M.; Bugert, J.J. Prophylactic Strategies to Control Chikungunya Virus Infection. Virus Genes 2021, 57, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Pal, P.; Fox, J.M.; Hawman, D.W.; Huang, Y.-J.S.; Messaoudi, I.; Kreklywich, C.; Denton, M.; Legasse, A.W.; Smith, P.P.; Johnson, S.; et al. Chikungunya Viruses That Escape Monoclonal Antibody Therapy Are Clinically Attenuated, Stable, and Not Purified in Mosquitoes. J. Virol. 2014, 88, 8213–8226. [Google Scholar] [CrossRef] [PubMed]
- Julander, J.G.; Anderson, N.; Haese, N.; Andoh, T.; Streblow, D.N.; Cortez, P.; Carter, K.; Marniquet, X.; Watson, H.; Mandron, M. Therapeutic and Prophylactic Treatment with a Virus-Specific Antibody Is Highly Effective in Rodent Models of Chikungunya Infection and Disease. Antiviral Res. 2022, 202, 105295. [Google Scholar] [CrossRef]
- Broeckel, R.; Fox, J.M.; Haese, N.; Kreklywich, C.N.; Sukulpovi-Petty, S.; Legasse, A.; Smith, P.P.; Denton, M.; Corvey, C.; Krishnan, S.; et al. Therapeutic Administration of a Recombinant Human Monoclonal Antibody Reduces the Severity of Chikungunya Virus Disease in Rhesus Macaques. PLoS Negl. Trop. Dis. 2017, 11, e0005637. [Google Scholar] [CrossRef] [PubMed]
- Fong, R.H.; Banik, S.S.R.; Mattia, K.; Barnes, T.; Tucker, D.; Liss, N.; Lu, K.; Selvarajah, S.; Srinivasan, S.; Mabila, M.; et al. Exposure of Epitope Residues on the Outer Face of the Chikungunya Virus Envelope Trimer Determines Antibody Neutralizing Efficacy. J. Virol. 2014, 88, 14364–14379. [Google Scholar] [CrossRef]
- Liu, J.L.; Shriver-Lake, L.C.; Zabetakis, D.; Anderson, G.P.; Goldman, E.R. Selection and Characterization of Protective Anti-Chikungunya Virus Single Domain Antibodies. Mol. Immunol. 2019, 105, 190–197. [Google Scholar] [CrossRef]
- Kim, A.S.; Kafai, N.M.; Winkler, E.S.; Gilliland, T.C.; Cottle, E.L.; Earnest, J.T.; Jethva, P.N.; Kaplonek, P.; Shah, A.P.; Fong, R.H.; et al. Pan-Protective Anti-Alphavirus Human Antibodies Target a Conserved E1 Protein Epitope. Cell 2021, 184, 4414–4429. [Google Scholar] [CrossRef]
- Diamond, M.S.; Pierson, T.C.; Fremont, D.H. The Structural Immunology of Antibody Protection against West Nile Virus. Immunol. Rev. 2008, 225, 212–225. [Google Scholar] [CrossRef]
- Lelli, D.; Moreno, A.; Brocchi, E.; Sozzi, E.; Capucci, L.; Canelli, E.; Barbieri, I.; Zeller, H.; Cordioli, P. West Nile Virus: Characterization and Diagnostic Applications of Monoclonal Antibodies. Virol. J. 2012, 9, 81. [Google Scholar] [CrossRef]
- Duan, T.; Ferguson, M.; Yuan, L.; Xu, F.; Li, G. Human Monoclonal Fab Antibodies Against West Nile Virus and Its Neutralizing Activity Analyzed in Vitro and in Vivo. J. Antivir. Antiretrovir. 2009, 01, 036–042. [Google Scholar] [CrossRef] [PubMed]
- Throsby, M.; Geuijen, C.; Goudsmit, J.; Bakker, A.Q.; Korimbocus, J.; Kramer, R.A.; Clijsters-van Der Horst, M.; De Jong, M.; Jongeneelen, M.; Thijsse, S.; et al. Isolation and Characterization of Human Monoclonal Antibodies from Individuals Infected with West Nile Virus. J. Virol. 2006, 80, 6982–6992. [Google Scholar] [CrossRef] [PubMed]
- Nybakken, G.E.; Oliphant, T.; Johnson, S.; Burke, S.; Diamond, M.S.; Fremont, D.H. Structural Basis of West Nile Virus Neutralization by a Therapeutic Antibody. Nature 2005, 437, 764–769. [Google Scholar] [CrossRef] [PubMed]
- Oliphant, T.; Engle, M.; Nybakken, G.E.; Doane, C.; Johnson, S.; Huang, L.; Gorlatov, S.; Mehlhop, E.; Marri, A.; Chung, K.M.; et al. Development of a Humanized Monoclonal Antibody with Therapeutic Potential against West Nile Virus. Nat. Med. 2005, 11, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Morrey, J.D.; Siddharthan, V.; Olsen, A.L.; Roper, G.Y.; Wang, H.; Baldwin, T.J.; Koenig, S.; Johnson, S.; Nordstrom, J.L.; Diamond, M.S. Humanized Monoclonal Antibody against West Nile Virus Envelope Protein Administered after Neuronal Infection Protects against Lethal Encephalitis in Hamsters. J. Infect. Dis. 2006, 194, 1300–1308. [Google Scholar] [CrossRef]
- Morrey, J.D.; Siddharthan, V.; Olsen, A.L.; Wang, H.; Julander, J.G.; Hall, J.O.; Li, H.; Nordstrom, J.L.; Koenig, S.; Johnson, S.; et al. Defining Limits of Treatment with Humanized Neutralizing Monoclonal Antibody for West Nile Virus Neurological Infection in a Hamster Model. Antimicrob. Agents Chemother. 2007, 51, 2396–2402. [Google Scholar] [CrossRef]
- Zhang, S.; Vogt, M.R.; Oliphant, T.; Engle, M.; Bovshik, E.I.; Diamond, M.S.; Beasley, D.W.C. Development of Resistance to Passive Therapy with a Potently Neutralizing Humanized Monoclonal Antibody against West Nile Virus. J. Infect. Dis. 2009, 200, 202–205. [Google Scholar] [CrossRef]
- Beigel, J.H.; Nordstrom, J.L.; Pillemer, S.R.; Roncal, C.; Goldwater, D.R.; Li, H.; Holland, P.C.; Johnson, S.; Stein, K.; Koenig, S. Safety and Pharmacokinetics of Single Intravenous Dose of MGAWN1, a Novel Monoclonal Antibody to West Nile Virus. Antimicrob. Agents Chemother. 2010, 54, 2431–2436. [Google Scholar] [CrossRef]
- Goo, L.; Debbink, K.; Kose, N.; Sapparapu, G.; Doyle, M.P.; Wessel, A.W.; Richner, J.M.; Burgomaster, K.E.; Larman, B.C.; Dowd, K.A.; et al. A Potently Neutralizing Human Monoclonal Antibody Targeting the West Nile Virus E Protein Preferentially Recognizes Mature Virions. Nat. Microbiol. 2019, 4, 71–77. [Google Scholar] [CrossRef]
- Vogt, M.R.; Moesker, B.; Goudsmit, J.; Jongeneelen, M.; Austin, S.K.; Oliphant, T.; Nelson, S.; Pierson, T.C.; Wilschut, J.; Throsby, M.; et al. Human Monoclonal Antibodies against West Nile Virus Induced by Natural Infection Neutralize at a Postattachment Step. J. Virol. 2009, 83, 6494–6507. [Google Scholar] [CrossRef]
- Kaufmann, B.; Vogt, M.R.; Goudsmit, J.; Holdaway, H.A.; Aksyuk, A.A.; Chipman, P.R.; Kuhn, R.J.; Diamond, M.S.; Rossmann, M.G. Neutralization of West Nile Virus by Cross-Linking of Its Surface Proteins with Fab Fragments of the Human Monoclonal Antibody CR4354. Proc. Natl. Acad. Sci. 2010, 107, 18950–18955. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, T.; Masaki, H.; Takasaki, T.; Aoyama, I.; Yumisashi, T.; Yamanaka, A.; Konishi, E.; Ohnuki, Y.; Muraguchi, A.; Kishi, H. Human Monoclonal Antibodies against West Nile Virus from Japanese Encephalitis-Vaccinated Volunteers. Antiviral Res. 2018, 154, 58–65. [Google Scholar] [CrossRef] [PubMed]
- A Trial to Evaluate the Safety of a Single Intravenous Infusion of MGAWN1 in Healthy Adults. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT00515385?term=nct00515385&draw=2&rank=1 (accessed on 7 September 2023).
- Treatment of West Nile Virus With MGAWN1 (PARADIGM). Available online: https://classic.clinicaltrials.gov/ct2/show/NCT00927953?term=NCT00927953&draw=2&rank=1 (accessed on 7 September 2023).
- Gould, L.H.; Sui, J.; Foellmer, H.; Oliphant, T.; Wang, T.; Ledizet, M.; Murakami, A.; Noonan, K.; Lambeth, C.; Kar, K.; et al. Protective and Therapeutic Capacity of Human Single-Chain Fv-Fc Fusion Proteins against West Nile Virus. J. Virol. 2005, 79, 14606–14613. [Google Scholar] [CrossRef] [PubMed]
- Tharakaraman, K.; Robinson, L.N.; Hatas, A.; Chen, Y.-L.; Siyue, L.; Raguram, S.; Sasisekharan, V.; Wogan, G.N.; Sasisekharan, R. Redesign of a Cross-Reactive Antibody to Dengue Virus with Broad-Spectrum Activity and Increased in Vivo Potency. Proc. Natl. Acad. Sci. 2013, 110. [Google Scholar] [CrossRef] [PubMed]
- Robinson, L.N.; Tharakaraman, K.; Rowley, K.J.; Costa, V.V.; Chan, K.R.; Wong, Y.H.; Ong, L.C.; Tan, H.C.; Koch, T.; Cain, D.; et al. Structure-Guided Design of an Anti-Dengue Antibody Directed to a Non-Immunodominant Epitope. Cell 2015, 162, 493–504. [Google Scholar] [CrossRef]
- Ong, E.Z.; Budigi, Y.; Tan, H.C.; Robinson, L.N.; Rowley, K.J.; Winnett, A.; Hobbie, S.; Shriver, Z.; Babcock, G.J.; Ooi, E.E. Preclinical Evaluation of VIS513, a Therapeutic Antibody against Dengue Virus, in Non-Human Primates. Antiviral Res. 2017, 144, 44–47. [Google Scholar] [CrossRef]
- Budigi, Y.; Ong, E.Z.; Robinson, L.N.; Ong, L.C.; Rowley, K.J.; Winnett, A.; Tan, H.C.; Hobbie, S.; Shriver, Z.; Babcock, G.J.; et al. Neutralization of Antibody-Enhanced Dengue Infection by VIS513, a Pan Serotype Reactive Monoclonal Antibody Targeting Domain III of the Dengue E Protein. PLoS Negl. Trop. Dis. 2018, 12, e0006209. [Google Scholar] [CrossRef]
- Magnani, D.M.; Rogers, T.F.; Beutler, N.; Ricciardi, M.J.; Bailey, V.K.; Gonzalez-Nieto, L.; Briney, B.; Sok, D.; Le, K.; Strubel, A.; et al. Neutralizing Human Monoclonal Antibodies Prevent Zika Virus Infection in Macaques. Sci. Transl. Med. 2017, 9, eaan8184. [Google Scholar] [CrossRef]
- Wang, J.; Bardelli, M.; Espinosa, D.A.; Pedotti, M.; Ng, T.-S.; Bianchi, S.; Simonelli, L.; Lim, E.X.Y.; Foglierini, M.; Zatta, F.; et al. A Human Bi-Specific Antibody against Zika Virus with High Therapeutic Potential. Cell 2017, 171, 229–241. [Google Scholar] [CrossRef]
- Arduin, E.; Arora, S.; Bamert, P.R.; Kuiper, T.; Popp, S.; Geisse, S.; Grau, R.; Calzascia, T.; Zenke, G.; Kovarik, J. Highly Reduced Binding to High and Low Affinity Mouse Fc Gamma Receptors by L234A/L235A and N297A Fc Mutations Engineered into Mouse IgG2a. Mol. Immunol. 2015, 63, 456–463. [Google Scholar] [CrossRef]
- Schlothauer, T.; Herter, S.; Koller, C.F.; Grau-Richards, S.; Steinhart, V.; Spick, C.; Kubbies, M.; Klein, C.; Umaña, P.; Mössner, E. Novel Human IgG1 and IgG4 Fc-Engineered Antibodies with Completely Abolished Immune Effector Functions. Protein Eng. Des. Sel. 2016, 29, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Magnani, D.M.; Rogers, T.F.; Maness, N.J.; Grubaugh, N.D.; Beutler, N.; Bailey, V.K.; Gonzalez-Nieto, L.; Gutman, M.J.; Pedreño-Lopez, N.; Kwal, J.M.; et al. Fetal Demise and Failed Antibody Therapy during Zika Virus Infection of Pregnant Macaques. Nat. Commun. 2018, 9, 1624. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).