Submitted:
13 May 2024
Posted:
14 May 2024
You are already at the latest version
Abstract
Keywords:
Introduction
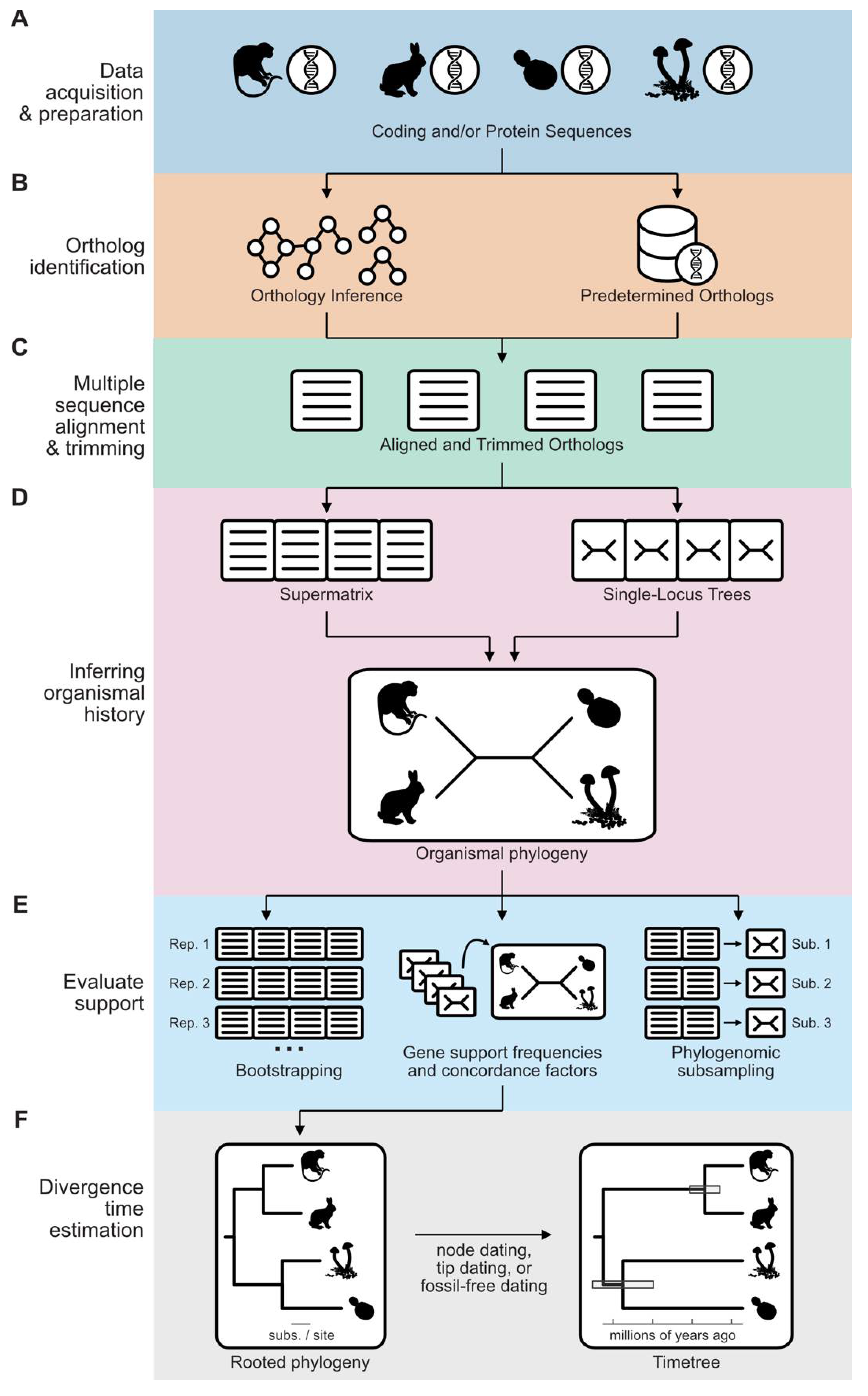
Overview of a Phylogenomic Workflow
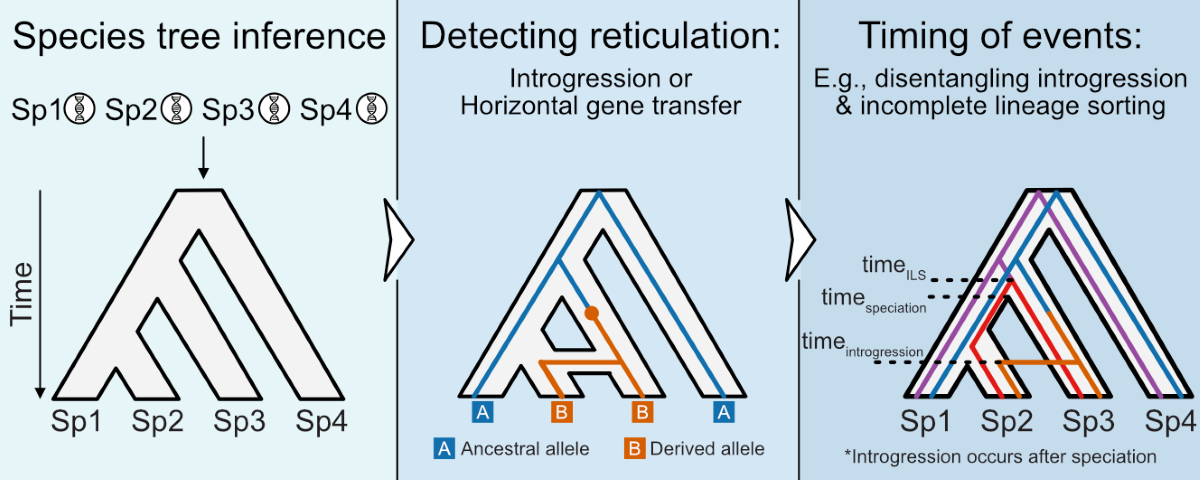
Reticulate Evolution: Identification and Relevance for Relative Divergences
Signatures of Hybridization/Introgression across the Genome, Gene Trees, and Sites
Coalescent Times Differ between Incomplete Lineage Sorting and Introgression
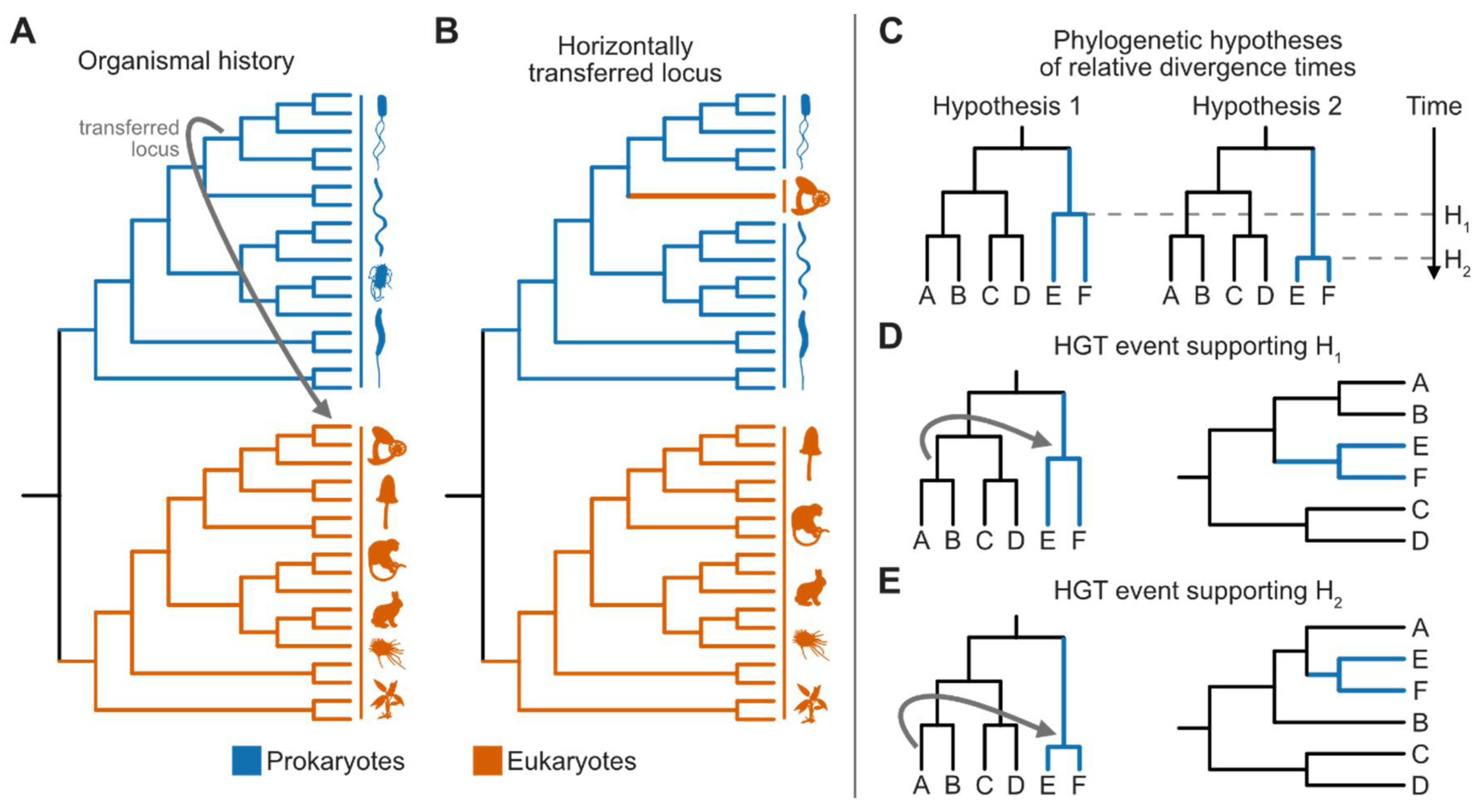
Horizontal Gene Transfer: High Throughput Screens and the Phylogenetic Gold Standard
Horizontal Gene Transfer Events Can Inform Relative Divergences
Time-Calibration of Inferred Phylogenetic Divergences
Conclusion
Funding
Acknowledgments
Competing Interests
References
- Abbott R., D. Albach, S. Ansell, J. W. Arntzen, S. J. E. Baird, et al., 2013 Hybridization and speciation. J. Evol. Biol. 26: 229–246. [CrossRef]
- Adavoudi R., and M. Pilot, 2021 Consequences of Hybridization in Mammals: A Systematic Review. Genes 13: 50. [CrossRef]
- Alexander W. G., J. H. Wisecaver, A. Rokas, and C. T. Hittinger, 2016 Horizontally acquired genes in early-diverging pathogenic fungi enable the use of host nucleosides and nucleotides. Proc. Natl. Acad. Sci. U.S.A. 113: 4116–4121. [CrossRef]
- Allen R., H. Ryan, B. W. Davis, C. King, L. Frantz, et al., 2020 A mitochondrial genetic divergence proxy predicts the reproductive compatibility of mammalian hybrids. Proc. R. Soc. B. 287: 20200690. [CrossRef]
- Álvarez-Carretero S., P. Kapli, and Z. Yang, 2023 Beginner’s Guide on the Use of PAML to Detect Positive Selection, (K. Crandall, Ed.). Molecular Biology and Evolution 40: msad041. [CrossRef]
- Andréoletti J., A. Zwaans, R. C. M. Warnock, G. Aguirre-Fernández, J. Barido-Sottani, et al., 2022 The Occurrence Birth–Death Process for Combined-Evidence Analysis in Macroevolution and Epidemiology, (S. Höhna, Ed.). Systematic Biology 71: 1440–1452. [CrossRef]
- Arnold B. J., I.-T. Huang, and W. P. Hanage, 2022 Horizontal gene transfer and adaptive evolution in bacteria. Nat Rev Microbiol 20: 206–218. [CrossRef]
- Ayres D. L., M. P. Cummings, G. Baele, A. E. Darling, P. O. Lewis, et al., 2019 BEAGLE 3: Improved Performance, Scaling, and Usability for a High-Performance Computing Library for Statistical Phylogenetics, (D. Posada, Ed.). Systematic Biology 68: 1052–1061. [CrossRef]
- Barba-Montoya J., M. Dos Reis, and Z. Yang, 2017 Comparison of different strategies for using fossil calibrations to generate the time prior in Bayesian molecular clock dating. Molecular Phylogenetics and Evolution 114: 386–400. [CrossRef]
- Barley A. J., J. M. Brown, and R. C. Thomson, 2018 Impact of Model Violations on the Inference of Species Boundaries Under the Multispecies Coalescent. Systematic Biology 67: 269–284. [CrossRef]
- Bautista C., S. Marsit, and C. R. Landry, 2021 Interspecific hybrids show a reduced adaptive potential under DNA damaging conditions. Evol Appl 14: 758–769. [CrossRef]
- Bergeron L. A., S. Besenbacher, J. Zheng, P. Li, M. F. Bertelsen, et al., 2023 Evolution of the germline mutation rate across vertebrates. Nature 615: 285–291. [CrossRef]
- Bringloe T. T., D. Zaparenkov, S. Starko, W. S. Grant, C. Vieira, et al., 2021 Whole-genome sequencing reveals forgotten lineages and recurrent hybridizations within the kelp genus Alaria (Phaeophyceae), (M. Coleman, Ed.). J. Phycol. 57: 1721–1738. [CrossRef]
- Buck R., D. Ortega-Del Vecchyo, C. Gehring, R. Michelson, D. Flores-Rentería, et al., 2023 Sequential hybridization may have facilitated ecological transitions in the Southwestern pinyon pine syngameon. New Phytologist 237: 2435–2449. [CrossRef]
- Buckner J. C., R. C. Sanders, B. C. Faircloth, and P. Chakrabarty, 2021 The critical importance of vouchers in genomics. eLife 10: e68264. [CrossRef]
- Capella-Gutiérrez S., M. Marcet-Houben, and T. Gabaldón, 2012 Phylogenomics supports microsporidia as the earliest diverging clade of sequenced fungi. BMC Biol 10: 47. [CrossRef]
- Chain F. J., J. Dushoff, and B. J. Evans, 2011 The odds of duplicate gene persistence after polyploidization. BMC Genomics 12: 599. [CrossRef]
- Chen L., J. Xu, X. Sun, and P. Xu, 2022 Research advances and future perspectives of genomics and genetic improvement in allotetraploid common carp. Reviews in Aquaculture 14: 957–978. [CrossRef]
- Cheon S., J. Zhang, and C. Park, 2020 Is Phylotranscriptomics as Reliable as Phylogenomics?, (E. Teeling, Ed.). Molecular Biology and Evolution 37: 3672–3683. [CrossRef]
- Chikina M., J. D. Robinson, and N. L. Clark, 2016 Hundreds of Genes Experienced Convergent Shifts in Selective Pressure in Marine Mammals. Mol Biol Evol 33: 2182–2192. [CrossRef]
- Coelho M. A., C. Gonçalves, J. P. Sampaio, and P. Gonçalves, 2013 Extensive Intra-Kingdom Horizontal Gene Transfer Converging on a Fungal Fructose Transporter Gene, (J. Heitman, Ed.). PLoS Genet 9: e1003587. [CrossRef]
- Coleman G. A., A. A. Davín, T. A. Mahendrarajah, L. L. Szánthó, A. Spang, et al., 2021 A rooted phylogeny resolves early bacterial evolution. Science 372: eabe0511. [CrossRef]
- Criscuolo A., and S. Gribaldo, 2010 BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol 10: 210. [CrossRef]
- Dannemann M., and J. Kelso, 2017 The Contribution of Neanderthals to Phenotypic Variation in Modern Humans. The American Journal of Human Genetics 101: 578–589. [CrossRef]
- Dannemann M., K. Prüfer, and J. Kelso, 2017 Functional implications of Neandertal introgression in modern humans. Genome Biol 18: 61. [CrossRef]
- Davín A. A., E. Tannier, T. A. Williams, B. Boussau, V. Daubin, et al., 2018 Gene transfers can date the tree of life. Nat Ecol Evol 2: 904–909. [CrossRef]
- Davín A. A., D. Schrempf, T. A. Williams, P. Hugenholtz, and G. J. Szöllősi, 2022 Relative Time Inference Using Lateral Gene Transfers, pp. 75–94 in Environmental Microbial Evolution, Methods in Molecular Biology. edited by Luo H. Springer US, New York, NY.
- Degnan J. H., and N. A. Rosenberg, 2009 Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends in Ecology & Evolution 24: 332–340. [CrossRef]
- Depotter J. R., M. F. Seidl, T. A. Wood, and B. P. Thomma, 2016 Interspecific hybridization impacts host range and pathogenicity of filamentous microbes. Current Opinion in Microbiology 32: 7–13. [CrossRef]
- Díaz-Tapia P., C. A. Maggs, J. A. West, and H. Verbruggen, 2017 Analysis of chloroplast genomes and a supermatrix inform reclassification of the Rhodomelaceae (Rhodophyta), (K. Müller, Ed.). J. Phycol. 53: 920–937. [CrossRef]
- Dobzhansky T., 1982 Genetics and the Origin of Species. Columbia university press.
- Doronina L., O. Reising, H. Clawson, D. A. Ray, and J. Schmitz, 2019 True Homoplasy of Retrotransposon Insertions in Primates, (E. Susko, Ed.). Systematic Biology 68: 482–493. [CrossRef]
- Dorrell R. G., A. Kuo, Z. Füssy, E. H. Richardson, A. Salamov, et al., 2023 Convergent evolution and horizontal gene transfer in Arctic Ocean microalgae. Life Sci. Alliance 6: e202201833. [CrossRef]
- Dos Reis M., P. C. J. Donoghue, and Z. Yang, 2016 Bayesian molecular clock dating of species divergences in the genomics era. Nat Rev Genet 17: 71–80. [CrossRef]
- Dos Reis M., G. F. Gunnell, J. Barba-Montoya, A. Wilkins, Z. Yang, et al., 2018 Using Phylogenomic Data to Explore the Effects of Relaxed Clocks and Calibration Strategies on Divergence Time Estimation: Primates as a Test Case, (S. Ho, Ed.). Systematic Biology 67: 594–615. [CrossRef]
- Douglas J., C. L. Jiménez-Silva, and R. Bouckaert, 2022 StarBeast3: Adaptive Parallelized Bayesian Inference under the Multispecies Coalescent, (R. Bell, Ed.). Systematic Biology 71: 901–916. [CrossRef]
- Drummond A. J., S. Y. W. Ho, M. J. Phillips, and A. Rambaut, 2006 Relaxed Phylogenetics and Dating with Confidence, (D. Penny, Ed.). PLoS Biol 4: e88. [CrossRef]
- Dunn C. W., A. Hejnol, D. Q. Matus, K. Pang, W. E. Browne, et al., 2008 Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452: 745–749. [CrossRef]
- Eaton D. A. R., A. L. Hipp, A. González-Rodríguez, and J. Cavender-Bares, 2015 Historical introgression among the American live oaks and the comparative nature of tests for introgression: INTROGRESSION IN THE AMERICAN LIVE OAKS. Evolution 69: 2587–2601. [CrossRef]
- Edelman N. B., P. B. Frandsen, M. Miyagi, B. Clavijo, J. Davey, et al., 2019 Genomic architecture and introgression shape a butterfly radiation. Science 366: 594–599. [CrossRef]
- Edgar R. C., 2022 Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat Commun 13: 6968. [CrossRef]
- Edger P. P., T. J. Poorten, R. VanBuren, M. A. Hardigan, M. Colle, et al., 2019 Origin and evolution of the octoploid strawberry genome. Nat Genet 51: 541–547. [CrossRef]
- Edwards S. V., 2016 Phylogenomic subsampling: a brief review. Zool Scr 45: 63–74. [CrossRef]
- Feijó A., D. Ge, Z. Wen, J. Cheng, L. Xia, et al., 2022 Mammalian diversification bursts and biotic turnovers are synchronous with Cenozoic geoclimatic events in Asia. Proc. Natl. Acad. Sci. U.S.A. 119: e2207845119. [CrossRef]
- Fernández R., T. Gabaldón, and C. Dessimoz, 2019 Orthology: definitions, inference, and impact on species phylogeny inference. [CrossRef]
- Fernández R., and T. Gabaldón, 2020 Gene gain and loss across the metazoan tree of life. Nat Ecol Evol 4: 524–533.
- Flouri T., X. Jiao, B. Rannala, and Z. Yang, 2018 Species Tree Inference with BPP Using Genomic Sequences and the Multispecies Coalescent, (A. D. Yoder, Ed.). Molecular Biology and Evolution 35: 2585–2593. [CrossRef]
- Flouri T., J. Huang, X. Jiao, P. Kapli, B. Rannala, et al., 2022 Bayesian Phylogenetic Inference using Relaxed-clocks and the Multispecies Coalescent, (R. Nielsen, Ed.). Molecular Biology and Evolution 39: msac161. [CrossRef]
- Foley N. M., V. C. Mason, A. J. Harris, K. R. Bredemeyer, J. Damas, et al., 2023 A genomic timescale for placental mammal evolution. Science 380: eabl8189. [CrossRef]
- Friedman R., and B. Ely, 2012 Codon Usage Methods for Horizontal Gene Transfer Detection Generate an Abundance of False Positive and False Negative Results. Curr Microbiol 65: 639–642. [CrossRef]
- Galindo L. J., P. López-García, G. Torruella, S. Karpov, and D. Moreira, 2021 Phylogenomics of a new fungal phylum reveals multiple waves of reductive evolution across Holomycota. Nat Commun 12: 4973. [CrossRef]
- Galtier N., 2007 A Model of Horizontal Gene Transfer and the Bacterial Phylogeny Problem, (M. Steel, Ed.). Systematic Biology 56: 633–642. [CrossRef]
- Gatesy J., R. W. Meredith, J. E. Janecka, M. P. Simmons, W. J. Murphy, et al., 2017 Resolution of a concatenation/coalescence kerfuffle: partitioned coalescence support and a robust family-level tree for Mammalia. Cladistics 33: 295–332. [CrossRef]
- Gladyshev E. A., M. Meselson, and I. R. Arkhipova, 2008 Massive Horizontal Gene Transfer in Bdelloid Rotifers. Science 320: 1210–1213. [CrossRef]
- Gonçalves C., J. H. Wisecaver, J. Kominek, M. S. Oom, M. J. Leandro, et al., 2018 Evidence for loss and reacquisition of alcoholic fermentation in a fructophilic yeast lineage. eLife 7: e33034. [CrossRef]
- Gonçalves C., and P. Gonçalves, 2019 Multilayered horizontal operon transfers from bacteria reconstruct a thiamine salvage pathway in yeasts. Proc. Natl. Acad. Sci. U.S.A. 116: 22219–22228. [CrossRef]
- Gonçalves P., and C. Gonçalves, 2022 Horizontal gene transfer in yeasts. Current Opinion in Genetics & Development 76: 101950. [CrossRef]
- Gophna U., and N. Altman-Price, 2022 Horizontal Gene Transfer in Archaea—From Mechanisms to Genome Evolution. Annu. Rev. Microbiol. 76: 481–502. [CrossRef]
- Green R. E., J. Krause, A. W. Briggs, T. Maricic, U. Stenzel, et al., 2010 A draft sequence of the Neandertal genome. science 328: 710–722. [CrossRef]
- Harvey M. G., G. A. Bravo, S. Claramunt, A. M. Cuervo, G. E. Derryberry, et al., 2020 The evolution of a tropical biodiversity hotspot. Science 370: 1343–1348. [CrossRef]
- Heath T. A., J. P. Huelsenbeck, and T. Stadler, 2014 The fossilized birth–death process for coherent calibration of divergence-time estimates. Proc. Natl. Acad. Sci. U.S.A. 111. [CrossRef]
- Hibbins M. S., and M. W. Hahn, 2022 Phylogenomic approaches to detecting and characterizing introgression, (M. Turelli, Ed.). Genetics 220: iyab173. [CrossRef]
- Ho S. Y. M., 2007 Calibrating molecular estimates of substitution rates and divergence times in birds. J Avian Biology 38: 409–414. [CrossRef]
- Ho S. Y. W., and M. J. Phillips, 2009 Accounting for Calibration Uncertainty in Phylogenetic Estimation of Evolutionary Divergence Times. Systematic Biology 58: 367–380. [CrossRef]
- Huang J., T. Flouri, and Z. Yang, 2020 A Simulation Study to Examine the Information Content in Phylogenomic Data Sets under the Multispecies Coalescent Model, (B. Su, Ed.). Molecular Biology and Evolution 37: 3211–3224. [CrossRef]
- Husnik F., and J. P. McCutcheon, 2018 Functional horizontal gene transfer from bacteria to eukaryotes. Nature Reviews Microbiology 16: 67–79.
- Huson D. H., T. Huson D. H., T. Klöpper, P. J. Lockhart, and M. A. Steel, 2005 Reconstruction of Reticulate Networks from Gene Trees, pp. 233–249 in Research in Computational Molecular Biology, Lecture Notes in Computer Science. edited by Miyano S., Mesirov J., Kasif S., Istrail S., Pevzner P. A., et al. Springer Berlin Heidelberg, Berlin, Heidelberg.
- Irwin N. A. T., A. A. Pittis, T. A. Richards, and P. J. Keeling, 2021 Systematic evaluation of horizontal gene transfer between eukaryotes and viruses. Nat Microbiol 7: 327–336. [CrossRef]
- Jaramillo V. D. A., S. A. Sukno, and M. R. Thon, 2015 Identification of horizontally transferred genes in the genus Colletotrichum reveals a steady tempo of bacterial to fungal gene transfer. BMC Genomics 16: 2. [CrossRef]
- Jiao X., T. Flouri, and Z. Yang, 2021 Multispecies coalescent and its applications to infer species phylogenies and cross-species gene flow. National Science Review 8: nwab127.
- Kainer D., and R. Lanfear, 2015 The Effects of Partitioning on Phylogenetic Inference. Molecular Biology and Evolution 32: 1611–1627. [CrossRef]
- Kapli P., Z. Yang, and M. J. Telford, 2020 Phylogenetic tree building in the genomic age. Nat Rev Genet 21: 428–444. [CrossRef]
- Katoh K., and D. M. Standley, 2013 MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30: 772–780. [CrossRef]
- King N., and A. Rokas, 2017 Embracing Uncertainty in Reconstructing Early Animal Evolution. Current Biology 27: R1081–R1088. [CrossRef]
- Kishino H., and M. Hasegawa, 1989 Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol 29: 170–179. [CrossRef]
- Koch N. M., and P. M. Carmona, 2024 Chronospaces: an R package for the statistical exploration of divergence times reveals extreme dependence on molecular clocks and gene choice.
- Kominek J., D. T. Doering, D. A. Opulente, X.-X. Shen, X. Zhou, et al., 2019 Eukaryotic Acquisition of a Bacterial Operon. Cell 176: 1356-1366.e10. [CrossRef]
- Koutsovoulos G. D., S. Granjeon Noriot, M. Bailly-Bechet, E. G. J. Danchin, and C. Rancurel, 2022 AvP: A software package for automatic phylogenetic detection of candidate horizontal gene transfers, (M. Ziemann, Ed.). PLoS Comput Biol 18: e1010686. [CrossRef]
- Kowalczyk A., W. K. Meyer, R. Partha, W. Mao, N. L. Clark, et al., 2019 RERconverge: an R package for associating evolutionary rates with convergent traits, (A. Valencia, Ed.). Bioinformatics 35: 4815–4817. [CrossRef]
- Lara M. C., J. L. Patton, and M. N. F. Da Silva, 1996 The Simultaneous Diversification of South American Echimyid Rodents (Hystricognathi) Based on Complete Cytochrome b Sequences. Molecular Phylogenetics and Evolution 5: 403–413. [CrossRef]
- Lenski R. E., 2017 Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. The ISME Journal 11: 2181–2194. [CrossRef]
- Lepage T., D. Bryant, H. Philippe, and N. Lartillot, 2007 A General Comparison of Relaxed Molecular Clock Models. Molecular Biology and Evolution 24: 2669–2680. [CrossRef]
- Li Z., A. R. De La Torre, L. Sterck, F. M. Cánovas, C. Avila, et al., 2017 Single-Copy Genes as Molecular Markers for Phylogenomic Studies in Seed Plants. Genome Biology and Evolution 9: 1130–1147. [CrossRef]
- Li Y., J. L. Steenwyk, Y. Chang, Y. Wang, T. Y. James, et al., 2021 A genome-scale phylogeny of the kingdom Fungi. Current Biology 31: 1653-1665.e5. [CrossRef]
- Li Y., Z. Liu, C. Liu, Z. Shi, L. Pang, et al., 2022a HGT is widespread in insects and contributes to male courtship in lepidopterans. Cell 185: 2975-2987.e10. [CrossRef]
- Li Y., H. Liu, J. L. Steenwyk, A. L. LaBella, M.-C. Harrison, et al., 2022b Contrasting modes of macro and microsynteny evolution in a eukaryotic subphylum. Current Biology S0960982222016700. [CrossRef]
- Lin X., S. Patel, A. P. Litvintseva, A. Floyd, T. G. Mitchell, et al., 2009 Diploids in the Cryptococcus neoformans Serotype A Population Homozygous for the α Mating Type Originate via Unisexual Mating, (A. Andrianopoulos, Ed.). PLoS Pathog 5: e1000283. [CrossRef]
- Liu L., D. K. Pearl, R. T. Brumfield, and S. V. Edwards, 2008 Estimating species trees using multiple-allele DNA sequence data. Evolution 62: 2080–2091.
- Liu L., L. Yu, L. Kubatko, D. K. Pearl, and S. V. Edwards, 2009a Coalescent methods for estimating phylogenetic trees. Molecular Phylogenetics and Evolution 53: 320–328. [CrossRef]
- Liu L., L. Yu, D. K. Pearl, and S. V. Edwards, 2009b Estimating Species Phylogenies Using Coalescence Times among Sequences. Systematic Biology 58: 468–477. [CrossRef]
- Liu H., J. L. Steenwyk, X. Zhou, D. T. Schultz, K. M. Kocot, et al., 2023 A genome-scale Opisthokonta tree of life: toward phylogenomic resolution of ancient divergences. Evolutionary Biology.
- Lutzoni F., M. D. Nowak, M. E. Alfaro, V. Reeb, J. Miadlikowska, et al., 2018 Contemporaneous radiations of fungi and plants linked to symbiosis. Nat Commun 9: 5451. [CrossRef]
- Mallet J., 2005 Hybridization as an invasion of the genome. Trends in Ecology & Evolution 20: 229–237. [CrossRef]
- Mallet J., 2008 Hybridization, ecological races and the nature of species: empirical evidence for the ease of speciation. Phil. Trans. R. Soc. B 363: 2971–2986. [CrossRef]
- Marcet-Houben M., and T. Gabaldón, 2015 Beyond the Whole-Genome Duplication: Phylogenetic Evidence for an Ancient Interspecies Hybridization in the Baker’s Yeast Lineage, (L. D. Hurst, Ed.). PLoS Biol 13: e1002220. [CrossRef]
- Mateo-Estrada V., L. Graña-Miraglia, G. López-Leal, and S. Castillo-Ramírez, 2019 Phylogenomics Reveals Clear Cases of Misclassification and Genus-Wide Phylogenetic Markers for Acinetobacter, (L. Delaye, Ed.). Genome Biology and Evolution 11: 2531–2541. [CrossRef]
- Minh B. Q., H. A. Schmidt, O. Chernomor, D. Schrempf, M. D. Woodhams, et al., 2020 IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era, (E. Teeling, Ed.). Molecular Biology and Evolution 37: 1530–1534. [CrossRef]
- Misof B., S. Liu, K. Meusemann, R. S. Peters, A. Donath, et al., 2014 Phylogenomics resolves the timing and pattern of insect evolution. Science 346: 763–767. [CrossRef]
- Mixão V., and T. Gabaldón, 2020 Genomic evidence for a hybrid origin of the yeast opportunistic pathogen Candida albicans. BMC Biol 18: 48. [CrossRef]
- Moi D., C. Moi D., C. Bernard, M. Steinegger, Y. Nevers, M. Langleib, et al., 2023 Structural phylogenetics unravels the evolutionary diversification of communication systems in gram-positive bacteria and their viruses. Bioinformatics.
- Mongiardino Koch N., 2021 Phylogenomic Subsampling and the Search for Phylogenetically Reliable Loci, (Y. Satta, Ed.). Molecular Biology and Evolution 38: 4025–4038. [CrossRef]
- Moran B. M., C. Payne, Q. Langdon, D. L. Powell, Y. Brandvain, et al., 2021 The genomic consequences of hybridization. eLife 10: e69016. [CrossRef]
- Muñoz-Gómez S. A., F. G. Mejía-Franco, K. Durnin, M. Colp, C. J. Grisdale, et al., 2017 The New Red Algal Subphylum Proteorhodophytina Comprises the Largest and Most Divergent Plastid Genomes Known. Current Biology 27: 1677-1684.e4. [CrossRef]
- Neafsey D. E., B. M. Barker, T. J. Sharpton, J. E. Stajich, D. J. Park, et al., 2010 Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res. 20: 938–946. [CrossRef]
- Nelsen M. P., C. S. Moreau, C. Kevin Boyce, and R. H. Ree, 2023 Macroecological diversification of ants is linked to angiosperm evolution. Evolution Letters 7: 79–87. [CrossRef]
- Oliveros C. H., D. J. Field, D. T. Ksepka, F. K. Barker, A. Aleixo, et al., 2019 Earth history and the passerine superradiation. Proc. Natl. Acad. Sci. U.S.A. 116: 7916–7925. [CrossRef]
- One Thousand Plant Transcriptomes Initiative, 2019 One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685. [CrossRef]
- Opulente D. A., A. L. Opulente D. A., A. L. LaBella, M.-C. Harrison, J. F. Wolters, C. Liu, et al., 2023 Genomic and ecological factors shaping specialism and generalism across an entire subphylum. Evolutionary Biology.
- Ortiz-Merino R. A., N. Kuanyshev, S. Braun-Galleani, K. P. Byrne, D. Porro, et al., 2017 Evolutionary restoration of fertility in an interspecies hybrid yeast, by whole-genome duplication after a failed mating-type switch, (L. Hurst, Ed.). PLoS Biol 15: e2002128. [CrossRef]
- Osmanski A. B., N. S. Paulat, J. Korstian, J. R. Grimshaw, M. Halsey, et al., 2023 Insights into mammalian TE diversity through the curation of 248 genome assemblies. Science 380: eabn1430. [CrossRef]
- Ozkan H., A. A. Levy, and M. Feldman, 2001 Allopolyploidy-Induced Rapid Genome Evolution in the Wheat ( Aegilops – Triticum ) Group. Plant Cell 13: 1735–1747. [CrossRef]
- Parey E., A. Louis, J. Montfort, O. Bouchez, C. Roques, et al., 2023 Genome structures resolve the early diversification of teleost fishes. Science 379: 572–575. [CrossRef]
- Parham J. F., P. C. J. Donoghue, C. J. Bell, T. D. Calway, J. J. Head, et al., 2012 Best Practices for Justifying Fossil Calibrations. Systematic Biology 61: 346–359. [CrossRef]
- Parks D. H., M. Chuvochina, D. W. Waite, C. Rinke, A. Skarshewski, et al., 2018 A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36: 996–1004. [CrossRef]
- Pease J. B., and M. W. Hahn, 2015 Detection and Polarization of Introgression in a Five-Taxon Phylogeny. Systematic Biology 64: 651–662. [CrossRef]
- Philippe H., D. M. D. Vienne, V. Ranwez, B. Roure, D. Baurain, et al., 2017 Pitfalls in supermatrix phylogenomics. EJT. [CrossRef]
- Phillips M. A., J. L. Steenwyk, X.-X. Shen, and A. Rokas, 2021 Examination of Gene Loss in the DNA Mismatch Repair Pathway and Its Mutational Consequences in a Fungal Phylum, (K. Wolfe, Ed.). Genome Biology and Evolution 13: evab219. [CrossRef]
- Pipes L., H. Wang, J. P. Huelsenbeck, and R. Nielsen, 2021 Assessing Uncertainty in the Rooting of the SARS-CoV-2 Phylogeny, (H. Malik, Ed.). Molecular Biology and Evolution 38: 1537–1543. [CrossRef]
- Qiao H., W. Liu, Y. Zhang, Y. Zhang, and Q. Q. Li, 2019 Genetic admixture accelerates invasion via provisioning rapid adaptive evolution. Mol Ecol 28: 4012–4027. [CrossRef]
- Racimo F., S. Sankararaman, R. Nielsen, and E. Huerta-Sánchez, 2015 Evidence for archaic adaptive introgression in humans. Nature Reviews Genetics 16: 359–371.
- Rieseberg L. H., S.-C. Kim, R. A. Randell, K. D. Whitney, B. L. Gross, et al., 2007 Hybridization and the colonization of novel habitats by annual sunflowers. Genetica 129: 149–165. [CrossRef]
- Rokas A., B. L. Williams, N. King, and S. B. Carroll, 2003 Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425: 798–804. [CrossRef]
- Rokas A., and S. B. Carroll, 2006 Bushes in the Tree of Life. PLoS Biol 4: e352. [CrossRef]
- S. Meseguer A., and F. L. Condamine, 2020 Ancient tropical extinctions at high latitudes contributed to the latitudinal diversity gradient*. Evolution 74: 1966–1987. [CrossRef]
- Sabrina Pankey M., D. C. Plachetzki, K. J. Macartney, M. Gastaldi, M. Slattery, et al., 2022 Cophylogeny and convergence shape holobiont evolution in sponge–microbe symbioses. Nat Ecol Evol 6: 750–762. [CrossRef]
- Salojärvi J., A. Rambani, Z. Yu, R. Guyot, S. Strickler, et al., 2024 The genome and population genomics of allopolyploid Coffea arabica reveal the diversification history of modern coffee cultivars. Nat Genet 56: 721–731. [CrossRef]
- Sanderson M. J., 2003 r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19: 301–302. [CrossRef]
- Sankararaman S., S. Mallick, N. Patterson, and D. Reich, 2016 The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Current Biology 26: 1241–1247.
- Sayyari E., and S. Mirarab, 2018 Testing for Polytomies in Phylogenetic Species Trees Using Quartet Frequencies. Genes 9: 132. [CrossRef]
- Scannell D. R., K. P. Byrne, J. L. Gordon, S. Wong, and K. H. Wolfe, 2006 Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440: 341–345. [CrossRef]
- Schenk J. J., 2016 Consequences of Secondary Calibrations on Divergence Time Estimates, (A. Janke, Ed.). PLoS ONE 11: e0148228. [CrossRef]
- Schönknecht G., W.-H. Chen, C. M. Ternes, G. G. Barbier, R. P. Shrestha, et al., 2013 Gene Transfer from Bacteria and Archaea Facilitated Evolution of an Extremophilic Eukaryote. Science 339: 1207–1210. [CrossRef]
- Schubert M., T. Marcussen, A. S. Meseguer, and S. Fjellheim, 2019 The grass subfamily Pooideae: Cretaceous–Palaeocene origin and climate-driven Cenozoic diversification, (G. Jordan, Ed.). Global Ecol Biogeogr geb.12923. [CrossRef]
- Schultz D. T., S. H. D. Haddock, J. V. Bredeson, R. E. Green, O. Simakov, et al., 2023 Ancient gene linkages support ctenophores as sister to other animals. Nature. [CrossRef]
- Session A. M., Y. Uno, T. Kwon, J. A. Chapman, A. Toyoda, et al., 2016 Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538: 336–343. [CrossRef]
- Session A. M., and D. S. Rokhsar, 2023 Transposon signatures of allopolyploid genome evolution. Nat Commun 14: 3180. [CrossRef]
- Shaul S., and D. Graur, 2002 Playing chicken ( Gallus gallus ): methodological inconsistencies of molecular divergence date estimates due to secondary calibration points. Gene 300: 59–61. [CrossRef]
- Shen X.-X., D. A. Opulente, J. Kominek, X. Zhou, J. L. Steenwyk, et al., 2018 Tempo and Mode of Genome Evolution in the Budding Yeast Subphylum. Cell 175: 1533-1545.e20. [CrossRef]
- Shen X.-X., J. L. Steenwyk, A. L. LaBella, D. A. Opulente, X. Zhou, et al., 2020 Genome-scale phylogeny and contrasting modes of genome evolution in the fungal phylum Ascomycota. Sci. Adv. 6: eabd0079. [CrossRef]
- Shimodaira H., and M. Hasegawa, 1999 Multiple Comparisons of Log-Likelihoods with Applications to Phylogenetic Inference. Molecular Biology and Evolution 16: 1114–1116. [CrossRef]
- Sierra-Patev S., B. Min, M. Naranjo-Ortiz, B. Looney, Z. Konkel, et al., 2023 A global phylogenomic analysis of the shiitake genus Lentinula. Proc. Natl. Acad. Sci. U.S.A. 120: e2214076120. [CrossRef]
- Sievers F., and D. G. Higgins, 2018 Clustal Omega for making accurate alignments of many protein sequences: Clustal Omega for Many Protein Sequences. Protein Science 27: 135–145. [CrossRef]
- Simion P., H. Philippe, D. Baurain, M. Jager, D. J. Richter, et al., 2017 A Large and Consistent Phylogenomic Dataset Supports Sponges as the Sister Group to All Other Animals. Current Biology 27: 958–967. [CrossRef]
- Simonti C. N., B. Vernot, L. Bastarache, E. Bottinger, D. S. Carrell, et al., 2016 The phenotypic legacy of admixture between modern humans and Neandertals. Science 351: 737–741. [CrossRef]
- Smith S. A., and B. C. O’Meara, 2012 treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28: 2689–2690. [CrossRef]
- Smith S. A., J. W. Brown, and J. F. Walker, 2018 So many genes, so little time: A practical approach to divergence-time estimation in the genomic era, (H. Escriva, Ed.). PLoS ONE 13: e0197433. [CrossRef]
- Song H., Y. Wang, H. Shao, Z. Li, P. Hu, et al., 2023 Scaphopoda is the sister taxon to Bivalvia: Evidence of ancient incomplete lineage sorting. Proc. Natl. Acad. Sci. U.S.A. 120: e2302361120. [CrossRef]
- Sousa F., J. Neiva, N. Martins, R. Jacinto, L. Anderson, et al., 2019 Increased evolutionary rates and conserved transcriptional response following allopolyploidization in brown algae: GENOME EVOLUTION IN ALLOPOLYPLOID ALGAE. Evolution 73: 59–72. [CrossRef]
- Stadler T., and Z. Yang, 2013 Dating Phylogenies with Sequentially Sampled Tips. Systematic Biology 62: 674–688. [CrossRef]
- Stadler T., O. G. Pybus, and M. P. H. Stumpf, 2021 Phylodynamics for cell biologists. Science 371: eaah6266. [CrossRef]
- Steenwyk J. L., D. A. Opulente, J. Kominek, X.-X. Shen, X. Zhou, et al., 2019a Extensive loss of cell-cycle and DNA repair genes in an ancient lineage of bipolar budding yeasts, (S. Kamoun, Ed.). PLoS Biol 17: e3000255. [CrossRef]
- Steenwyk J. L., X.-X. Shen, A. L. Lind, G. H. Goldman, and A. Rokas, 2019b A Robust Phylogenomic Time Tree for Biotechnologically and Medically Important Fungi in the Genera Aspergillus and Penicillium, (J. P. Boyle, Ed.). mBio 10: e00925-19. [CrossRef]
- Steenwyk J. L., A. L. Lind, L. N. A. Ries, T. F. dos Reis, L. P. Silva, et al., 2020a Pathogenic Allodiploid Hybrids of Aspergillus Fungi. Current Biology 30: 2495-2507.e7. [CrossRef]
- Steenwyk J. L., T. J. Buida, Y. Li, X.-X. Shen, and A. Rokas, 2020b ClipKIT: A multiple sequence alignment trimming software for accurate phylogenomic inference, (A. Hejnol, Ed.). PLoS Biol 18: e3001007. [CrossRef]
- Steenwyk J. L., T. J. Buida, A. L. Labella, Y. Li, X.-X. Shen, et al., 2021 PhyKIT: a broadly applicable UNIX shell toolkit for processing and analyzing phylogenomic data, (R. Schwartz, Ed.). Bioinformatics 37: 2325–2331. [CrossRef]
- Steenwyk J. L., M. A. Phillips, F. Yang, S. S. Date, T. R. Graham, et al., 2022a An orthologous gene coevolution network provides insight into eukaryotic cellular and genomic structure and function. Sci. Adv. 8: eabn0105. [CrossRef]
- Steenwyk J. L., C. Steenwyk J. L., C. Balamurugan, H. A. Raja, C. Gonçalves, N. Li, et al., 2022b Phylogenomics reveals extensive misidentification of fungal strains from the genus Aspergillus. Evolutionary Biology.
- Steenwyk J. L., and A. Rokas, 2023 The dawn of relaxed phylogenetics. PLoS Biol 21: e3001998. [CrossRef]
- Steenwyk J. L., Y. Li, X. Zhou, X.-X. Shen, and A. Rokas, 2023a Incongruence in the phylogenomics era. Nature Reviews Genetics.
- Steenwyk J. L., S. L. Steenwyk J. L., S. L. Knowles, R. Bastos, C. Balamurugan, D. Rinker, et al., 2023b Evolutionary origin, population diversity, and diagnostics for a cryptic hybrid pathogen. Evolutionary Biology.
- Steenwyk J., and N. King, 2023 From Genes to Genomes: Opportunities and Challenges for Synteny-based Phylogenies. Preprints. [CrossRef]
- Stukenbrock E. H., 2016 The Role of Hybridization in the Evolution and Emergence of New Fungal Plant Pathogens. Phytopathology® 106: 104–112. [CrossRef]
- Suárez-Menéndez M., M. Bérubé, F. Furni, V. E. Rivera-León, M.-P. Heide-Jørgensen, et al., 2023 Wild pedigrees inform mutation rates and historic abundance in baleen whales. Science 381: 990–995. [CrossRef]
- Suvorov A., B. Y. Kim, J. Wang, E. E. Armstrong, D. Peede, et al., 2022 Widespread introgression across a phylogeny of 155 Drosophila genomes. Current Biology 32: 111-123.e5. [CrossRef]
- Swofford D. L., P. J. Waddell, J. P. Huelsenbeck, P. G. Foster, P. O. Lewis, et al., 2001 Bias in Phylogenetic Estimation and Its Relevance to the Choice between Parsimony and Likelihood Methods. Systematic Biology 50: 525–539. [CrossRef]
- Tahon G., P. Geesink, and T. J. G. Ettema, 2021 Expanding Archaeal Diversity and Phylogeny: Past, Present, and Future. Annu. Rev. Microbiol. 75: 359–381. [CrossRef]
- Talavera G., and J. Castresana, 2007 Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments, (K. Kjer, R. Page, and J. Sullivan, Eds.). Systematic Biology 56: 564–577. [CrossRef]
- Tan G., M. Muffato, C. Ledergerber, J. Herrero, N. Goldman, et al., 2015 Current Methods for Automated Filtering of Multiple Sequence Alignments Frequently Worsen Single-Gene Phylogenetic Inference. Syst Biol 64: 778–791. [CrossRef]
- Tao Q., K. Tao Q., K. Tamura, and S. Kumar, 2020 Efficient Methods for Dating Evolutionary Divergences, pp. 197–219 in The Molecular Evolutionary Clock, edited by Ho S. Y. W. Springer International Publishing, Cham.
- Tiley G. P., J. W. Poelstra, M. Dos Reis, Z. Yang, and A. D. Yoder, 2020 Molecular Clocks without Rocks: New Solutions for Old Problems. Trends in Genetics 36: 845–856. [CrossRef]
- Tiley G. P., T. Flouri, X. Jiao, J. W. Poelstra, B. Xu, et al., 2023 Estimation of species divergence times in presence of cross-species gene flow, (B. Carstens, Ed.). Systematic Biology 72: 820–836. [CrossRef]
- Title P. O., S. Singhal, M. C. Grundler, G. C. Costa, R. A. Pyron, et al., 2024 The macroevolutionary singularity of snakes. Science 383: 918–923. [CrossRef]
- Tomlin C. M., S. Rajaraman, J. T. Sebesta, A.-C. Scheen, M. Bendiksby, et al., 2024 Allopolyploid origin and diversification of the Hawaiian endemic mints. Nat Commun 15: 3109. [CrossRef]
- Turnbull R., J. L. Steenwyk, S. J. Mutch, P. Scholten, V. W. Salazar, et al., 2023 OrthoFlow: phylogenomic analysis and diagnostics with one command. In Review.
- Upham N. S., J. A. Esselstyn, and W. Jetz, 2019 Inferring the mammal tree: Species-level sets of phylogenies for questions in ecology, evolution, and conservation, (A. J. Tanentzap, Ed.). PLoS Biol 17: e3000494. [CrossRef]
- Upham N. S., J. A. Esselstyn, and W. Jetz, 2021 Molecules and fossils tell distinct yet complementary stories of mammal diversification. Current Biology 31: 4195-4206.e3. [CrossRef]
- Van Etten J., and D. Bhattacharya, 2020 Horizontal Gene Transfer in Eukaryotes: Not if, but How Much? Trends in Genetics 36: 915–925. [CrossRef]
- Volz E. M., K. Koelle, and T. Bedford, 2013 Viral Phylodynamics, (S. Wodak, Ed.). PLoS Comput Biol 9: e1002947. [CrossRef]
- Wang M.-S., G. G. R. Murray, D. Mann, P. Groves, A. O. Vershinina, et al., 2022 A polar bear paleogenome reveals extensive ancient gene flow from polar bears into brown bears. Nat Ecol Evol 6: 936–944. [CrossRef]
- Wei W., W.-C. Ho, M. G. Behringer, S. F. Miller, G. Bcharah, et al., 2022 Rapid evolution of mutation rate and spectrum in response to environmental and population-genetic challenges. Nat Commun 13: 4752. [CrossRef]
- Wickett N. J., S. Mirarab, N. Nguyen, T. Warnow, E. Carpenter, et al., 2014 Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. U.S.A. 111. [CrossRef]
- Williams T. A., C. J. Cox, P. G. Foster, G. J. Szöllősi, and T. M. Embley, 2019 Phylogenomics provides robust support for a two-domains tree of life. Nat Ecol Evol 4: 138–147. [CrossRef]
- Worobey M., T. D. Watts, R. A. McKay, M. A. Suchard, T. Granade, et al., 2016 1970s and ‘Patient 0’ HIV-1 genomes illuminate early HIV/AIDS history in North America. Nature 539: 98–101. [CrossRef]
- Yang Z., and B. Rannala, 2010 Bayesian species delimitation using multilocus sequence data. Proceedings of the National Academy of Sciences 107: 9264–9269.
- Yu Y., R. M. Barnett, and L. Nakhleh, 2013 Parsimonious Inference of Hybridization in the Presence of Incomplete Lineage Sorting. Systematic Biology 62: 738–751. [CrossRef]
- Yuan L., H. Lu, F. Li, J. Nielsen, and E. J. Kerkhoven, 2023 HGTphyloDetect: facilitating the identification and phylogenetic analysis of horizontal gene transfer. Briefings in Bioinformatics 24: bbad035. [CrossRef]
- Yue J., X. Hu, H. Sun, Y. Yang, and J. Huang, 2012 Widespread impact of horizontal gene transfer on plant colonization of land. Nat Commun 3: 1152. [CrossRef]
- Zanewich K. P., D. W. Pearce, and S. B. Rood, 2018 Heterosis in poplar involves phenotypic stability: cottonwood hybrids outperform their parental species at suboptimal temperatures. Tree Physiology 38: 789–800. [CrossRef]
- Zeberg H., and S. Pääbo, 2020 The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature 587: 610–612. [CrossRef]
- Zhang R., H.-Y. Ou, F. Gao, and H. Luo, 2014a Identification of Horizontally-transferred Genomic Islands and Genome Segmentation Points by Using the GC Profile Method. CG 15: 113–121. [CrossRef]
- Zhang G., C. Li, Q. Li, B. Li, D. M. Larkin, et al., 2014b Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346: 1311–1320. [CrossRef]
- Zhang C., M. Rabiee, E. Sayyari, and S. Mirarab, 2018 ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19: 153. [CrossRef]
- Zheng Y., and A. Janke, 2018 Gene flow analysis method, the D-statistic, is robust in a wide parameter space. BMC bioinformatics 19: 1–19.
- Zhou X., X.-X. Shen, C. T. Hittinger, and A. Rokas, 2018 Evaluating Fast Maximum Likelihood-Based Phylogenetic Programs Using Empirical Phylogenomic Data Sets. Molecular Biology and Evolution 35: 486–503. [CrossRef]
- Zhu Q., M. Kosoy, and K. Dittmar, 2014 HGTector: an automated method facilitating genome-wide discovery of putative horizontal gene transfers. BMC Genomics 15: 717. [CrossRef]
- Zhu Q., U. Mai, W. Pfeiffer, S. Janssen, F. Asnicar, et al., 2019 Phylogenomics of 10,575 genomes reveals evolutionary proximity between domains Bacteria and Archaea. Nat Commun 10: 5477. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



