Submitted:
06 September 2023
Posted:
07 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
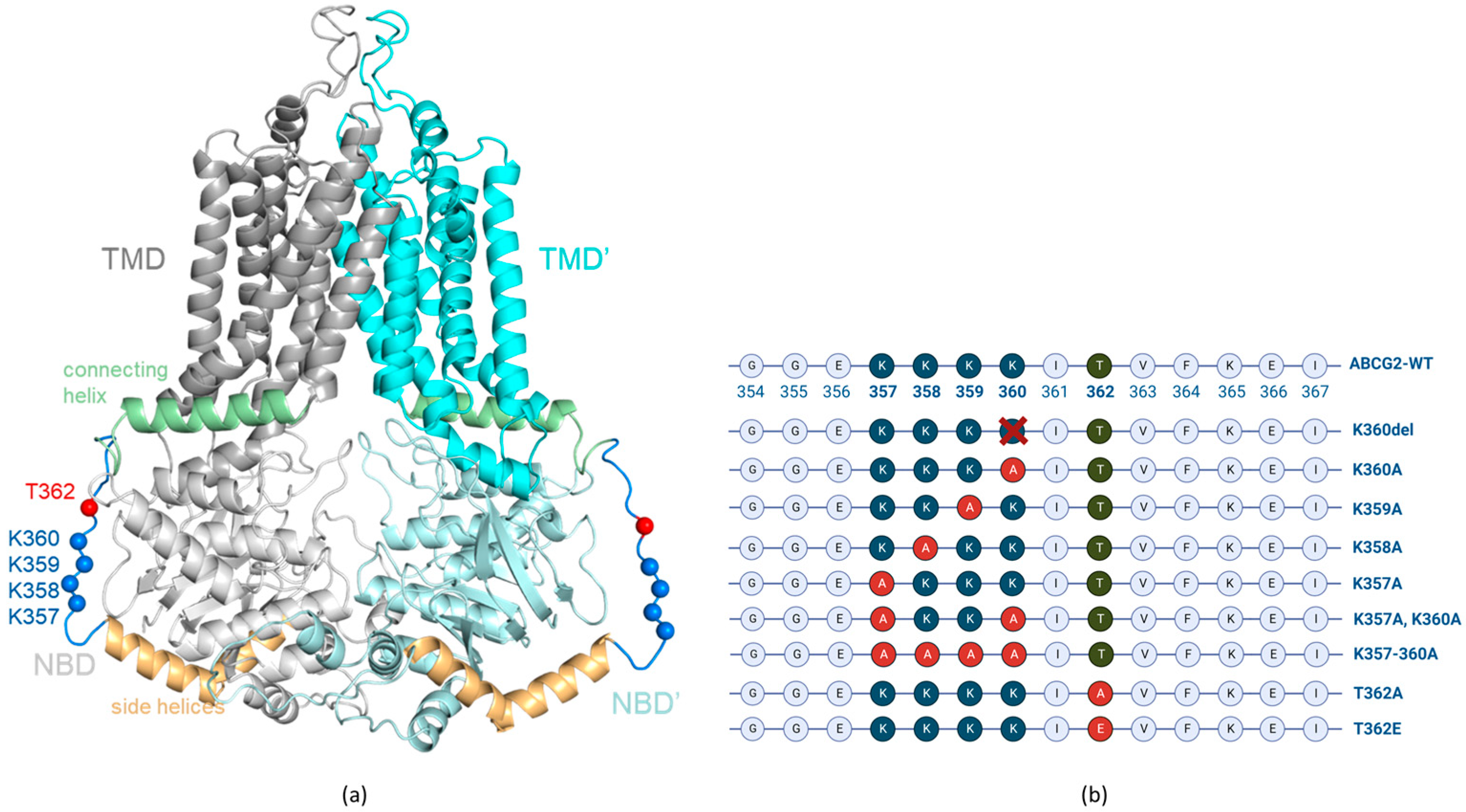
2. Materials and Methods
2.1. Vector constructs
2.2. Cell culture and transfection
2.3. Generation of stable cell lines
2.4. Hoechst33342 uptake measurements
2.5. Plasma membrane expression measurement
2.6. Western blotting
2.7. Immunostaining and confocal imaging of MDCKII cells
2.8. Confocal microscopy imaging and kinetic analysis
2.9. Molecular dynamics simulations
2.10. Statistical analysis
3. Results
3.1. Expression, membrane localization and function of the K357-K360 and T362 ABCG2 mutant variants
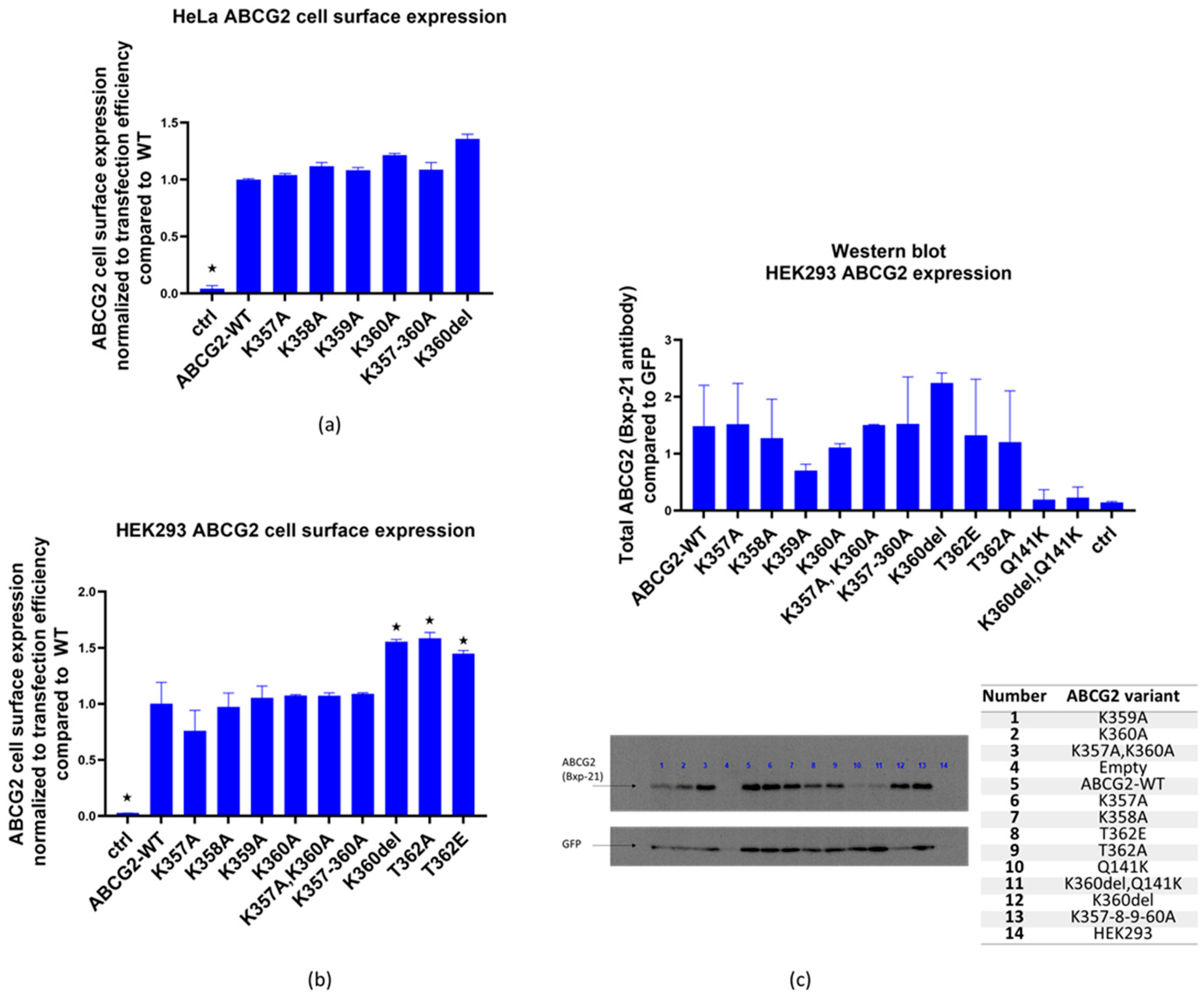
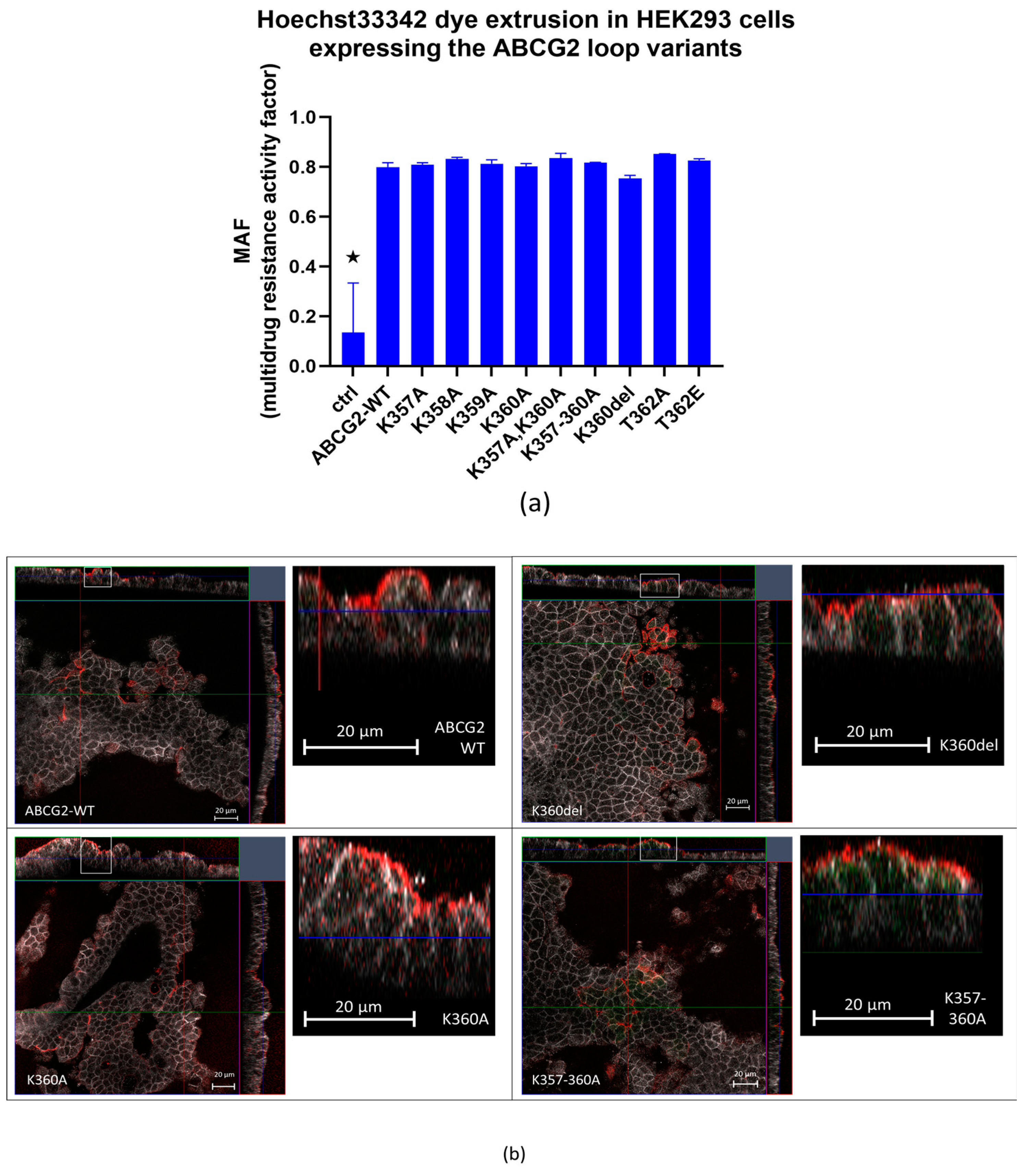
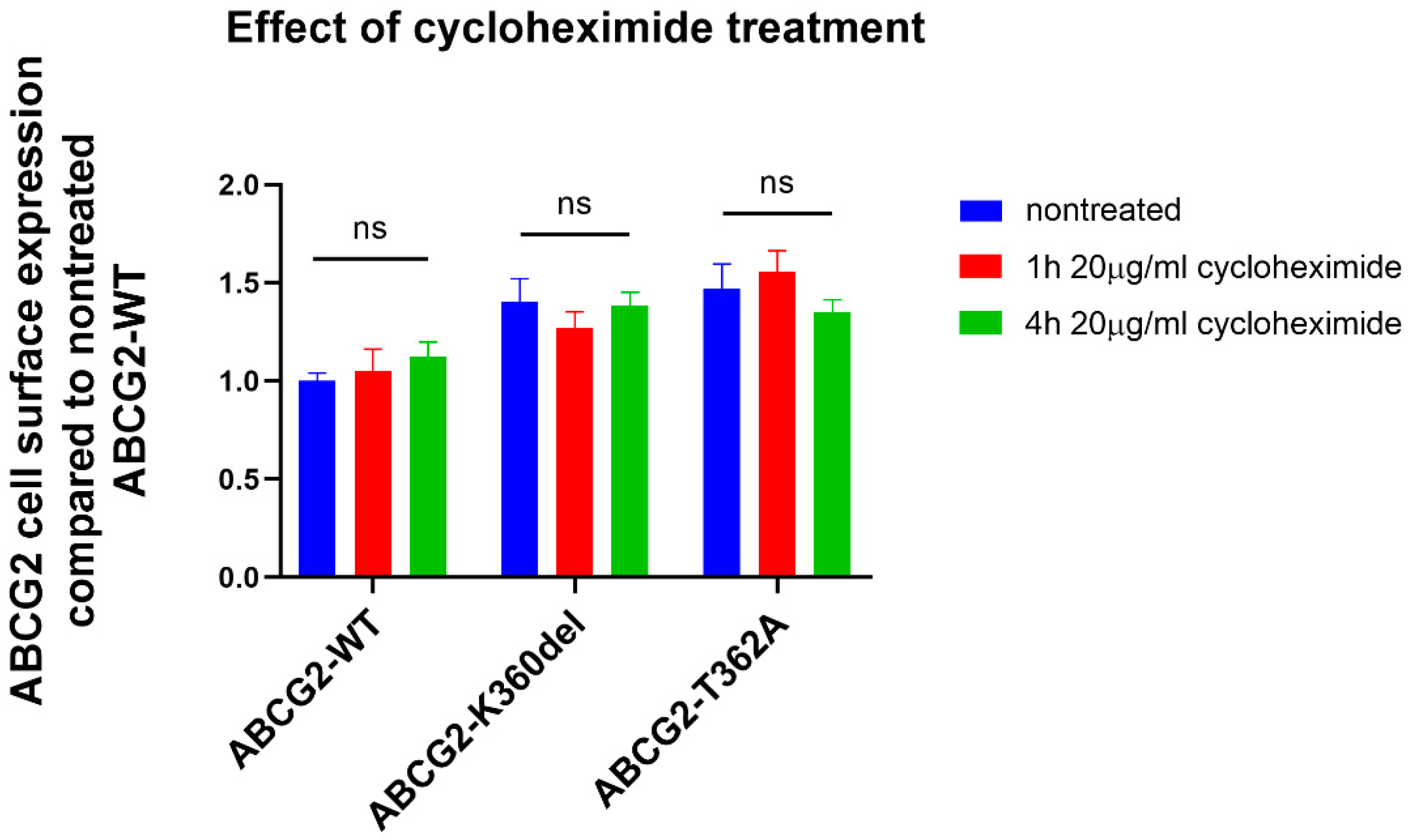
3.2. Effects of inhibition of protein synthesis and proteasomal degradation on the expression of the ABCG2 loop mutant variants
3.3. Application of the RUSH system to study the cellular trafficking of the K360del and K360A ABCG2 mutant variants
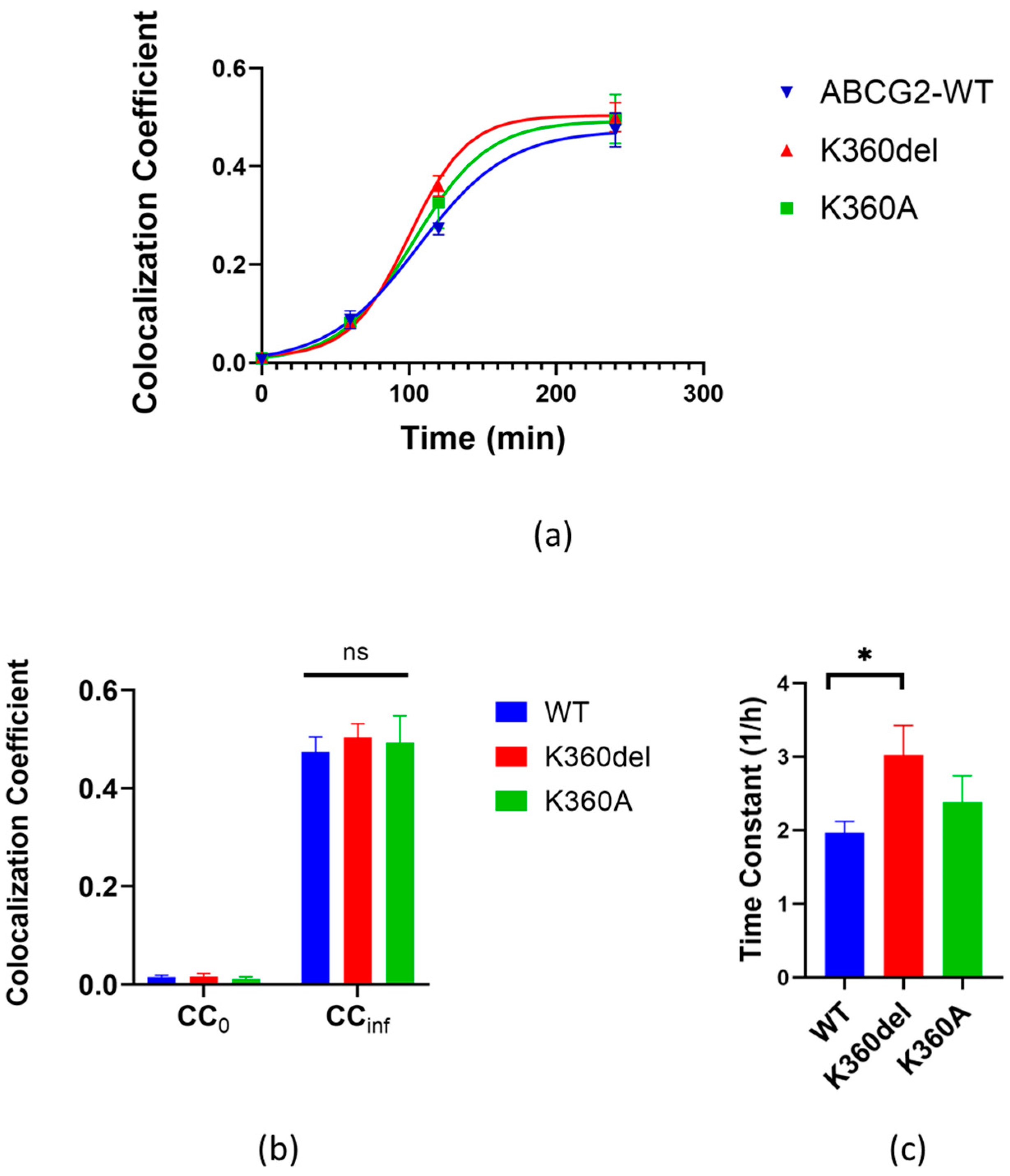
3.4. Molecular dynamics simulation of the K360del intracellular ABCG2 loop (a.a. 354-367) variant
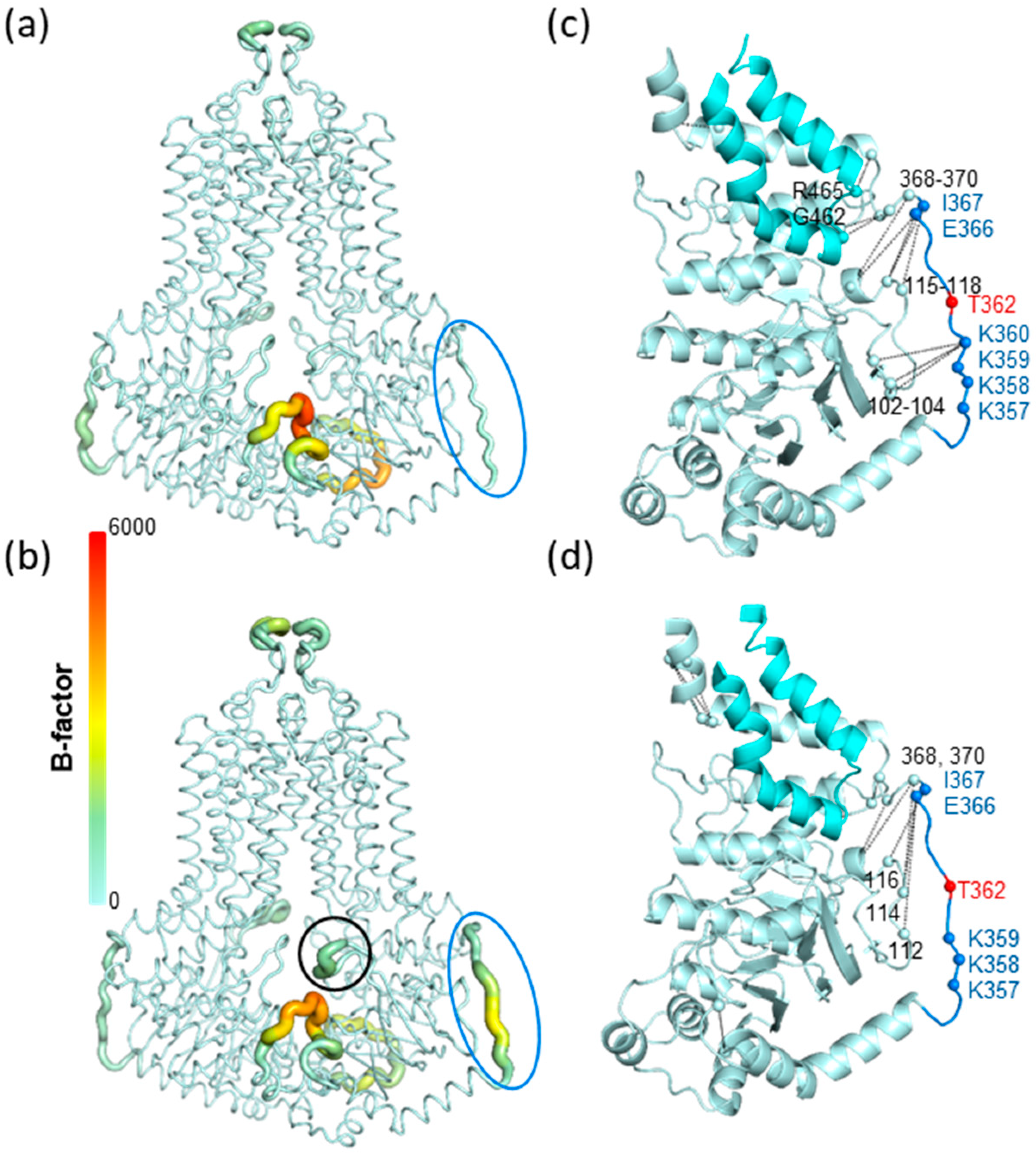
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Woodward, O.M.; Kottgen, A.; Coresh, J.; Boerwinkle, E.; Guggino, W.B.; Kottgen, M. Identification of a Urate Transporter, ABCG2, with a Common Functional Polymorphism Causing Gout. Proceedings of the National Academy of Sciences 2009, 106, 10338–10342. [Google Scholar] [CrossRef]
- Szakács, G.; Váradi, A.; Özvegy-Laczka, C.; Sarkadi, B.; Ozvegy-Laczka, C.; Sarkadi, B. The Role of ABC Transporters in Drug Absorption, Distribution, Metabolism, Excretion and Toxicity (ADME–Tox). Drug Discov Today 2008, 13, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, K.; Xie, Y.; Baer, M.R.; Ross, D.D. Role of Breast Cancer Resistance Protein (BCRP/ABCG2) in Cancer Drug Resistance. Biochem Pharmacol 2012, 83, 1084–1103. [Google Scholar] [CrossRef] [PubMed]
- Robey, R.W.; To, K.K.K.; Polgar, O.; Dohse, M.; Fetsch, P.; Dean, M.; Bates, S.E. ABCG2: A Perspective. Adv Drug Deliv Rev 2009, 61, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Sarkadi, B.; Homolya, L.; Hegedűs, T. The ABCG2/BCRP Transporter and Its Variants – from Structure to Pathology. FEBS Lett 2020, 594, 4012–4034. [Google Scholar] [CrossRef] [PubMed]
- Homolya, L. Medically Important Alterations in Transport Function and Trafficking of ABCG2. Int. J. Mol. Sci 2021, 22, 2786. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, H.; Ichida, K.; Takada, T.; Nakayama, A.; Nakashima, H.; Nakamura, T.; Kawamura, Y.; Takada, Y.; Yamamoto, K.; Inoue, H.; et al. Common Dysfunctional Variants in ABCG2 Are a Major Cause of Early-Onset Gout. Sci Rep 2013, 3, 2014. [Google Scholar] [CrossRef]
- Stiburkova, B.; Pavelcova, K.; Pavlikova, M.; Ješina, P.; Pavelka, K. The Impact of Dysfunctional Variants of ABCG2 on Hyperuricemia and Gout in Pediatric-Onset Patients. Arthritis Res Ther 2019, 21, 1–10. [Google Scholar] [CrossRef]
- Stornaiuolo, M.; Lotti, L. V; Borgese, N.; Torrisi, M.-R.; Mottola, G.; Martire, G.; Bonatti, S. KDEL and KKXX Retrieval Signals Appended to the Same Reporter Protein Determine Different Trafficking between Endoplasmic Reticulum, Intermediate Compartment, and Golgi Complex. Mol Biol Cell 2003, 14, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Dunbrack, R.L.; Lee, S.; Strominger, J.L. Signals for Retention of Transmembrane Proteins in the Endoplasmic Reticulum Studied with CD4 Truncation Mutants (Retention Motif/Cytoplasmic Domain/C-Terminal Sequence). Proc. Nad. Acad. Sci. USA 1991, 88, 1918–1922. [Google Scholar] [CrossRef]
- Matheson, L.A.; Hanton, S.L.; Brandizzi, F. Traffic between the Plant Endoplasmic Reticulum and Golgi Apparatus: To the Golgi and Beyond. Curr Opin Plant Biol 2006, 9, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Stiburkova, B.; Pavelcova, K.; Zavada, J.; Petru, L.; Simek, P.; Cepek, P.; Pavlikova, M.; Matsuo, H.; Merriman, T.R.; Pavelka, K. Functional Non-Synonymous Variants of ABCG2 and Gout Risk. Rheumatology 2017, 56, 1982–1992. [Google Scholar] [CrossRef] [PubMed]
- Zámbó, B.; Mózner, O.; Bartos, Z.; Török, G.; Várady, G.; Telbisz, Á.; Homolya, L.; Orbán, T.I.; Sarkadi, B. Cellular Expression and Function of Naturally Occurring Variants of the Human ABCG2 Multidrug Transporter. Cellular and Molecular Life Sciences 2020, 77, 365–378. [Google Scholar] [CrossRef]
- Toyoda; Mančíková; Krylov; Morimoto; Pavelcová; Bohatá; Pavelka; Pavlíková; Suzuki; Matsuo; et al. Functional Characterization of Clinically-Relevant Rare Variants in ABCG2 Identified in a Gout and Hyperuricemia Cohort. Cells 2019, 8, 363. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Xu, K.; Linn, D.E.; Yang, X.; Guo, Z.; Shimelis, H.; Nakanishi, T.; Ross, D.D.; Chen, H.; Fazli, L.; et al. The 44-KDa Pim-1 Kinase Phosphorylates BCRP/ABCG2 and Thereby Promotes Its Multimerization and Drug-Resistant Activity in Human Prostate Cancer Cells. Journal of Biological Chemistry 2008, 283, 3349–3356. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lin, N.; Li, Y. The PI3K/AKT Signaling Pathway Regulates ABCG2 Expression and Confers Resistance to Chemotherapy in Human Multiple Myeloma. Oncol Rep 2019, 41, 1678–1690. [Google Scholar] [CrossRef]
- Boncompain, G.; Divoux, S.; Gareil, N.; de Forges, H.; Lescure, A.; Latreche, L.; Mercanti, V.; Jollivet, F.; Raposo, G.; Perez, F. Synchronization of Secretory Protein Traffic in Populations of Cells. Nat Methods 2012, 9, 493–498. [Google Scholar] [CrossRef]
- Bartos, Z.; Homolya, L. Identification of Specific Trafficking Defects of Naturally Occurring Variants of the Human ABCG2 Transporter. Front Cell Dev Biol 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.J.; Bonito, C.A.; Cordeiro, M.N.D.S.; Ferreira, M.J.U.; Dos Santos, D.J.V.A. Structure-Function Relationships in ABCG2: Insights from Molecular Dynamics Simulations and Molecular Docking Studies. Sci Rep 2017, 7. [Google Scholar] [CrossRef]
- Mátés, L.; Chuah, M.K.L.; Belay, E.; Jerchow, B.; Manoj, N.; Acosta-Sanchez, A.; Grzela, D.P.; Schmitt, A.; Becker, K.; Matrai, J.; et al. Molecular Evolution of a Novel Hyperactive Sleeping Beauty Transposase Enables Robust Stable Gene Transfer in Vertebrates. Nat Genet 2009, 41, 753–761. [Google Scholar] [CrossRef]
- Homolya, L.; Holló, Z.; Müller, M.; Mechetner, E.B.; Sarkadi, B. A New Method for Quantitative Assessment of P-Glycoprotein-Related Multidrug Resistance in Tumour Cells. Br J Cancer 1996, 73, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Fiser, A.; Gian do, R.K.; Sali, A. Modeling of Loops in Protein Structures. Protein Science 2000, 9, 1753–1773. [Google Scholar] [CrossRef] [PubMed]
- Lomize, M.A.; Lomize, A.L.; Pogozheva, I.D.; Mosberg, H.I. OPM: Orientations of Proteins in Membranes Database. Bioinformatics 2006, 22, 623–625. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J Comput Chem 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward Realistic Biological Membrane Simulations. J Comput Chem 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Kohnke, B.; Kutzner, C.; Grubmüller, H. A GPU-Accelerated Fast Multipole Method for GROMACS: Performance and Accuracy. J Chem Theory Comput 2020, 16, 6938–6949. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; De Groot, B.L.; Grubmüller, H.; Mackerell, A.D. Charmm36M: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat Methods 2016, 14, 71–73. [Google Scholar] [CrossRef]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A Toolkit for the Analysis of Molecular Dynamics Simulations. J Comput Chem 2011, 32, 2319–2327. [Google Scholar] [CrossRef] [PubMed]
- Hunter John, D. Matplotib: A 2D Graphics Environment. Comput Sci Eng 2007. [Google Scholar] [CrossRef]
- Özvegy-Laczka, C.; Laczko, R.; Hegedus, C.; Litman, T.; Várady, G.; Goda, K.; Hegedus, T.; Dokholyan, N. V.; Sorrentino, B.P.; Váradi, A.; et al. Interaction with the 5D3 Monoclonal Antibody Is Regulated by Intramolecular Rearrangements but Not by Covalent Dimer Formation of the Human ABCG2 Multidrug Transporter. Journal of Biological Chemistry 2008, 283, 26059–26070. [Google Scholar] [CrossRef] [PubMed]
- Krahn, M.P. Phospholipids of the Plasma Membrane – Regulators or Consequence of Cell Polarity? Front Cell Dev Biol 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Qi, J.; Dong, Z.; Zhang, J.T. Dynamic vs Static ABCG2 Inhibitors to Sensitize Drug Resistant Cancer Cells. PLoS One 2010, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Orbán, T.I.; Seres, L.; Özvegy-Laczka, C.; Elkind, N.B.; Sarkadi, B.; Homolya, L. Combined Localization and Real-Time Functional Studies Using a GFP-Tagged ABCG2 Multidrug Transporter. Biochem Biophys Res Commun 2008, 367, 667–673. [Google Scholar] [CrossRef]
- Bi, Y.; Mann, E.; Whitfield, C.; Zimmer, J. Architecture of a Channel-Forming O-Antigen Polysaccharide ABC Transporter. Nature 2018, 553, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Caffalette, C.A.; Corey, R.A.; Sansom, M.S.P.; Stansfeld, P.J.; Zimmer, J. A Lipid Gating Mechanism for the Channel-Forming O Antigen ABC Transporter. Nat Commun 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hou, W.T.; Fan, T.; Liu, B.; Pan, T.; Li, Y.H.; Jiang, Y.L.; Wen, W.; Chen, Z.P.; Sun, L.; et al. Cryo-Electron Microscopy Structure and Transport Mechanism of a Wall Teichoic Acid ABC Transporter. mBio 2020, 11. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




