Submitted:
05 September 2023
Posted:
07 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. History and state of the art of herbaria
1.2. Herbarium genomics
1.3. Challeneges in the use of herbaria and ancient samples for genomics studies
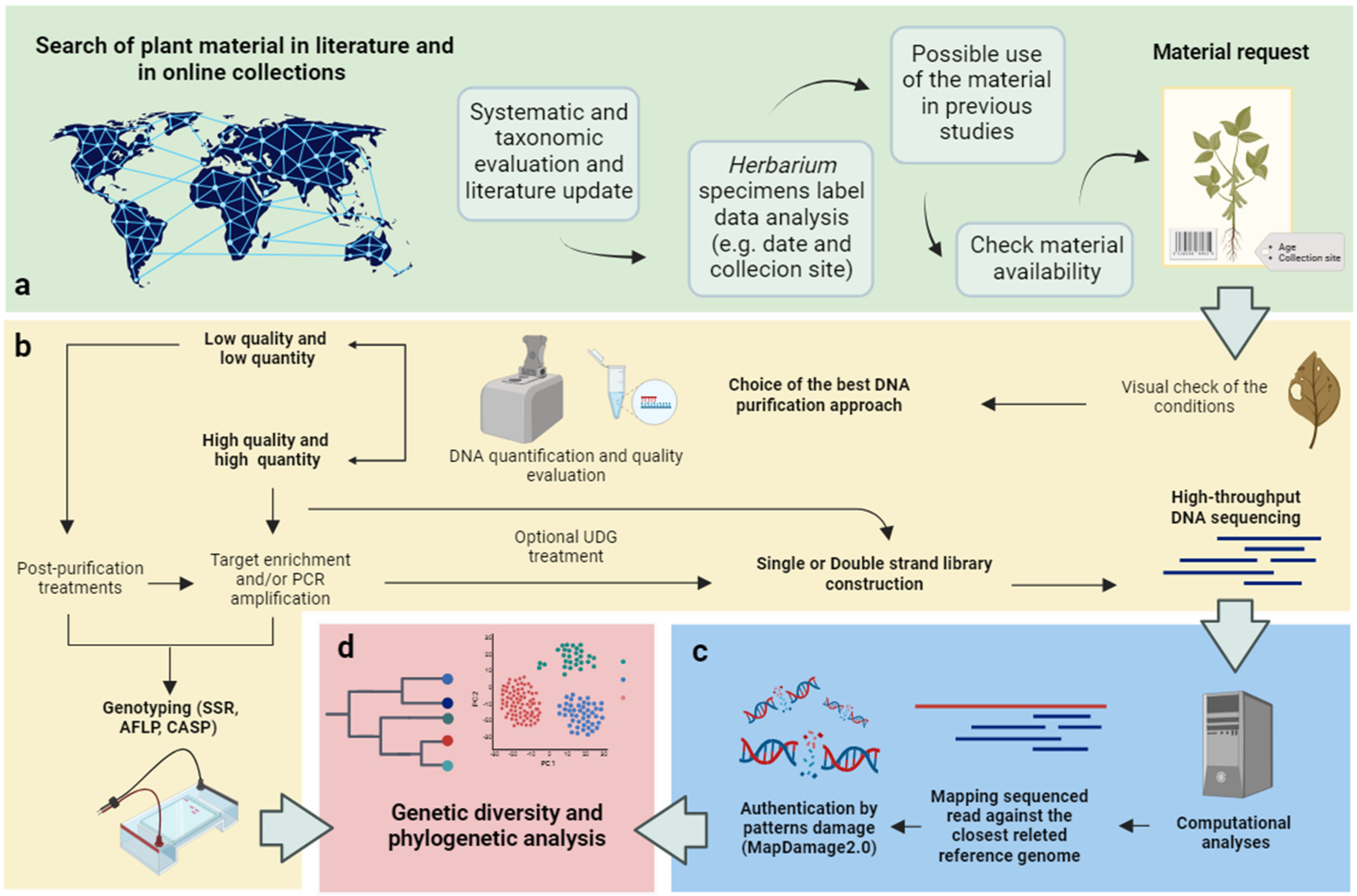
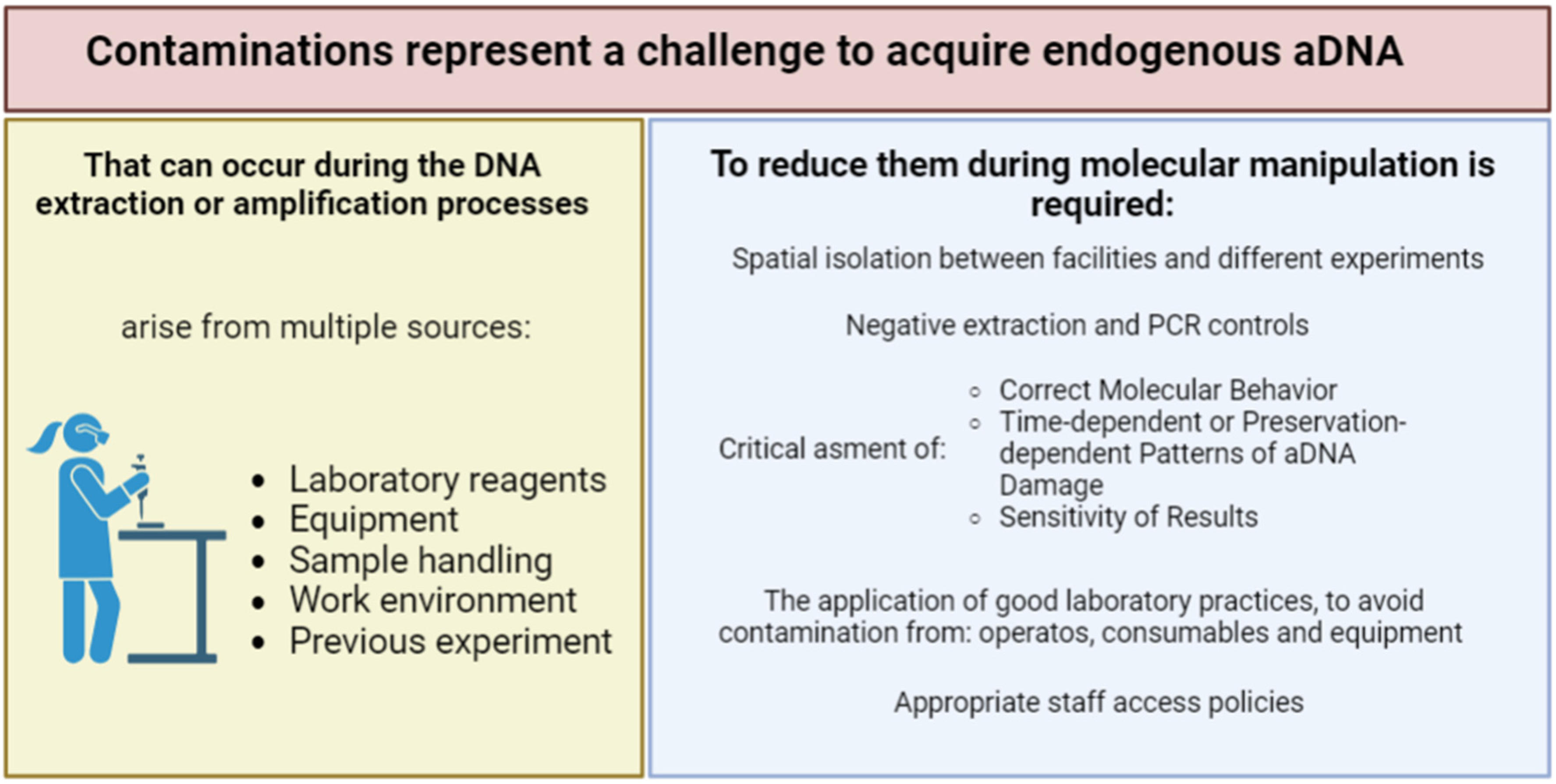
2. Extraction and purification of aDNA from Herbarium specimens
3. DNA amplification by PCR from Herbarium specimens
4. Next-generation sequencing (NGS) and genotyping on Herbarium specimens to disentangle relevant aspects of the evolutionary history of a species
5. Conclusion
Author Contributions
Funding
References
- Bakker: F. T. (2018). Herbarium Genomics: Plant Archival DNA Explored (pp. 205–224). https://doi.org/10.1007/13836_2018_40. [CrossRef]
- Barrett, C. F., Huebner, C. D., Bender, Z. A., Budinsky, T. A., Corbett, C. W., Latvis, M., McKain, M. R., Motley, M., Skibicki, S. v., Thixton, H. L., Santee, M. v., & Cumberledge, A. N. (2022). Digitized collections elucidate invasion history and patterns of awn polymorphism in Microstegium vimineum. American Journal of Botany, 109(5), 689–705. https://doi.org/10.1002/ajb2.1852. [CrossRef]
- Bebber, D. P., Carine, M. A., Wood, J. R. I., Wortley, A. H., Harris, D. J., Prance, G. T., Davidse, G., Paige, J., Pennington, T. D., Robson, N. K. B., & Scotland, R. W. (2010). Herbaria are a major frontier for species discovery. Proceedings of the National Academy of Sciences of the United States of America, 107(51), 22169–22171. https://doi.org/10.1073/pnas.1011841108. [CrossRef]
- Beck, J. B., & Semple, J. C. (2015). Next-Generation Sampling: Pairing Genomics with Herbarium Specimens Provides Species-Level Signal in Solidago (Asteraceae). Applications in Plant Sciences, 3(6), 1500014. https://doi.org/10.3732/apps.1500014. [CrossRef]
- Bellorini C. (2016). The World of PlanTs in renaissance Tuscany.
- Bellucci, E., Benazzo, A., Xu, C., Bitocchi, E., Rodriguez, M., Alseekh, S., di Vittori, V., Gioia, T., Neumann, K., Cortinovis, G., Frascarelli, G., Murube, E., Trucchi, E., Nanni, L., Ariani, A., Logozzo, G., Shin, J. H., Liu, C., Jiang, L., … Papa, R. (2022). Selection and adaptive introgression guided the complex evolutionary history of the European common bean. BioRxiv 2022.09.28.509856. https://doi.org/10.1101/2022.09.28.509856. [CrossRef]
- Bennett, E. A., Massilani, D., Lizzo, G., Daligault, J., Geigl, E. M., & Grange, T. (2014). Library construction for ancient genomics: Single strand or double strand? BioTechniques, 56(6), 289–300. https://doi.org/10.2144/000114176. [CrossRef]
- Bi, K., Linderoth, T., Vanderpool, D., Good, J. M., Nielsen, R., & Moritz, C. (2013). Unlocking the vault: Next-generation museum population genomics. Molecular Ecology, 22(24), 6018–6032. https://doi.org/10.1111/mec.12516. [CrossRef]
- Binladen, J., & Willerslev, E. (2010). Why study ancient DNA damage? In Journal of Nordic Archaeological Science (Vol. 17).
- Briggs, A. W., & Heyn, P. (2012). Preparation of next-generation sequencing libraries from damaged DNA. Methods in Molecular Biology, 840, 143–154. https://doi.org/10.1007/978-1-61779-516-9_18. [CrossRef]
- Burrell, A. S., Disotell, T. R., & Bergey, C. M. (2015). The use of museum specimens with high-throughput DNA sequencers. Journal of Human Evolution, 79, 35–44. https://doi.org/10.1016/j.jhevol.2014.10.015. [CrossRef]
- Butler, S. L., & Falke, J. J. (1996). Effects of protein stabilizing agents on thermal backbone motions: A disulfide trapping study. Biochemistry, 35(33), 10595–10600. https://doi.org/10.1021/bi961107v. [CrossRef]
- Champlot, S., Berthelot, C., Pruvost, M., Andrew Bennett, E., Grange, T., & Geigl, E. M. (2010). An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications. PLoS ONE, 5(9). https://doi.org/10.1371/journal.pone.0013042. [CrossRef]
- Cooper A., & Poinar N. H. (2000). Science’s compass letters. SCIENCE, 289(1139).
- Cortinovis, G., di Vittori, V., Bellucci, E., Bitocchi, E., & Papa, R. (2020). Adaptation to novel environments during crop diversification. In Current Opinion in Plant Biology (Vol. 56, pp. 203–217). Elsevier Ltd. https://doi.org/10.1016/j.pbi.2019.12.011. [CrossRef]
- Cota-Sánchez, J. H., Remarchuk, K., & Ubayasena, K. (2006). Ready-to-Use DNA Extracted with a CTAB Method Adapted for Herbarium Specimens and Mucilaginous Plant Tissue.
- Cozzolino, S., Cafasso, D., Pellegrino, G., Musacchio, A., & Widmer, A. (2007). Genetic variation in time and space: The use of herbarium specimens to reconstruct patterns of genetic variation in the endangered orchid Anacamptis palustris. Conservation Genetics, 8(3), 629–639. https://doi.org/10.1007/s10592-006-9209-7. [CrossRef]
- Cristofolini, G., & Mossetti, U. (1993). Pre-linnean herbaria in Bologna: some newly discovered collections from the time of Ulisse Aldrovandi. https://www.researchgate.net/publication/260179625.
- Cronn, R., Knaus, B. J., Liston, A., Maughan, P. J., Parks, M., Syring, J. v., & Udall, J. (2012). Targeted enrichment strategies for next-generation plant biology. American Journal of Botany, 99(2), 291–311. https://doi.org/10.3732/ajb.1100356. [CrossRef]
- Damerval, C., ben Othman, W., Manicacci, D., & Jabbour, F. (2018). Distribution area of the two floral morphs of Nigella damascena L. (Ranunculaceae): a diachronic study using herbarium specimens collected in France. Botany Letters, 165(3–4), 396–403. https://doi.org/10.1080/23818107.2017.1422437. [CrossRef]
- Davis, C. C., Willis, C. G., Connolly, B., Kelly, C., & Ellison, A. M. (2015). Herbarium records are reliable sources of phenological change driven by climate and provide novel insights into species’ phenological cueing mechanisms. American Journal of Botany, 102(10), 1599–1609. https://doi.org/10.3732/ajb.1500237. [CrossRef]
- Doyle, J. J., & Dickson, E. E. (1987). Preservation of Plant Samples for DNA Restriction Endonuclease Analysis. In Source (Vol. 36, Issue 4).
- Drabkova, L., Kirschner, J., & Vl~k, C. (2002). Comparison of Seven DNA Extraction and Amplification Protocols in Historical Herbarium Specimens of Juncaceae. In International Society for Plant Molecular Biology (Vol. 20).
- Drábková, L. Z. (2014). DNA extraction from herbarium specimens. Methods in Molecular Biology, 1115, 69–84. https://doi.org/10.1007/978-1-62703-767-9_4. [CrossRef]
- Exposito-Alonso, M., Becker, C., Schuenemann, V. J., Reiter, E., Setzer, C., Slovak, R., Brachi, B., Hagmann, J., Grimm, D. G., Chen, J., Busch, W., Bergelson, J., Ness, R. W., Krause, J., Burbano, H. A., & Weigel, D. (2018). The rate and potential relevance of new mutations in a colonizing plant lineage. PLoS Genetics, 14(2). https://doi.org/10.1371/journal.pgen.1007155. [CrossRef]
- Freedman, J., Dorp, V., & Brace, L. &. (2018). Title: Destructive sampling natural science collections: An overview for museum professionals and researchers. http://www.natsca.orgURL:http://www.natsca.org/article/2440.
- Gansauge, M. T., & Meyer, M. (2013). Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nature Protocols, 8(4), 737–748. https://doi.org/10.1038/nprot.2013.038. [CrossRef]
- Gibbs, R. A. (1990). PERSPECTIVE: ANALYTICAL BIOTECHNOLOGY DNA Amplification by the Polymerase Chain Reaction. In Anal. Chem (Vol. 62). https://pubs.acs.org/sharingguidelines.
- Gilbert, M. T. P., Moore, W., Melchior, L., & Worebey, M. (2007). DNA extraction from dry museum beetles without conferring external morphological damage. PLoS ONE, 2(3). https://doi.org/10.1371/journal.pone.0000272. [CrossRef]
- Ginolhac, A., Rasmussen, M., Gilbert, M. T. P., Willerslev, E., & Orlando, L. (2011). mapDamage: Testing for damage patterns in ancient DNA sequences. Bioinformatics, 27(15), 2153–2155. https://doi.org/10.1093/bioinformatics/btr347. [CrossRef]
- Gutaker, R. M., & Burbano, H. A. (2017). Reinforcing plant evolutionary genomics using ancient DNA. In Current Opinion in Plant Biology (Vol. 36, pp. 38–45). Elsevier Ltd. https://doi.org/10.1016/j.pbi.2017.01.002. [CrossRef]
- Gutaker, R. M., Reiter, E., Furtwängler, A., Schuenemann, V. J., & Burbano, H. A. (2017b). Extraction of ultrashort DNA molecules from herbarium specimens. BioTechniques, 62(2), 76–79. https://doi.org/10.2144/000114517. [CrossRef]
- Hart, M. L., Forrest, L. L., Nicholls, J. A., & Kidner, C. A. (2016a). Retrieval of hundreds of nuclear loci from herbarium specimens. Taxon, 65(5), 1081–1092. https://doi.org/10.12705/655.9. [CrossRef]
- Hart, M. L., Forrest, L. L., Nicholls, J. A., & Kidner, C. A. (2016b). Retrieval of hundreds of nuclear loci from herbarium specimens. Taxon, 65(5), 1081–1092. https://doi.org/10.12705/655.9. [CrossRef]
- Heberling, J. M. (2022). HERBARIA AS BIG DATA SOURCES OF PLANT TRAITS. International Journal of Plant Sciences, 183(2), 87–118. https://doi.org/10.1086/717623. [CrossRef]
- Heberling, J. M., Prather, L. A., & Tonsor, S. J. (2019). The Changing Uses of Herbarium Data in an Era of Global Change: An Overview Using Automated Content Analysis. In BioScience (Vol. 69, Issue 10, pp. 812–822). Oxford University Press. https://doi.org/10.1093/biosci/biz094. [CrossRef]
- Jankowiak, K., Buczkowska, K., & Szweykowska-Kulinska, Z. (2005). Successful extraction of DNA from 100-year-old herbarium specimens of the liverwort Bazzania trilobata. Taxon, 54(2), 335–336. https://doi.org/10.2307/25065361. [CrossRef]
- Jobes, D. v, Hurley, D. L., & Thien, L. B. (1995). Plant DNA Isolation: A Method to Efficiently Remove Polyphenolics, Polysaccharides, and RNA. In Source (Vol. 44, Issue 3).
- Jónsson, H., Ginolhac, A., Schubert, M., Johnson, P. L. F., & Orlando, L. (2013). MapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics, 29(13), 1682–1684. https://doi.org/10.1093/bioinformatics/btt193. [CrossRef]
- Kistler, L., Bieker, V. C., Martin, M. D., Pedersen, M. W., Ramos Madrigal, J., & Wales, N. (2020). Ancient Plant Genomics in Archaeology, Herbaria, and the Environment. https://doi.org/10.1146/annurev-arplant-081519. [CrossRef]
- Knapp, M., Clarke, A. C., Horsburgh, K. A., & Matisoo-Smith, E. A. (2012). Setting the stage—Building and working in an ancient DNA laboratory. Annals of Anatomy, 194(1), 3–6. https://doi.org/10.1016/j.aanat.2011.03.008. [CrossRef]
- Kreader, C. A. (1996). Relief of Amplification Inhibition in PCR with Bovine Serum Albumin or T4 Gene 32 Protein. In APPLIED AND ENVIRONMENTAL MICROBIOLOGY (Vol. 62, Issue 3).
- Krinitsina, A. A., Sizova, T. v., Zaika, M. A., Speranskaya, A. S., & Sukhorukov, A. P. (2015). A rapid and cost-effective method for DNA extraction from archival herbarium specimens. Biochemistry (Moscow), 80(11), 1478–1484. https://doi.org/10.1134/S0006297915110097. [CrossRef]
- Kumar Shasany, A., & Darokar, M. (1999). Rapid Isolation of DNA from Dry and Fresh Samples of Plants Producing Large Amounts of Secondary Metabolites and Essential Oils. https://doi.org/10.1023/A:1007528101452. [CrossRef]
- Lambertini, C., Frydenberg, J., Gustafsson, M. H. G., & Brix, H. (2008). Herbarium specimens as a source of DNA for AFLP fingerprinting of Phragmites (Poaceae): Possibilities and limitations. Plant Systematics and Evolution, 272(1–4), 223–231. https://doi.org/10.1007/s00606-007-0633-z. [CrossRef]
- Lister, D. L., Bower, M. A., Howe, C. J., & Jones, M. K. (2008). Extraction and amplification of nuclear DNA from herbarium specimens of emmer wheat: a method for assessing DNA preservation by maximum amplicon length recovery. In TAXON (Vol. 57, Issue 1). https://doi.org/10.2307/25065966. [CrossRef]
- Liu, L., Li, Y., Li, S., Hu, N., He, Y., Pong, R., Lin, D., Lu, L., & Law, M. (2012). Comparison of next-generation sequencing systems. In Journal of Biomedicine and Biotechnology (Vol. 2012). https://doi.org/10.1155/2012/251364. [CrossRef]
- Malenica, N., Šimon, S., Besendorfer, V., Maletic, E., Kontić, J. K., & Pejić, I. (2011). Whole genome amplification and microsatellite genotyping of herbarium DNA revealed the identity of an ancient grapevine cultivar. Naturwissenschaften, 98(9), 763–772. https://doi.org/10.1007/s00114-011-0826-8. [CrossRef]
- Marinček, P., Wagner, N. D., & Tomasello, S. (2022). Ancient DNA extraction methods for herbarium specimens: When is it worth the effort? Applications in Plant Sciences, 10(3). https://doi.org/10.1002/aps3.11477. [CrossRef]
- Martin, M. D., Quiroz-Claros, E., Brush, G. S., & Zimmer, E. A. (2018). Herbarium collection-based phylogenetics of the ragweeds (Ambrosia, Asteraceae). Molecular Phylogenetics and Evolution, 120, 335–341. https://doi.org/10.1016/j.ympev.2017.12.023. [CrossRef]
- Monroe, C., Grier, C., & Kemp, B. M. (2013). Evaluating the efficacy of various thermo-stable polymerases against co-extracted PCR inhibitors in ancient DNA samples. Forensic Science International, 228(1–3), 142–153. https://doi.org/10.1016/j.forsciint.2013.02.029. [CrossRef]
- Müller-Wille, S. (2006). Linnaeus’ herbarium cabinet: a piece of furniture and its function. In Endeavour (Vol. 30, Issue 2, pp. 60–64). https://doi.org/10.1016/j.endeavour.2006.03.001. [CrossRef]
- Myers, J. R., Formiga, A. K., & Janick, J. (2022). Iconography of Beans and Related Legumes Following the Columbian Exchange. Frontiers in Plant Science, 13. https://doi.org/10.3389/fpls.2022.851029. [CrossRef]
- Nelson, G., Paul, D., Riccardi, G., & Mast, A. R. (2012). Five task clusters that enable efficient and effective digitization of biological collections. ZooKeys, 209, 19–45. https://doi.org/10.3897/zookeys.209.3135. [CrossRef]
- Nepi Chiara. (2007). La “slegatura” dell’erbario di Andrea Cesalpino (1525-1603). MUSEOLOGIA SCIENTIFICA Nuova Serie, 1, 50–54.
- Orlando, L., Allaby, R., Skoglund, P., der Sarkissian, C., Stockhammer, P. W., Ávila-Arcos, M. C., Fu, Q., Krause, J., Willerslev, E., Stone, A. C., & Warinner, C. (2021). Ancient DNA analysis. In Nature Reviews Methods Primers (Vol. 1, Issue 1). Springer Nature. https://doi.org/10.1038/s43586-020-00011-0. [CrossRef]
- Pääbo S., Irwin D. M., Wilson A. C., (1990) DNA damage promotes jumping between templates during enzymatic amplification. Journal of Biological Chemistry. Volume 265, Issue 8,. https://doi.org/10.1016/S0021-9258(19)39621-8. [CrossRef]
- Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. Genetic analyses from ancient DNA. Annu Rev Genet. 2004;38:645-79. doi: 10.1146/annurev.genet.37.110801.143214. PMID: 15568989. [CrossRef] [PubMed]
- Pont, C., Wagner, S., Kremer, A., Orlando, L., Plomion, C., & Salse, J. (2019). Paleogenomics: Reconstruction of plant evolutionary trajectories from modern and ancient DNA. In Genome Biology (Vol. 20, Issue 1). BioMed Central Ltd. https://doi.org/10.1186/s13059-019-1627-1. [CrossRef]
- Porebski, S., Bailey, L. G., & Baum, B. R. (1997). Modification of a CTAB DNA Extraction Protocol for Plants Containing High Polysaccharide and Polyphenol Components. In Plant Molecular Biology Reporter (Vol. 15, Issue 1).
- Pruvost M, Grange T, Geigl EM. Minimizing DNA contamination by using UNG-coupled quantitative real-time PCR on degraded DNA samples: application to ancient DNA studies. Biotechniques. 2005 Apr;38(4):569-75. doi: 10.2144/05384ST03. PMID: 15884675. [CrossRef] [PubMed]
- Prüfer, K., Stenzel, U., Hofreiter, M., Pääbo, S., Kelso, J., & Green, R. E. (2010). Open Access METHOD Computational challenges in the analysis of ancient DNA. In Genome Biology (Vol. 11). http://genomebiology.com/2010/11/5/R47.
- Psonis, N., Vassou, D., & Kafetzopoulos, D. (2021). Testing a series of modifications on genomic library preparation methods for ancient or degraded DNA. Analytical Biochemistry, 623. https://doi.org/10.1016/j.ab.2021.114193. [CrossRef]
- Riahi, M., Zarre, S., Maassoumi, A. A., Attar, F., & Kazempour Osaloo, S. (2010). An inexpensive and rapid method for extracting papilionoid genomic DNA from herbarium specimens. Genetics and Molecular Research : GMR, 9(3), 1334–1342. https://doi.org/10.4238/vol9-3gmr839. [CrossRef]
- Ribeiro, R., & Lovato, M. (2007a). DNA extraction in fresh and herbarium specimens of Dalbergia Comparative analysis of different DNA extraction protocols in fresh and herbarium specimens of the genus Dalbergia. In Genetics and Molecular Research (Vol. 6, Issue 1). www.funpecrp.com.brwww.funpecrp.com.br.
- Ribeiro, R., & Lovato, M. (2007b). DNA extraction in fresh and herbarium specimens of Dalbergia Comparative analysis of different DNA extraction protocols in fresh and herbarium specimens of the genus Dalbergia. In Genetics and Molecular Research (Vol. 6, Issue 1). www.funpecrp.com.brwww.funpecrp.com.br.
- Rogers, S. O., & Bendich, A. J. (1985). Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. In Plant Molecular Biology (Vol. 5).
- Rosche, C., Baasch, A., Runge, K., Brade, P., Träger, S., Parisod, C., & Hensen, I. (2022). Tracking population genetic signatures of local extinction with herbarium specimens. Annals of Botany, 129(7), 857–868. https://doi.org/10.1093/aob/mcac061. [CrossRef]
- Roullier, C., Benoit, L., Mckey, D. B., & Lebot, V. (2013). Historical collections reveal patterns of diffusion of sweet potato in Oceania obscured by modern plant movements and recombination. PNAS. https://doi.org/10.5061/dryad. [CrossRef]
- Rowe, K. C., Singhal, S., Macmanes, M. D., Ayroles, J. F., Morelli, T. L., Rubidge, E. M., Bi, K., & Moritz, C. C. (2011). Museum genomics: Low-cost and high-accuracy genetic data from historical specimens. Molecular Ecology Resources, 11(6), 1082–1092. https://doi.org/10.1111/j.1755-0998.2011.03052.x. [CrossRef]
- Samarakoon, T., Wang, S. Y., & Alford, M. H. (2013). Enhancing PCR Amplification of DNA from Recalcitrant Plant Specimens Using a Trehalose-Based Additive. Applications in Plant Sciences, 1(1), 1200236. https://doi.org/10.3732/apps.1200236. [CrossRef]
- Santos, D., Ribeiro, G. C., Cabral, A. D., & Sperança, M. A. (2018). A non-destructive enzymatic method to extract DNA from arthropod specimens: Implications for morphological and molecular studies. PLoS ONE, 13(2). https://doi.org/10.1371/journal.pone.0192200. [CrossRef]
- Särkinen, T., Staats, M., Richardson, J. E., Cowan, R. S., & Bakker, F. T. (2012). How to Open the Treasure Chest? Optimising DNA Extraction from Herbarium Specimens. PLoS ONE, 7(8). https://doi.org/10.1371/journal.pone.0043808. [CrossRef]
- Saville, A. C., Martin, M. D., & Ristaino, J. B. (2016). Historic late blight outbreaks caused by a widespread dominant lineage of Phytophthora infestans (Mont.) de Bary. In PLoS ONE (Vol. 11, Issue 12). Public Library of Science. https://doi.org/10.1371/journal.pone.0168381. [CrossRef]
- Savolainen, V., Cui~noud, P., Sp1chiger, R., Martinez, M. D. P., Cri~vecoeur, M., & Manen, J.-F. (1995). Systematics and Evolution The use of herbarium specimens in DNA phylogenetics: evaluation and improvement. In P1. Syst. Evol (Vol. 197).
- Sawyer, S., Krause, J., Guschanski, K., Savolainen, V., & Pääbo, S. (2012). Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS ONE, 7(3). https://doi.org/10.1371/journal.pone.0034131. [CrossRef]
- Shapiro, B., Barlow, A., Heintzman, P. D., Hofreiter, M., Paijmans, J. L. A., & Soares Editors, A. E. R. (n.d.). Ancient DNA Methods and Protocols Second Edition Methods in Molecular Biology 1963. http://www.springer.com/series/7651.
- Shepherd, L. D. (2017). A non-destructive DNA sampling technique for herbarium specimens. PLoS ONE, 12(8). https://doi.org/10.1371/journal.pone.0183555. [CrossRef]
- Signorini, M. A. (1993). SULLE PIANTE DIPINTE DAL BACHIACCA NELLO SCRITTOIO DI COSIMO I A PALAZZO VECCHI. https://www.jstor.org/stable/27654359.
- Smith, J. P. (2017). The Herbarium. https://digitalcommons.humboldt.edu/botany_jps/79.
- Staats, M., Cuenca, A., Richardson, J. E., Ginkel, R. V. van, Petersen, G., Seberg, O., & Bakker, F. T. (2011). DNA damage in plant herbarium tissue. PLoS ONE, 6(12). https://doi.org/10.1371/journal.pone.0028448. [CrossRef]
- Stearn, W. T. (1961). A New Photographic Record of the Linnaean Herbarium. In Source (Vol. 10, Issue 1).
- Stefanaki, A., Porck, H., Grimaldi, I. M., Thurn, N., Pugliano, V., Kardinaal, A., Salemink, J., Thijsse, G., Chavannes-Mazel, C., Kwakkel, E., & van Andel, T. (2018). Breaking the silence of the 500-year-old smiling garden of everlasting flowers: The En Tibi book herbarium. PLoS ONE, 14(6). https://doi.org/10.1371/journal.pone.0217779. [CrossRef]
- Sugita, N., Ebihara, A., Hosoya, T., Jinbo, U., Kaneko, S., Kurosawa, T., Nakae, M., & Yukawa, T. (2020). Non-destructive DNA extraction from herbarium specimens: a method particularly suitable for plants with small and fragile leaves. Journal of Plant Research, 133(1), 133–141. https://doi.org/10.1007/s10265-019-01152-4. [CrossRef]
- Taylor, W., & Swan, E. C. (1994). DNA from Herbarium Specimens. In: Herrmann, B., Hummel, S. (eds) Ancient DNA. https://doi.org/10.1007/978-1-4612-4318-2_11. [CrossRef]
- Thiers, B. M., Tulig, M. C., & Watson, K. A. (2016). Digitization of The New York Botanical Garden Herbarium. Brittonia, 68(3), 324–333. https://doi.org/10.1007/s12228-016-9423-7. [CrossRef]
- Thiers M. (2021). The World’s Herbaria 2020: A Summary Report Based on Data from Index Herbariorum. http://sweetgum.nybg.org/science/ih/.
- Thomas, M., Gilbert, P., Wilson, A. S., Bunce, M., Hansen, A. J., Willerslev, E., Shapiro, B., Higham, T. F. G., Richards, M. P., O’connell, T. C., Tobin, D. J., Janaway, R. C., & Cooper, A. (2004). Ancient mito-chondrial DNA from hair. http://www.current-.
- Thornhill, A. H., Baldwin, B. G., Freyman, W. A., Nosratinia, S., Kling, M. M., Morueta-Holme, N., Madsen, T. P., Ackerly, D. D., & Mishler, B. D. (2017). Spatial phylogenetics of the native California flora. BMC Biology, 15(1). https://doi.org/10.1186/s12915-017-0435-x. [CrossRef]
- Trucchi, E., Benazzo, A., Lari, M., Iob, A., Vai, S., Nanni, L., Bellucci, E., Bitocchi, E., Raffini, F., Xu, C., Jackson, S. A., Lema, V., Babot, P., Oliszewski, N., Gil, A., Neme, G., Michieli, C. T., de Lorenzi, M., Calcagnile, L., … Bertorelle, G. (2021). Ancient genomes reveal early Andean farmers selected common beans while preserving diversity. Nature Plants, 7(2), 123–128. https://doi.org/10.1038/s41477-021-00848-7. [CrossRef]
- Von Engelhardt, D. (2011). LUCA GHINI (1490-1556) IL PADRE FONDATORE DELLA BOTANICA MODERNA NEL CONTESTO DEI RAPPORTI SCIENTIFICI EUROPEI DEL SEDICESIMO SECOLO (1) (Vol. 27).
- Wales, N., Andersen, K., Cappellini, E., Ávila-Arcos, M. C., & Gilbert, M. T. P. (2014). Optimization of DNA recovery and amplification from non-carbonized archaeobotanical remains. PLoS ONE, 9(1). https://doi.org/10.1371/journal.pone.0086827. [CrossRef]
- Wang, W. (2018). A primer to the use of herbarium specimens in plant phylogenetics. Botany Letters, 165(3–4), 404–408. https://doi.org/10.1080/23818107.2018.1438311. [CrossRef]
- Willerslev, E., & Cooper, A. (2005). Ancient DNA. In Proceedings of the Royal Society B: Biological Sciences (Vol. 272, Issue 1558, pp. 3–16). Royal Society. https://doi.org/10.1098/rspb.2004.2813. [CrossRef]
- Willis, C. G., Ellwood, E. R., Primack, R. B., Davis, C. C., Pearson, K. D., Gallinat, A. S., Yost, J. M., Nelson, G., Mazer, S. J., Rossington, N. L., Sparks, T. H., & Soltis, P. S. (2017). Old Plants, New Tricks: Phenological Research Using Herbarium Specimens. In Trends in Ecology and Evolution (Vol. 32, Issue 7, pp. 531–546). Elsevier Ltd. https://doi.org/10.1016/j.tree.2017.03.015. [CrossRef]
- Yoshida, K., Schuenemann, V. J., Cano, L. M., Pais, M., Mishra, B., Sharma, R., Lanz, C., Martin, F. N., Kamoun, S., Krause, J., Thines, M., Weigel, D., & Burbano, H. A. (2013). The rise and fall of the Phytophthora infestans lineage that triggered the Irish potato famine. ELife, 2013(2). https://doi.org/10.7554/eLife.00731. [CrossRef]
- Zedane, L., Hong-wa, C., Murienne, O., Eline Jeziorski, C., Baldwin, B. G., & Besnard, G. (2015). Museomics illuminate the history of an extinct, paleoendemic plant lineage (Hesperelaea, Oleaceae) known from an 1875 collection from Guadalupe Island, Mexico. https://academic.oup.com/biolinnean/article/117/1/44/2440216.
- Zeng, C. X., Hollingsworth, P. M., Yang, J., He, Z. S., Zhang, Z. R., Li, D. Z., & Yang, J. B. (2018). Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods, 14(1). https://doi.org/10.1186/s13007-018-0300-0. [CrossRef]
- Zunic, L., Skrbo, A., & Dobraca, A. (2017). Historical Contribution of Pharmaceutics to Botany and Pharmacognosy Development. Materia Socio Medica, 29(4), 291. https://doi.org/10.5455/msm.2017.29.291-300. [CrossRef]

| DNA extraction/purification protocol | Source of plant material | Timing of sampling | Suitable for extracting Herbarium DNA | Quality evaluation approach | Reference |
|---|---|---|---|---|---|
| CTAB according to the protocol of Doyle and Doyle (1990) | Juncus and Luzula genera (Juncaceae) | * | Yes | PCR amplification | Drabkova et al., 2002 |
| CTAB + pre-wash with a sorbitol-containing buffer | Lafoensia spp. | N.A. | Yes | PCR amplification | Inglis et al., 2018 |
| Modified CTAB | Agropyronjunceum (Gramineae), Poa juncifolia (Gramineae), Poa palustris, Triticum aestivum (Gramineae), Vicia faba (Fabaceae), Zea mays ssp. mays | N.A. | Yes | Restriction enzymes | Rogers & Bendich 1985 |
| Juncus and Luzula genera (Juncaceae) | N.A. | Yes, but may present CTAB contamination | PCR amplification | Drabkova et al., 2002 | |
| N.A. | ≥60 years | Yes | PCR amplification | Cota-Sánchez et al., 2006 | |
| Species from nine genera of the Papilionoideae | N.A. | Yes | PCR amplification | Riahi et al. (2010) | |
| DNeasy Plant Mini Kit (QIAgen) | Juncus and Luzula genera (Juncaceae) | N.A. | Yes | PCR amplification | Drabkova et al., 2002 |
| DNA extraction with phenol purification and liquid nitrogen | Juncus and Luzula genera (Juncaceae) | N.A. | No | PCR amplification | Drabkova et al., 2002 |
| Long term precipitation in isopropanol and CsCl gradient | Juncus and Luzula genera (Juncaceae) | N.A. | No | PCR amplification | Drabkova et al., 2002 |
| Proteinase K and sodium dodecyl sulfate (SDS) | Scripus hattorianus | 1934 | Yes | PCR amplification | Sugita et al. 2020 |
| N-phenacylthiazolium bromide (PTB)—dithiothreitol (DTT) | Arabidopsis thaliana | Between 1839 and 1898 | Yes | NGS | Gutaker et al. 2017 |
| Phenol-chloroform and silica spin column purification | Herbarium grape leaf tissue (unpublished data) | N.A. | Yes | PCR amplification | Wales et al. 2019 |
| Polyvinylpyrrolidone PVP | genus Dalbergia | N.A. | Yes | PCR amplification | Ribeiro & Lovato 2007 |
| AMPure XP magnetic beads/PEG 8000-containing buffer | genus Scorzonera | Between 1920 and 1960 | Yes | PCR amplification | Krinitsina et al. 2015 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





