Submitted:
08 February 2024
Posted:
09 February 2024
You are already at the latest version
Abstract
Keywords:
“A comparison of the different gene arrangements in the same chromosome may, in certain cases, throw light on the historical relationships of these structures, and consequently on the history of the species as a whole.” Dobzhansky & Sturtevant, 1938, Genetics [1].
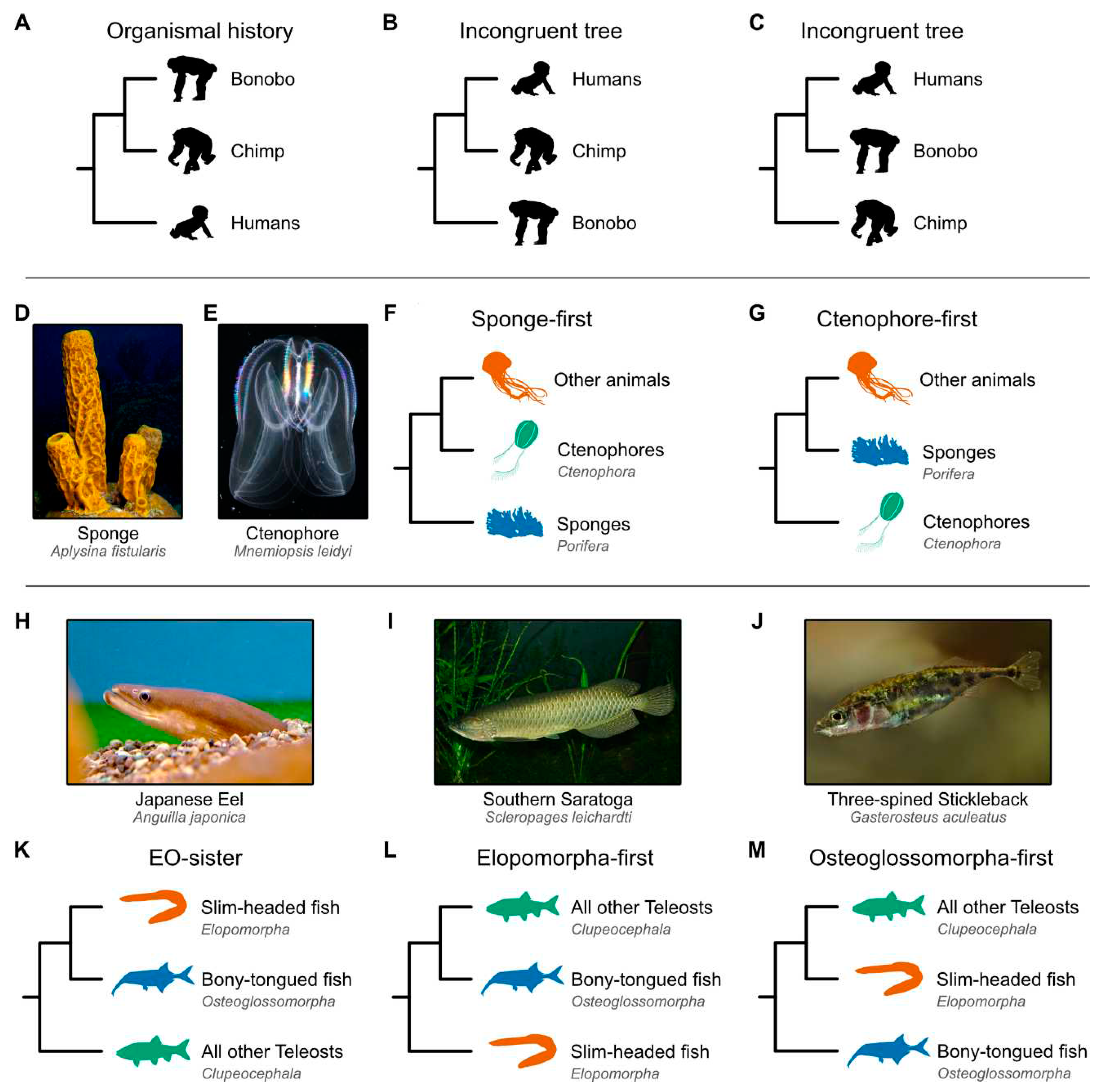
Tangled branches in the Tree of Life
Rare genomic changes as phylogenomic markers
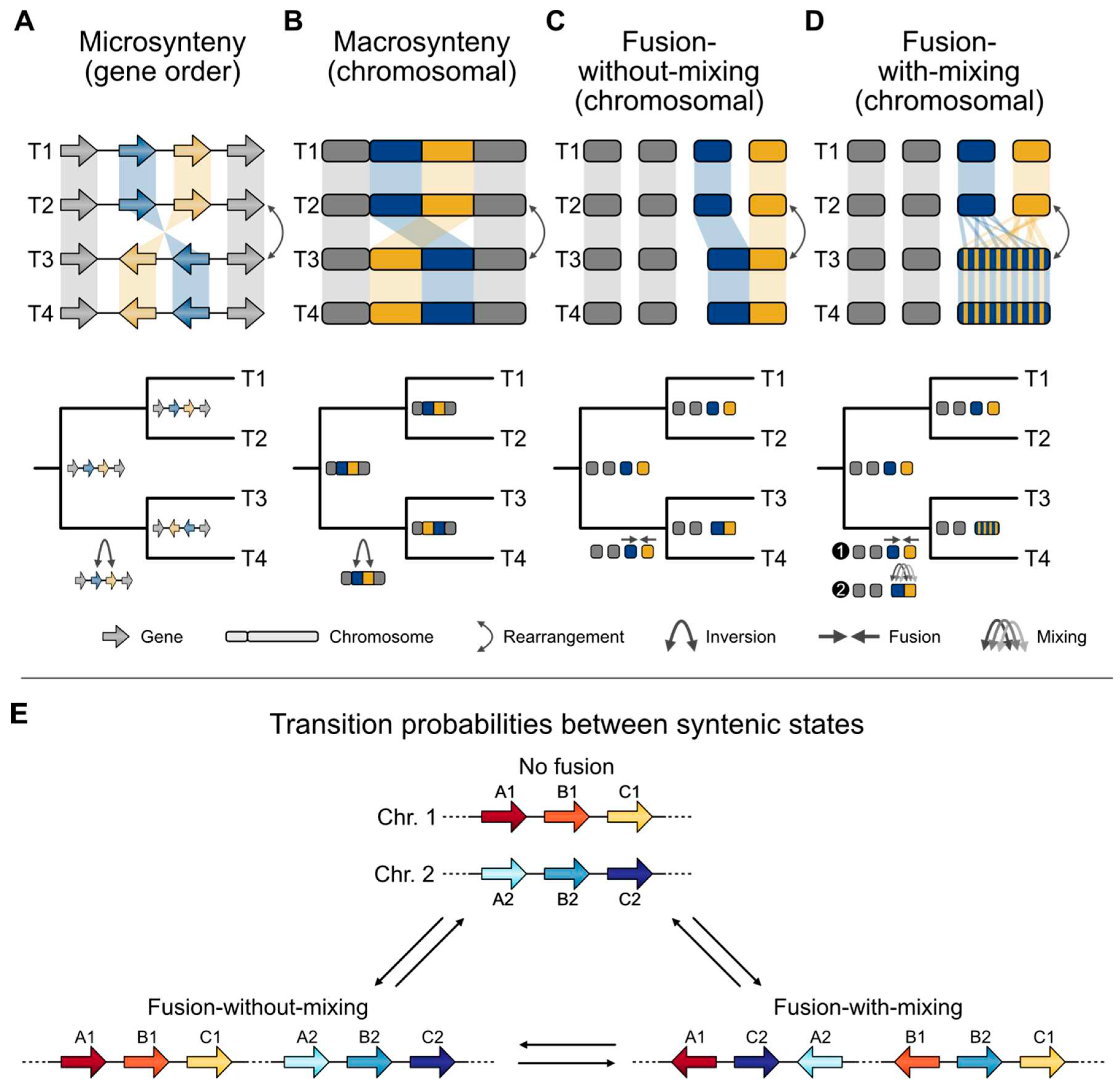
Synteny emerges in the phylogenomic era
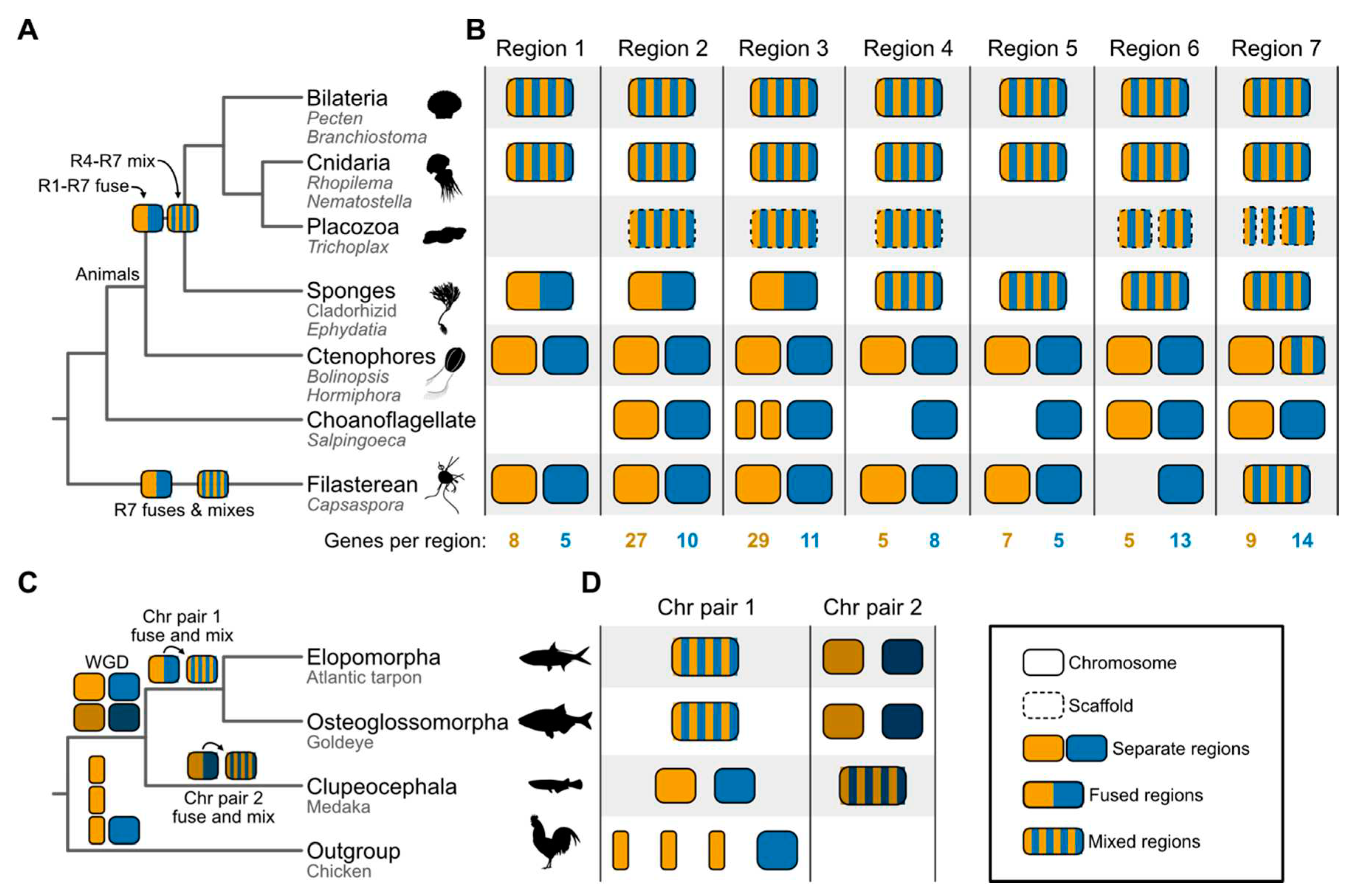
Synteny brings fresh perspectives to the Tree of Life
Toward high-quality synteny-based Tree of Life reconstructions
A roadmap to infer synteny-based phylogenies.
Research opportunities using synteny data and species trees.
Conclusion
Funding
Acknowledgments
Conflicts of interest
References
- Dobzhansky T, Sturtevant AH. INVERSIONS IN THE CHROMOSOMES OF DROSOPHILA PSEUDOOBSCURA. Genetics. 1938, 23, 28–64. [CrossRef]
- Fitch WM, Margoliash E. Construction of Phylogenetic Trees: A method based on mutation distances as estimated from cytochrome c sequences is of general applicability. Science. 1967, 155, 279–284. [CrossRef]
- Haggerty LS, Martin FJ, Fitzpatrick DA, McInerney JO. Gene and genome trees conflict at many levels. Phil Trans R Soc B. 2009, 364, 2209–2219. [CrossRef]
- Steenwyk JL, Li Y, Zhou X. Incongruence in the phylogenomics era. Nature Reviews Genetics. 2023. [Google Scholar] [CrossRef] [PubMed]
- Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003, 425, 798–804. [CrossRef]
- Kapli P, Yang Z, Telford MJ. Phylogenetic tree building in the genomic age. Nat Rev Genet. 2020, 21, 428–444. [CrossRef]
- Philippe H, Lartillot N, Brinkmann H. Multigene Analyses of Bilaterian Animals Corroborate the Monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Molecular Biology and Evolution. 2005, 22, 1246–1253. [CrossRef]
- Giribet G, Edgecombe GD. Current Understanding of Ecdysozoa and its Internal Phylogenetic Relationships. Integrative and Comparative Biology. 2017, 57, 455–466. [CrossRef]
- Crotty SM, Minh BQ, Bean NG; et al. GHOST: Recovering Historical Signal from Heterotachously Evolved Sequence Alignments. Systematic Biology 2019, (syz051). [Google Scholar] [CrossRef]
- Williams TA, Cox CJ, Foster PG, Szöllősi GJ, Embley TM. Phylogenomics provides robust support for a two-domains tree of life. Nat Ecol Evol. 2019, 4, 138–147. [CrossRef]
- Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014, 346, 1320–1331. [CrossRef]
- Choi B, Crisp MD, Cook LG, Meusemann K, Edwards RD, Toon A, et al. Identifying genetic markers for a range of phylogenetic utility–From species to family level. Brewer MS, editor. PLoS ONE. 2019, 14, e0218995. [CrossRef]
- Debray K, Marie-Magdelaine J, Ruttink T, Clotault J, Foucher F, Malécot V. Identification and assessment of variable single-copy orthologous (SCO) nuclear loci for low-level phylogenomics: a case study in the genus Rosa (Rosaceae). BMC Evol Biol. 2019, 19, 152. [CrossRef]
- Parey E, Louis A, Montfort J, Bouchez O, Roques C, Iampietro C, et al. Genome structures resolve the early diversification of teleost fishes. Science. 2023, 379, 572–575. [CrossRef]
- Schultz DT, Haddock SHD, Bredeson JV, Green RE, Simakov O, Rokhsar DS. Ancient gene linkages support ctenophores as sister to other animals. Nature. 2023 [cited ]. 21 May. [CrossRef]
- Wainright PO, Hinkle G, Sogin ML, Stickel SK. Monophyletic Origins of the Metazoa: an Evolutionary Link with Fungi. Science. 1993, 260, 340–342. [CrossRef]
- Brusca RC, Brusca GJ. Invertebrates. Sinauer Associates Incorporated; 2002.
- Collins, AG. Evaluating multiple alternative hypotheses for the origin of Bilateria: An analysis of 18S rRNA molecular evidence. Proc Natl Acad Sci USA. 1998, 95, 15458–15463. [Google Scholar] [CrossRef]
- Medina M, Collins AG, Silberman JD, Sogin ML. Evaluating hypotheses of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc Natl Acad Sci USA. 2001, 98, 9707–9712. [CrossRef]
- Podar M, Haddock SHD, Sogin ML, Harbison GR. A Molecular Phylogenetic Framework for the Phylum Ctenophora Using 18S rRNA Genes. Molecular Phylogenetics and Evolution. 2001, 21, 218–230. [CrossRef]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008, 452, 745–749. [CrossRef]
- Philippe H, Derelle R, Lopez P, Pick K, Borchiellini C, Boury-Esnault N, et al. Phylogenomics Revives Traditional Views on Deep Animal Relationships. Current Biology. 2009, 19, 706–712. [CrossRef]
- Simion P, Philippe H, Baurain D, Jager M, Richter DJ, Di Franco A, et al. A Large and Consistent Phylogenomic Dataset Supports Sponges as the Sister Group to All Other Animals. Current Biology. 2017, 27, 958–967. [CrossRef]
- Shen X-X, Hittinger CT, Rokas A. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat Ecol Evol. 2017, 1, 0126. [CrossRef]
- King N, Rokas A. Embracing Uncertainty in Reconstructing Early Animal Evolution. Current Biology. 2017, 27, R1081–R1088. [CrossRef]
- Whelan NV, Kocot KM, Moroz TP, Mukherjee K, Williams P, Paulay G, et al. Ctenophore relationships and their placement as the sister group to all other animals. Nat Ecol Evol. 2017, 1, 1737–1746. [CrossRef]
- Li Y, Shen X-X, Evans B, Dunn CW, Rokas A. Rooting the Animal Tree of Life. Tamura K, editor. Molecular Biology and Evolution. 2021, 38, 4322–4333. [CrossRef]
- Whelan NV, Halanych KM. Available data do not rule out Ctenophora as the sister group to all other Metazoa. Nat Commun. 2023, 14, 711. [CrossRef]
- Le HLV, Lecointre G, Perasso R. A 28S rRNA-Based Phylogeny of the Gnathostomes: First Steps in the Analysis of Conflict and Congruence with Morphologically Based Cladograms. Molecular Phylogenetics and Evolution. 1993, 2, 31–51. [CrossRef]
- Dornburg A, Near TJ. The emerging phylogenetic perspective on the evolution of actinopterygian fishes. Annual Review of Ecology, Evolution, and Systematics. 2021, 52, 427–452.
- Rokas A, Holland PWH. Rare genomic changes as a tool for phylogenetics. Trends in Ecology & Evolution. 2000, 15, 454–459. [CrossRef]
- Castresana J, Feldmaier-Fuchs G, Yokobori S, Satoh N, Pääbo S. The Mitochondrial Genome of the Hemichordate Balanoglossus carnosus and the Evolution of Deuterostome Mitochondria. Genetics. 1998, 150, 1115–1123. [CrossRef]
- Venkatesh B, Ning Y, Brenner S. Late changes in spliceosomal introns define clades in vertebrate evolution. Proc Natl Acad Sci USA. 1999, 96, 10267–10271. [CrossRef]
- Rokas A, Kathirithamby J, Holland PWH. Intron insertion as a phylogenetic character: the engrailed homeobox of Strepsiptera does not indicate affinity with Diptera. Insect Mol Biol. 1999, 8, 527–530. [CrossRef]
- Steenwyk JL, Opulente DA, Kominek J, Shen X-X, Zhou X, Labella AL, et al. Extensive loss of cell-cycle and DNA repair genes in an ancient lineage of bipolar budding yeasts. Kamoun S, editor. PLoS Biol. 2019, 17, e3000255. [CrossRef]
- Sturtevant AH, Dobzhansky Th. Inversions in the Third Chromosome of Wild Races of Drosophila Pseudoobscura, and Their Use in the Study of the History of the Species. Proc Natl Acad Sci USA. 1936, 22, 448–450. [CrossRef]
- Steenwyk JL, Soghigian JS, Perfect JR, Gibbons JG. Copy number variation contributes to cryptic genetic variation in outbreak lineages of Cryptococcus gattii from the North American Pacific Northwest. BMC Genomics. 2016, 17, 700. [CrossRef]
- Lee Y-L, Bosse M, Mullaart E, Groenen MAM, Veerkamp RF, Bouwman AC. Functional and population genetic features of copy number variations in two dairy cattle populations. BMC Genomics. 2020, 21, 89. [CrossRef]
- Brown KH, Dobrinski KP, Lee AS, Gokcumen O, Mills RE, Shi X, et al. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci USA. 2012, 109, 529–534. [CrossRef]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015, 526, 75–81. [CrossRef]
- Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, et al. Lineage-Specific Gene Duplication and Loss in Human and Great Ape Evolution. Chris Tyler-Smith, editor. PLoS Biol. 2004, 2, e207. [CrossRef]
- Mühlhausen S, Schmitt HD, Pan K-T, Plessmann U, Urlaub H, Hurst LD, et al. Endogenous Stochastic Decoding of the CUG Codon by Competing Ser- and Leu-tRNAs in Ascoidea asiatica. Current Biology. 2018, 28, 2046–2057e5. [CrossRef]
- Mathews S, Donoghue MJ. The Root of Angiosperm Phylogeny Inferred from Duplicate Phytochrome Genes. Science. 1999, 286, 947–950. [CrossRef]
- One Thousand Plant Transcriptomes Initiative. One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019, 574, 679–685. [CrossRef]
- Doronina L, Reising O, Clawson H, Ray DA, Schmitz J. True Homoplasy of Retrotransposon Insertions in Primates. Susko E, editor. Systematic Biology. 2019, 68, 482–493. [CrossRef]
- Cloutier A, Sackton TB, Grayson P, Clamp M, Baker AJ, Edwards SV. Whole-Genome Analyses Resolve the Phylogeny of Flightless Birds (Palaeognathae) in the Presence of an Empirical Anomaly Zone. Faircloth B, editor. Systematic Biology. 2019, 68, 937–955. [CrossRef]
- Murphy WJ, Foley NM, Bredemeyer KR, Gatesy J, Springer MS. Phylogenomics and the Genetic Architecture of the Placental Mammal Radiation. Annu Rev Anim Biosci. 2021, 9, 29–53. [CrossRef]
- Takahashi K, Terai Y, Nishida M, Okada N. A novel family of short interspersed repetitive elements (SINEs) from cichlids: the patterns of insertion of SINEs at orthologous loci support the proposed monophyly of four major groups of cichlid fishes in Lake Tanganyika. Molecular Biology and Evolution. 1998, 15, 391–407. [CrossRef]
- Takahashi K, Nishida M, Yuma M, Okada N. Retroposition of the AFC Family of SINEs (Short Interspersed Repetitive Elements) Before and During the Adaptive Radiation of Cichlid Fishes in Lake Malawi and Related Inferences About Phylogeny. Journal of Molecular Evolution. 2001, 53, 496–507. [CrossRef]
- Springer MS, Molloy EK, Sloan DB, Simmons MP, Gatesy J. ILS-Aware Analysis of Low-Homoplasy Retroelement Insertions: Inference of Species Trees and Introgression Using Quartets. Murphy W, editor. Journal of Heredity. 2020, 111, 147–168. [CrossRef]
- Martín-Durán JM, Ryan JF, Vellutini BC, Pang K, Hejnol A. Increased taxon sampling reveals thousands of hidden orthologs in flatworms. Genome Res. 2017, 27, 1263–1272. [CrossRef]
- Krassowski T, Coughlan AY, Shen X-X, Zhou X, Kominek J, Opulente DA, et al. Evolutionary instability of CUG-Leu in the genetic code of budding yeasts. Nat Commun. 2018, 9, 1887. [CrossRef]
- Dellaporta SL, Xu A, Sagasser S, Jakob W, Moreno MA, Buss LW, et al. Mitochondrial genome of Trichoplax adhaerens supports Placozoa as the basal lower metazoan phylum. Proc Natl Acad Sci USA. 2006, 103, 8751–8756. [CrossRef]
- Laumer CE, Gruber-Vodicka H, Hadfield MG, Pearse VB, Riesgo A, Marioni JC, et al. Support for a clade of Placozoa and Cnidaria in genes with minimal compositional bias. eLife. 2018, 7, e36278. [CrossRef]
- Ding Y-M, Pang X-X, Cao Y, Zhang W-P, Renner SS, Zhang D-Y, et al. Genome structure-based Juglandaceae phylogenies contradict alignment-based phylogenies and substitution rates vary with DNA repair genes. Nat Commun. 2023, 14, 617. [CrossRef]
- Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004, 20, 3643–3646.
- Proost S, Fostier J, De Witte D, Dhoedt B, Demeester P, Van de Peer Y, et al. i-ADHoRe 3.0—fast and sensitive detection of genomic homology in extremely large data sets. Nucleic acids research. 2012, 40, e11–e11.
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic acids research. 2012, 40, e49–e49.
- Drillon G, Carbone A, Fischer G. SynChro: a fast and easy tool to reconstruct and visualize synteny blocks along eukaryotic chromosomes. PloS one. 2014, 9, e92621.
- Almeida-Silva F, Zhao T, Ullrich KK, Schranz ME, Van De Peer Y. syntenet: an R/Bioconductor package for the inference and analysis of synteny networks. Martelli PL, editor. Bioinformatics. 2023, 39, btac806. [CrossRef]
- Mackintosh A, De La Rosa PMG, Martin SH, Lohse K, Laetsch DR. Inferring inter-chromosomal rearrangements and ancestral linkage groups from synteny. Evolutionary Biology; 2023 Sep. [CrossRef]
- Hane JK, Rouxel T, Howlett BJ, Kema GH, Goodwin SB, Oliver RP. A novel mode of chromosomal evolution peculiar to filamentous Ascomycete fungi. Genome Biol. 2011, 12, R45. [CrossRef]
- Robberecht C, Voet T, Esteki MZ, Nowakowska BA, Vermeesch JR. Nonallelic homologous recombination between retrotransposable elements is a driver of de novo unbalanced translocations. Genome Res. 2013, 23, 411–418. [CrossRef]
- Ma J, Bennetzen JL. Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc Natl Acad Sci USA. 2006, 103, 383–388. [CrossRef]
- Liu P, Lacaria M, Zhang F, Withers M, Hastings PJ, Lupski JR. Frequency of Nonallelic Homologous Recombination Is Correlated with Length of Homology: Evidence that Ectopic Synapsis Precedes Ectopic Crossing-Over. The American Journal of Human Genetics. 2011, 89, 580–588. [CrossRef]
- Ferguson S, Jones A, Murray K, Schwessinger B, Borevitz JO. Interspecies genome divergence is predominantly due to frequent small scale rearrangements in Eucalyptus. Molecular Ecology. 2023, 32, 1271–1287. [CrossRef]
- Li Y, Liu H, Steenwyk JL, LaBella AL, Harrison M-C, Groenewald M, et al. Contrasting modes of macro and microsynteny evolution in a eukaryotic subphylum. Current Biology. 2022, S0960982222016700. [CrossRef]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005, 6, 361–375. [CrossRef]
- Zheng C, Sankoff D. Gene order in rosid phylogeny, inferred from pairwise syntenies among extant genomes. BMC Bioinformatics. 2012, 13, S9. [CrossRef]
- Drillon G, Champeimont R, Oteri F, Fischer G, Carbone A. Phylogenetic Reconstruction Based on Synteny Block and Gene Adjacencies. Battistuzzi FU, editor. Molecular Biology and Evolution. 2020, 37, 2747–2762. [CrossRef]
- Zhao T, Zwaenepoel A, Xue J-Y, Kao S-M, Li Z, Schranz ME, et al. Whole-genome microsynteny-based phylogeny of angiosperms. Nat Commun. 2021, 12, 3498. [CrossRef]
- Therman E, Susman B, Denniston C. The nonrandom participation of human acrocentric chromosomes in Robertsonian translocations. Annals of Human Genetics. 1989, 53, 49–65. [CrossRef]
- Fairclough SR, Chen Z, Kramer E, Zeng Q, Young S, Robertson HM, et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013, 14, R15. [CrossRef]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008, 451, 783–788. [CrossRef]
- Ocaña-Pallarès E, Williams TA, López-Escardó D, Arroyo AS, Pathmanathan JS, Bapteste E, et al. Divergent genomic trajectories predate the origin of animals and fungi. Nature. 2022, 609, 747–753. [CrossRef]
- Torruella G, Derelle R, Paps J, Lang BF, Roger AJ, Shalchian-Tabrizi K, et al. Phylogenetic Relationships within the Opisthokonta Based on Phylogenomic Analyses of Conserved Single-Copy Protein Domains. Molecular Biology and Evolution. 2012, 29, 531–544. [CrossRef]
- Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A Phylogenomic Investigation into the Origin of Metazoa. Molecular Biology and Evolution. 2008, 25, 664–672. [CrossRef]
- Richter DJ, Fozouni P, Eisen MB, King N. Gene family innovation, conservation and loss on the animal stem lineage. eLife. 2018, 7, e34226. [CrossRef]
- Whelan NV, Kocot KM, Moroz LL, Halanych KM. Error, signal, and the placement of Ctenophora sister to all other animals. Proc Natl Acad Sci USA. 2015, 112, 5773–5778. [CrossRef]
- Sperling EA, Pisani D, Peterson KJ. Poriferan paraphyly and its implications for Precambrian palaeobiology. SP. 2007, 286, 355–368. [CrossRef]
- Borchiellini C, Manuel M, Alivon E, Boury-Esnault N, Vacelet J, Le Parco Y. Sponge paraphyly and the origin of Metazoa: Sponge paraphyly. Journal of Evolutionary Biology. 2001, 14, 171–179. [CrossRef]
- Kenny NJ, Francis WR, Rivera-Vicéns RE, Juravel K, De Mendoza A, Díez-Vives C, et al. Tracing animal genomic evolution with the chromosomal-level assembly of the freshwater sponge Ephydatia muelleri. Nat Commun. 2020, 11, 3676. [CrossRef]
- Hughes LC, Ortí G, Huang Y, Sun Y, Baldwin CC, Thompson AW, et al. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc Natl Acad Sci USA. 2018, 115, 6249–6254. [CrossRef]
- Faircloth BC, Sorenson L, Santini F, Alfaro ME. A Phylogenomic Perspective on the Radiation of Ray-Finned Fishes Based upon Targeted Sequencing of Ultraconserved Elements (UCEs). Moreau CS, editor. PLoS ONE. 2013, 8, e65923. [CrossRef]
- Pollock DD, Zwickl DJ, McGuire JA, Hillis DM. Increased Taxon Sampling Is Advantageous for Phylogenetic Inference. Crandall K, editor. Systematic Biology. 2002, 51, 664–671. [CrossRef]
- Aberer AJ, Krompass D, Stamatakis A. Pruning Rogue Taxa Improves Phylogenetic Accuracy: An Efficient Algorithm and Webservice. Systematic Biology. 2013, 62, 162–166. [CrossRef]
- Shen X-X, Zhou X, Kominek J, Kurtzman CP, Hittinger CT, Rokas A. Reconstructing the Backbone of the Saccharomycotina Yeast Phylogeny Using Genome-Scale Data. G3 Genes|Genomes|Genetics. 2016, 6, 3927–3939. [CrossRef]
- Riley R, Haridas S, Wolfe KH, Lopes MR, Hittinger CT, Göker M, et al. Comparative genomics of biotechnologically important yeasts. Proc Natl Acad Sci USA. 2016, 113, 9882–9887. [CrossRef]
- Shen X-X, Opulente DA, Kominek J, Zhou X, Steenwyk JL, Buh KV, et al. Tempo and Mode of Genome Evolution in the Budding Yeast Subphylum. Cell. 2018, 175, 1533–1545e20. [CrossRef]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006, 440, 341–345. [CrossRef]
- Pisani D, Pett W, Dohrmann M, Feuda R, Rota-Stabelli O, Philippe H, et al. Genomic data do not support comb jellies as the sister group to all other animals. Proc Natl Acad Sci USA. 2015, 112, 15402–15407. [CrossRef]
- Brinkmann H, Van Der Giezen M, Zhou Y, De Raucourt GP, Philippe H. An Empirical Assessment of Long-Branch Attraction Artefacts in Deep Eukaryotic Phylogenomics. Hedin M, editor. Systematic Biology. 2005, 54, 743–757. [CrossRef]
- Marx, V. Method of the year: long-read sequencing. Nat Methods. 2023, 20, 6–11. [Google Scholar] [CrossRef]
- Giani AM, Gallo GR, Gianfranceschi L, Formenti G. Long walk to genomics: History and current approaches to genome sequencing and assembly. Computational and Structural Biotechnology Journal. 2020, 18, 9–19. [CrossRef]
- Belton J-M, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi–C: A comprehensive technique to capture the conformation of genomes. Methods. 2012, 58, 268–276. [CrossRef]
- Liu D, Hunt M, Tsai IJ. Inferring synteny between genome assemblies: a systematic evaluation. BMC Bioinformatics. 2018, 19, 26. [CrossRef]
- Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, et al. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Molecular Biology and Evolution. 2018, 35, 543–548. [CrossRef]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, et al. Towards complete and error-free genome assemblies of all vertebrate species. Nature. 2021, 592, 737–746. [CrossRef]
- Weisman CM, Murray AW, Eddy SR. Mixing genome annotation methods in a comparative analysis inflates the apparent number of lineage-specific genes. Current Biology. 2022, 32, 2632–2639e2. [CrossRef]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, R7. [CrossRef]
- Rosato M, Hoelscher B, Lin Z, Agwu C, Xu F. Transcriptome analysis provides genome annotation and expression profiles in the central nervous system of Lymnaea stagnalis at different ages. BMC Genomics. 2021, 22, 637. [CrossRef]
- Fernández R, Gabaldón T, Dessimoz C. Orthology: definitions, inference, and impact on species phylogeny inference. 2019 [cited ]. 25 May. [CrossRef]
- Philippe H, Vienne DMD, Ranwez V, Roure B, Baurain D, Delsuc F. Pitfalls in supermatrix phylogenomics. EJT. 2017 [cited 5 Dec 2023]. [CrossRef]
- Stolzer M, Lai H, Xu M, Sathaye D, Vernot B, Durand D. Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics. 2012, 28, i409–i415. [CrossRef]
- Minkin I, Medvedev P. Scalable multiple whole-genome alignment and locally collinear block construction with SibeliaZ. Nat Commun. 2020, 11, 6327. [CrossRef]
- Armstrong J, Hickey G, Diekhans M, Fiddes IT, Novak AM, Deran A, et al. Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature. 2020, 587, 246–251. [CrossRef]
- Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Wörheide G, et al. Resolving Difficult Phylogenetic Questions: Why More Sequences Are Not Enough. Penny D, editor. PLoS Biol. 2011, 9, e1000602. [CrossRef]
- Maddison WP, Knowles LL. Inferring Phylogeny Despite Incomplete Lineage Sorting. Collins T, editor. Systematic Biology. 2006, 55, 21–30. [CrossRef]
- Feng S, Bai M, Rivas-González I, Li C, Liu S, Tong Y, et al. Incomplete lineage sorting and phenotypic evolution in marsupials. Cell. 2022, 185, 1646–1660e18. [CrossRef]
- Avise JC, Robinson TJ. Hemiplasy: A New Term in the Lexicon of Phylogenetics. Kubatko L, editor. Systematic Biology. 2008, 57, 503–507. [CrossRef]
- Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, et al. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun. 2017, 8, 14061. [CrossRef]
- Steenwyk J, Rokas A. Extensive Copy Number Variation in Fermentation-Related Genes Among Saccharomyces cerevisiae Wine Strains. G3 Genes|Genomes|Genetics. 2017, 7, 1475–1485. [CrossRef]
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, et al. Hybridization and speciation. J Evol Biol. 2013, 26, 229–246. [CrossRef]
- Irisarri I, Singh P, Koblmüller S, Torres-Dowdall J, Henning F, Franchini P, et al. Phylogenomics uncovers early hybridization and adaptive loci shaping the radiation of Lake Tanganyika cichlid fishes. Nat Commun. 2018, 9, 3159. [CrossRef]
- Bjornson S, Upham N, Verbruggen H, Steenwyk J. Phylogenomic Inference, Divergence-Time Calibration, and Methods for Characterizing Reticulate Evolution. Biology and Life Sciences; 2023 Sep. [CrossRef]
- Arnold BJ, Huang I-T, Hanage WP. Horizontal gene transfer and adaptive evolution in bacteria. Nat Rev Microbiol. 2022, 20, 206–218. [CrossRef]
- Gophna U, Altman-Price N. Horizontal Gene Transfer in Archaea—From Mechanisms to Genome Evolution. Annu Rev Microbiol. 2022, 76, 481–502. [CrossRef]
- Buck R, Ortega-Del Vecchyo D, Gehring C, Michelson R, Flores-Rentería D, Klein B, et al. Sequential hybridization may have facilitated ecological transitions in the Southwestern pinyon pine syngameon. New Phytologist. 2023, 237, 2435–2449. [CrossRef]
- Goulet BE, Roda F, Hopkins R. Hybridization in Plants: Old Ideas, New Techniques. Plant Physiol. 2017, 173, 65–78. [CrossRef]
- Rieseberg LH, Kim S-C, Randell RA, Whitney KD, Gross BL, Lexer C, et al. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica. 2007, 129, 149–165. [CrossRef]
- Steenwyk JL, Lind AL, Ries LNA, dos Reis TF, Silva LP, Almeida F, et al. Pathogenic Allodiploid Hybrids of Aspergillus Fungi. Current Biology. 2020, 30, 2495–2507e7. [CrossRef]
- Marcet-Houben M, Gabaldón T. Beyond the Whole-Genome Duplication: Phylogenetic Evidence for an Ancient Interspecies Hybridization in the Baker’s Yeast Lineage. Hurst LD, editor. PLoS Biol. 2015, 13, e1002220. [CrossRef]
- Adavoudi R, Pilot M. Consequences of Hybridization in Mammals: A Systematic Review. Genes. 2021, 13, 50. [CrossRef]
- League GP, Slot JC, Rokas A. The ASP3 locus in Saccharomyces cerevisiae originated by horizontal gene transfer from Wickerhamomyces. FEMS Yeast Res. 2012, 12, 859–863. [CrossRef]
- Kominek J, Doering DT, Opulente DA, Shen X-X, Zhou X, DeVirgilio J, et al. Eukaryotic Acquisition of a Bacterial Operon. Cell. 2019, 176, 1356–1366e10. [CrossRef]
- Davín AA, Tannier E, Williams TA, Boussau B, Daubin V, Szöllősi GJ. Gene transfers can date the tree of life. Nat Ecol Evol. 2018, 2, 904–909. [CrossRef]
- Jukes TH, Cantor CR. Evolution of Protein Molecules. Mammalian Protein Metabolism. Elsevier; 1969. pp. 21–132. [CrossRef]
- Tavaré, S. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci (Am Math Soc). 1986, 17, 57–86. [Google Scholar]
- Philippe H, Zhou Y, Brinkmann H, Rodrigue N, Delsuc F. Heterotachy and long-branch attraction in phylogenetics. BMC Evol Biol. 2005, 5, 50. [CrossRef]
- Liao W-W, Asri M, Ebler J, Doerr D, Haukness M, Hickey G, et al. A draft human pangenome reference. Nature. 2023, 617, 312–324. [CrossRef]
- Svedberg J, Hosseini S, Chen J, Vogan AA, Mozgova I, Hennig L, et al. Convergent evolution of complex genomic rearrangements in two fungal meiotic drive elements. Nat Commun. 2018, 9, 4242. [CrossRef]
- Jain Y, Chandradoss KR, A. V. A, Bhattacharya J, Lal M, Bagadia M, et al. Convergent evolution of a genomic rearrangement may explain cancer resistance in hystrico- and sciuromorpha rodents. npj Aging Mech Dis. 2021, 7, 20. [CrossRef]
- Mezzasalma M, Streicher JW, Guarino FM, Jones MEH, Loader SP, Odierna G, et al. Microchromosome fusions underpin convergent evolution of chameleon karyotypes. Gaitan-Espitia JD, Chapman T, editors. Evolution. 2023, 77, 1930–1944. [CrossRef]
- Steenwyk JL, Shen X-X, Lind AL, Goldman GH, Rokas A. A Robust Phylogenomic Time Tree for Biotechnologically and Medically Important Fungi in the Genera Aspergillus and Penicillium. Boyle JP, editor. mBio. 2019, 10, e00925–19. [CrossRef]
- Liu L, Zhang J, Rheindt FE, Lei F, Qu Y, Wang Y, et al. Genomic evidence reveals a radiation of placental mammals uninterrupted by the KPg boundary. Proc Natl Acad Sci USA. 2017, 114. [CrossRef]
- Smith SA, Brown JW, Walker JF. So many genes, so little time: A practical approach to divergence-time estimation in the genomic era. Escriva H, editor. PLoS ONE. 2018, 13, e0197433. [CrossRef]
- Phillips MJ, Penny D. The root of the mammalian tree inferred from whole mitochondrial genomes. Molecular Phylogenetics and Evolution. 2003, 28, 171–185. [CrossRef]
- Ortiz-Merino RA, Kuanyshev N, Braun-Galleani S, Byrne KP, Porro D, Branduardi P, et al. Evolutionary restoration of fertility in an interspecies hybrid yeast, by whole-genome duplication after a failed mating-type switch. Hurst L, editor. PLoS Biol. 2017, 15, e2002128. [CrossRef]
- Clark JW, Donoghue PCJ. Whole-Genome Duplication and Plant Macroevolution. Trends in Plant Science. 2018, 23, 933–945. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




