Submitted:
17 August 2023
Posted:
18 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Patients and methods
2.1. Patient enrollment
2.2. Formalin-Fixed Paraffin-embedded (FFPE) tissues
2.3. TLR and VDR genotyping in blood samples
2.4. Whole Exome Sequencing (WES)
2.5. Bioinformatic analysis
2.6. Statistical analysis
3. Results
3.1. Patients
3.2. Annotated variants for each position
3.3. Identification of mutated genes
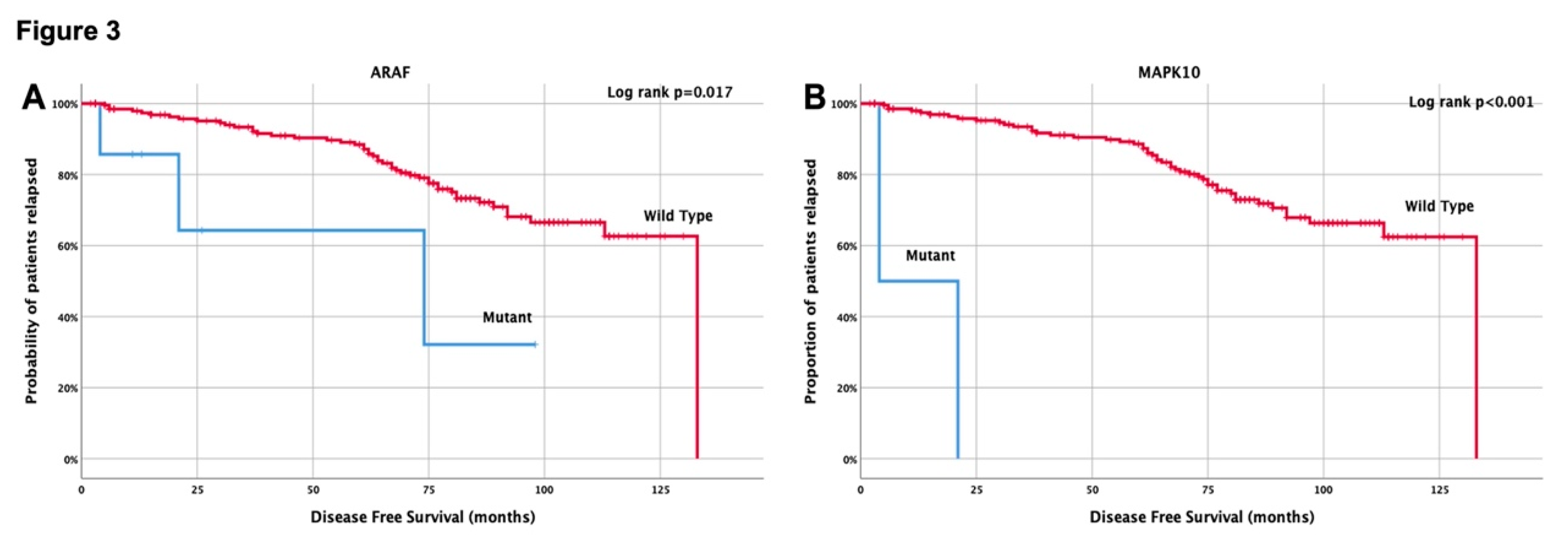
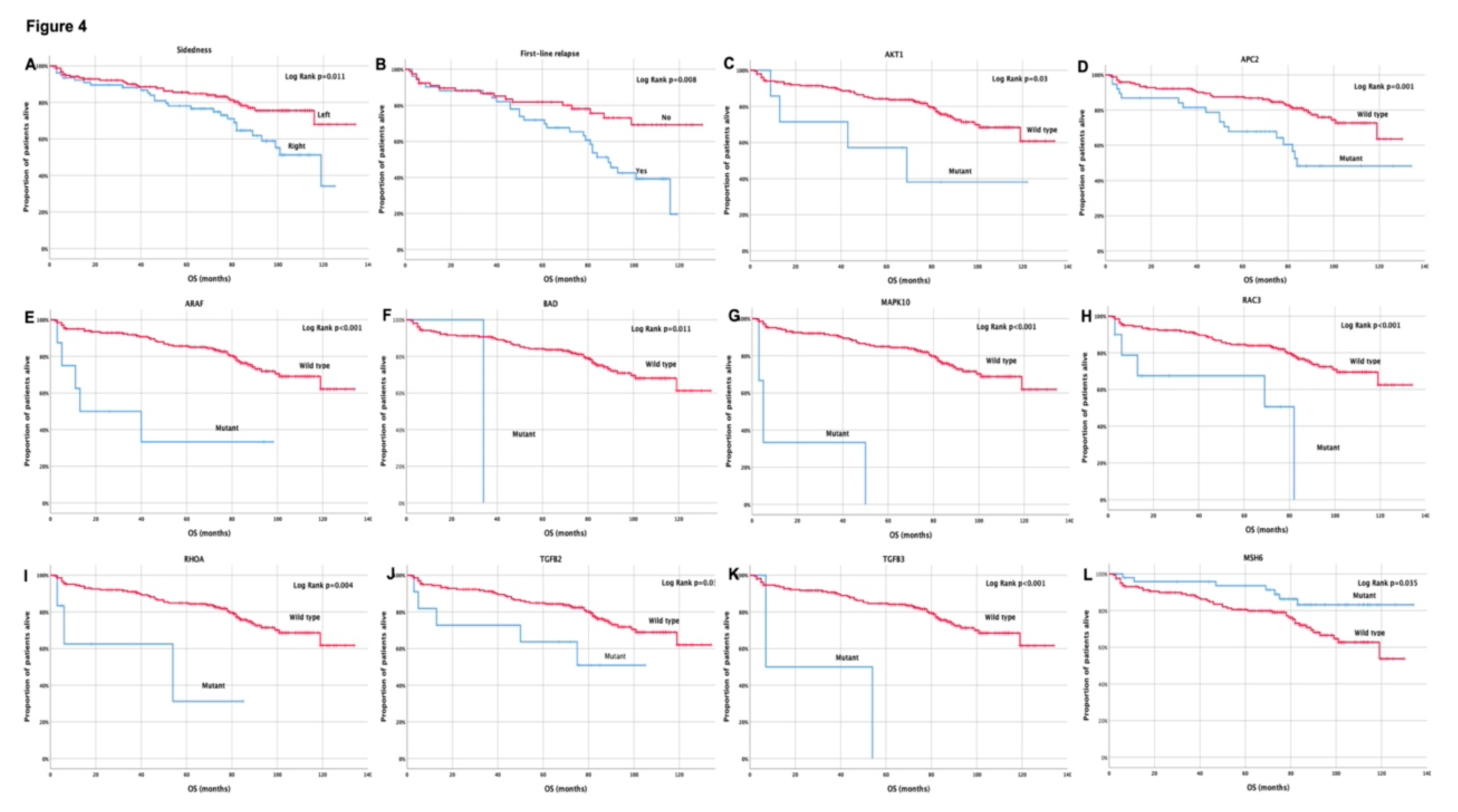
3.4. Clinical outcome based on molecular profile and patients’ characteristics
3.5. Univariate and multivariate Cox-regression analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgement
Conflicts of Interest
References
- Yu, H.; Hemminki, K. Genetic epidemiology of colorectal cancer and associated cancers. Mutagenesis 2020, 35, 207–219. [Google Scholar] [CrossRef]
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedzwiedzka, E.; Arlukowicz, T.; Przybylowicz, K.E. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef]
- Andre, T.; Boni, C.; Navarro, M.; Tabernero, J.; Hickish, T.; Topham, C.; Bonetti, A.; Clingan, P.; Bridgewater, J.; Rivera, F.; et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol 2009, 27, 3109–3116. [Google Scholar] [CrossRef]
- Haller, D.G.; Tabernero, J.; Maroun, J.; de Braud, F.; Price, T.; Van Cutsem, E.; Hill, M.; Gilberg, F.; Rittweger, K.; Schmoll, H.J. Capecitabine plus oxaliplatin compared with fluorouracil and folinic acid as adjuvant therapy for stage III colon cancer. J Clin Oncol 2011, 29, 1465–1471. [Google Scholar] [CrossRef]
- Sobrero, A.; Grothey, A.; Iveson, T.; Labianca, R.; Yoshino, T.; Taieb, J.; Maughan, T.; Buyse, M.; Andre, T.; Meyerhardt, J.; et al. The hard road to data interpretation: 3 or 6 months of adjuvant chemotherapy for patients with stage III colon cancer? Ann Oncol 2018, 29, 1099–1107. [Google Scholar] [CrossRef] [PubMed]
- Grothey, A.; Sobrero, A.F.; Shields, A.F.; Yoshino, T.; Paul, J.; Taieb, J.; Souglakos, J.; Shi, Q.; Kerr, R.; Labianca, R.; et al. Duration of Adjuvant Chemotherapy for Stage III Colon Cancer. N Engl J Med 2018, 378, 1177–1188. [Google Scholar] [CrossRef]
- Angell, H.K.; Bruni, D.; Barrett, J.C.; Herbst, R.; Galon, J. The Immunoscore: Colon Cancer and Beyond. Clin Cancer Res 2020, 26, 332–339. [Google Scholar] [CrossRef]
- Messaritakis, I.; Koulouridi, A.; Boukla, E.; Sfakianaki, M.; Vogiatzoglou, K.; Karagianni, M.; Gouvas, N.; Tsiaoussis, J.; Xynos, E.; Athanasakis, E.; et al. Investigation of Microbial Translocation, TLR and VDR Gene Polymorphisms, and Recurrence Risk in Stage III Colorectal Cancer Patients. Cancers (Basel) 2022, 14. [Google Scholar] [CrossRef]
- Messaritakis, I.; Vogiatzoglou, K.; Tsantaki, K.; Ntretaki, A.; Sfakianaki, M.; Koulouridi, A.; Tsiaoussis, J.; Mavroudis, D.; Souglakos, J. The Prognostic Value of the Detection of Microbial Translocation in the Blood of Colorectal Cancer Patients. Cancers (Basel) 2020, 12. [Google Scholar] [CrossRef]
- Messaritakis, I.; Koulouridi, A.; Sfakianaki, M.; Vogiatzoglou, K.; Gouvas, N.; Athanasakis, E.; Tsiaoussis, J.; Xynos, E.; Mavroudis, D.; Tzardi, M.; et al. The Role of Vitamin D Receptor Gene Polymorphisms in Colorectal Cancer Risk. Cancers (Basel) 2020, 12. [Google Scholar] [CrossRef]
- Available online: https://support.illumina.com/sequencing/sequencing_software/igenome.html.
- Geraldine A. Van der Auwera, B.D.O.C. Genomics in the cloud: Using docker, gatk, and wdl in terra, 1st ed.; 2020.
- Available online: https://www.genome.jp/pathway/hsa05210.
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010, 38, e164. [Google Scholar] [CrossRef]
- Deng, N.; Zhou, H.; Fan, H.; Yuan, Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget 2017, 8. [Google Scholar]
- Brenner, H.; Chen, C. The colorectal cancer epidemic: challenges and opportunities for primary, secondary and tertiary prevention. Br J Cancer 2018, 119, 785–792. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Obuch, J.C.; Ahnen, D.J. Colorectal Cancer: Genetics is Changing Everything. Gastroenterol Clin North Am 2016, 45, 459–476. [Google Scholar] [CrossRef]
- Sullivan, B.A.; Noujaim, M.; Roper, J. Cause, Epidemiology, and Histology of Polyps and Pathways to Colorectal Cancer. Gastrointest Endosc Clin N Am 2022, 32, 177–194. [Google Scholar] [CrossRef]
- Uhlitz, F.; Bischoff, P.; Peidli, S.; Sieber, A.; Trinks, A.; Luthen, M.; Obermayer, B.; Blanc, E.; Ruchiy, Y.; Sell, T.; et al. Mitogen-activated protein kinase activity drives cell trajectories in colorectal cancer. EMBO Mol Med 2021, 13, e14123. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Chan, D.K.H.; Buczacki, S.J.A. Tumour heterogeneity and evolutionary dynamics in colorectal cancer. Oncogenesis 2021, 10, 53. [Google Scholar] [CrossRef]
- Fennell, L.; Kane, A.; Liu, C.; McKeone, D.; Hartel, G.; Su, C.; Bond, C.; Bettington, M.; Leggett, B.; Whitehall, V. Braf mutation induces rapid neoplastic transformation in the aged and aberrantly methylated intestinal epithelium. Gut 2022, 71, 1127–1140. [Google Scholar] [CrossRef]
- Chen, T.; Zeineldin, M.; Johnson, B.A.; Dong, Y.; Narkar, A.; Li, T.; Zhu, J.; Li, R.; Larman, T.C. Colonic epithelial adaptation to EGFR-independent growth induces chromosomal instability and is accelerated by prior injury. Neoplasia 2021, 23, 488–501. [Google Scholar] [CrossRef]
- Lee-Six, H.; Olafsson, S.; Ellis, P.; Osborne, R.J.; Sanders, M.A.; Moore, L.; Georgakopoulos, N.; Torrente, F.; Noorani, A.; Goddard, M.; et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature 2019, 574, 532–537. [Google Scholar] [CrossRef]
- Norgaard, K.; Muller, C.; Christensen, N.; Chiloeches, M.L.; Madsen, C.L.; Nielsen, S.S.; Thingholm, T.E.; Belcheva, A. Loss of mismatch repair signaling impairs the WNT-bone morphogenetic protein crosstalk and the colonic homeostasis. J Mol Cell Biol 2020, 12, 410–423. [Google Scholar] [CrossRef]
- Eklof, V.; Wikberg, M.L.; Edin, S.; Dahlin, A.M.; Jonsson, B.A.; Oberg, A.; Rutegard, J.; Palmqvist, R. The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. Br J Cancer 2013, 108, 2153–2163. [Google Scholar] [CrossRef]
- Fu, X.; Wang, X.; Duanmu, J.; Li, T.; Jiang, Q. KRAS mutations are negatively correlated with immunity in colon cancer. Aging (Albany NY) 2020, 13, 750–768. [Google Scholar] [CrossRef]
- Sun, J.; Yao, N.; Lu, P.; Wang, Y. Effects of mFOLFOX6 regimen combined with carrelizumab on immune function and prognosis in patients with microsatellite instability colorectal cancer. Cell Mol Biol (Noisy-le-grand) 2022, 67, 356–362. [Google Scholar] [CrossRef]
- Ratovomanana, T.; Cohen, R.; Svrcek, M.; Renaud, F.; Cervera, P.; Siret, A.; Letourneur, Q.; Buhard, O.; Bourgoin, P.; Guillerm, E.; et al. Performance of Next-Generation Sequencing for the Detection of Microsatellite Instability in Colorectal Cancer With Deficient DNA Mismatch Repair. Gastroenterology 2021, 161, 814–826. [Google Scholar] [CrossRef]
- Xiao, J.; Li, W.; Huang, Y.; Huang, M.; Li, S.; Zhai, X.; Zhao, J.; Gao, C.; Xie, W.; Qin, H.; et al. A next-generation sequencing-based strategy combining microsatellite instability and tumor mutation burden for comprehensive molecular diagnosis of advanced colorectal cancer. BMC Cancer 2021, 21, 282. [Google Scholar] [CrossRef]
- Huang, J.; Liu, H.; Zhao, Y.; Luo, T.; Liu, J.; Liu, J.; Pan, X.; Tang, W. MicroRNAs Expression Patterns Predict Tumor Mutational Burden in Colorectal Cancer. Front Oncol 2020, 10, 550986. [Google Scholar] [CrossRef]
- Bae, J.M.; Yoo, S.Y.; Kim, J.H.; Kang, G.H. Immune landscape and biomarkers for immuno-oncology in colorectal cancers. J Pathol Transl Med 2020, 54, 351–360. [Google Scholar] [CrossRef]
- Saller, J.; Qin, D.; Felder, S.; Coppola, D. Microsatellite Stable Colorectal Cancer With an Immunogenic Phenotype: Challenges in Diagnosis and Treatment. Clin Colorectal Cancer 2020, 19, 123–131. [Google Scholar] [CrossRef]
- Available online: https://www.ncbi.nlm.nih.gov/gene/324.
- Available online: https://www.ncbi.nlm.nih.gov/gene/10297.
- Sun, Y.; Tian, H.; Xu, X.; Wang, L. Low expression of adenomatous polyposis coli 2 correlates with aggressive features and poor prognosis in colorectal cancer. Bioengineered 2020, 11, 1027–1033. [Google Scholar] [CrossRef]
- Geng, Y.; Zheng, X.; Hu, W.; Wang, Q.; Xu, Y.; He, W.; Wu, C.; Zhu, D.; Wu, C.; Jiang, J. Hsa_circ_0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin Sci (Lond) 2019, 133, 1197–1213. [Google Scholar] [CrossRef]
- Geng, Y.; Zheng, X.; Hu, W.; Wang, Q.; Xu, Y.; He, W.; Wu, C.; Zhu, D.; Jiang, J. Hsa_circ_0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin Sci (Lond) 2019, 133, 1197–1213. [Google Scholar] [CrossRef]
- Marcel, V.; Perrier, S.; Aoubala, M.; Ageorges, S.; Groves, M.J.; Diot, A.; Fernandes, K.; Tauro, S.; Bourdon, J.C. Delta160p53 is a novel N-terminal p53 isoform encoded by Delta133p53 transcript. FEBS Lett 2010, 584, 4463–4468. [Google Scholar] [CrossRef]
- Yin, Y.; Stephen, C.W.; Luciani, M.G.; Fåhraeus, R. p53 stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nature Cell Biology 2002, 4, 462–467. [Google Scholar] [CrossRef]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb Perspect Med 2016, 6, a026104. [Google Scholar] [CrossRef]
- Andreyev, H.J.; Norman, A.R.; Cunningham, D.; Oates, J.; Dix, B.R.; Iacopetta, B.J.; Young, J.; Walsh, T.; Ward, R.; Hawkins, N.; et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer 2001, 85, 692–696. [Google Scholar] [CrossRef]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010, 11, 753–762. [Google Scholar] [CrossRef]
- Biscaglia, G.; Latiano, A.; Castellana, S.; Fontana, R.; Gentile, A.; Latiano, T.; Corritore, G.; Panza, A.; Nardella, M.; Martino, G.; et al. Germline Alterations in Patients With IBD-associated Colorectal Cancer. Inflamm Bowel Dis 2022, 28, 447–454. [Google Scholar] [CrossRef]
- Xie, Z.; Ke, Y.; Chen, J.; Li, Z.; Wang, C.; Chen, Y.; Ding, H.; Cheng, L. Prevalence and Spectrum of Predisposition Genes With Germline Mutations Among Chinese Patients With Bowel Cancer. Front Genet 2021, 12, 755629. [Google Scholar] [CrossRef]
- Perne, C.; Peters, S.; Cartolano, M.; Horpaopan, S.; Grimm, C.; Altmuller, J.; Sommer, A.K.; Hillmer, A.M.; Thiele, H.; Odenthal, M.; et al. Variant profiling of colorectal adenomas from three patients of two families with MSH3-related adenomatous polyposis. PLoS One 2021, 16, e0259185. [Google Scholar] [CrossRef]
- Takebe, N.; Harris, P.J.; Warren, R.Q.; Ivy, S.P. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol 2011, 8, 97–106. [Google Scholar] [CrossRef]
- Medema, J.P. Cancer stem cells: the challenges ahead. Nat Cell Biol 2013, 15, 338–344. [Google Scholar] [CrossRef]
- Pattabiraman, D.R.; Weinberg, R.A. Tackling the cancer stem cells—what challenges do they pose? Nat Rev Drug Discov 2014, 13, 497–512. [Google Scholar] [CrossRef]
- Bienz, M.; Clevers, H. Linking colorectal cancer to Wnt signaling. Cell 2000, 103, 311–320. [Google Scholar] [CrossRef]
- Segditsas, S.; Tomlinson, I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene 2006, 25, 7531–7537. [Google Scholar] [CrossRef]
- Hernandez-Aya, L.F.; Gonzalez-Angulo, A.M. Targeting the phosphatidylinositol 3-kinase signaling pathway in breast cancer. Oncologist 2011, 16, 404–414. [Google Scholar] [CrossRef]
- Messaritakis, I.; Stogiannitsi, M.; Koulouridi, A.; Sfakianaki, M.; Voutsina, A.; Sotiriou, A.; Athanasakis, E.; Xynos, E.; Mavroudis, D.; Tzardi, M.; et al. Evaluation of the detection of Toll-like receptors (TLRs) in cancer development and progression in patients with colorectal cancer. PLoS One 2018, 13, e0197327. [Google Scholar] [CrossRef]
- Deng, J.; Zhou, S.; Wang, Z.; Huang, G.; Zeng, J.; Li, X. Comparison of Prognosis and Lymph Node Metastasis in T1-Stage Colonic and Rectal Carcinoma: A Retrospective Study. Int J Gen Med 2022, 15, 3651–3662. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, K.; Cho, S.H.; Chun, S.M.; Tak, E.; Hong, Y.S.; Kim, J.E.; Kim, T.W. Longitudinal change of genetic variations in cetuximab-treated metastatic colorectal cancer. Cancer Genet 2021, 258-259, 27–36. [Google Scholar] [CrossRef]
- Ohnami, S.; Maruyama, K.; Chen, K.; Takahashi, Y.; Hatakeyama, K.; Ohshima, K.; Shimoda, Y.; Sakai, A.; Kamada, F.; Nakatani, S.; et al. BMP4 and PHLDA1 are plausible drug-targetable candidate genes for KRAS G12A-, G12D-, and G12V-driven colorectal cancer. Mol Cell Biochem 2021, 476, 3469–3482. [Google Scholar] [CrossRef] [PubMed]
- Borakati, A.; Froghi, F.; Shetye, A.; Fusai, G.K.; Davidson, B.R.; Mirnezami, R. Assessing the Impact of Primary Tumour Location on Survival After Resection of Colorectal Liver Metastases: A Propensity Weighted Retrospective Cohort Study. World J Surg 2022, 46, 1734–1755. [Google Scholar] [CrossRef] [PubMed]
- Il, J.H.; Young, C.Y.; Jun, B.M.; Ho, B.S.; Byung, B.S.; Jun, J.D.; Yong, K.S.; Soo, L.M.; Sik, C.M.; Ho, K.C. Expression of RhoA in Colorectal Cancers and Its Clinicopathological Significance. J Korean Soc Coloproctol 2008, 24, 460–466. [Google Scholar] [CrossRef]
- Bonadona, V.; Bonaïti, B.; Olschwang, S.; Grandjouan, S.; Huiart, L.; Longy, M.; Guimbaud, R.; Buecher, B.; Bignon, Y.J.; Caron, O.; et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. Jama 2011, 305, 2304–2310. [Google Scholar] [CrossRef]
- Ramsoekh, D.; Wagner, A.; van Leerdam, M.E.; Dooijes, D.; Tops, C.M.; Steyerberg, E.W.; Kuipers, E.J. Cancer risk in MLH1, MSH2 and MSH6 mutation carriers; different risk profiles may influence clinical management. Hered Cancer Clin Pract 2009, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Skoulidis, F.; Heymach, J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer 2019, 19, 495–509. [Google Scholar] [CrossRef]
- Sfakianaki, M.; Papadaki, C.; Tzardi, M.; Trypaki, M.; Alam, S.; Lagoudaki, E.D.; Messaritakis, I.; Zoras, O.; Mavroudis, D.; Georgoulias, V.; et al. Loss of LKB1 Protein Expression Correlates with Increased Risk of Recurrence and Death in Patients with Resected, Stage II or III Colon Cancer. Cancer Res Treat 2019, 51, 1518–1526. [Google Scholar] [CrossRef]
- He, K.; Wang, Y.; Zhong, Y.; Pan, X.; Si, L.; Lu, J. KRAS Codon 12 Mutation is Associated with More Aggressive Invasiveness in Synchronous Metastatic Colorectal Cancer (mCRC): Retrospective Research. Onco Targets Ther 2020, 13, 12601–12613. [Google Scholar] [CrossRef]
- Scott, A.; Goffredo, P.; Ginader, T.; Hrabe, J.; Gribovskaja-Rupp, I.; Kapadia, M.R.; Weigel, R.J.; Hassan, I. The Impact of KRAS Mutation on the Presentation and Prognosis of Non-Metastatic Colon Cancer: an Analysis from the National Cancer Database. J Gastrointest Surg 2020, 24, 1402–1410. [Google Scholar] [CrossRef]
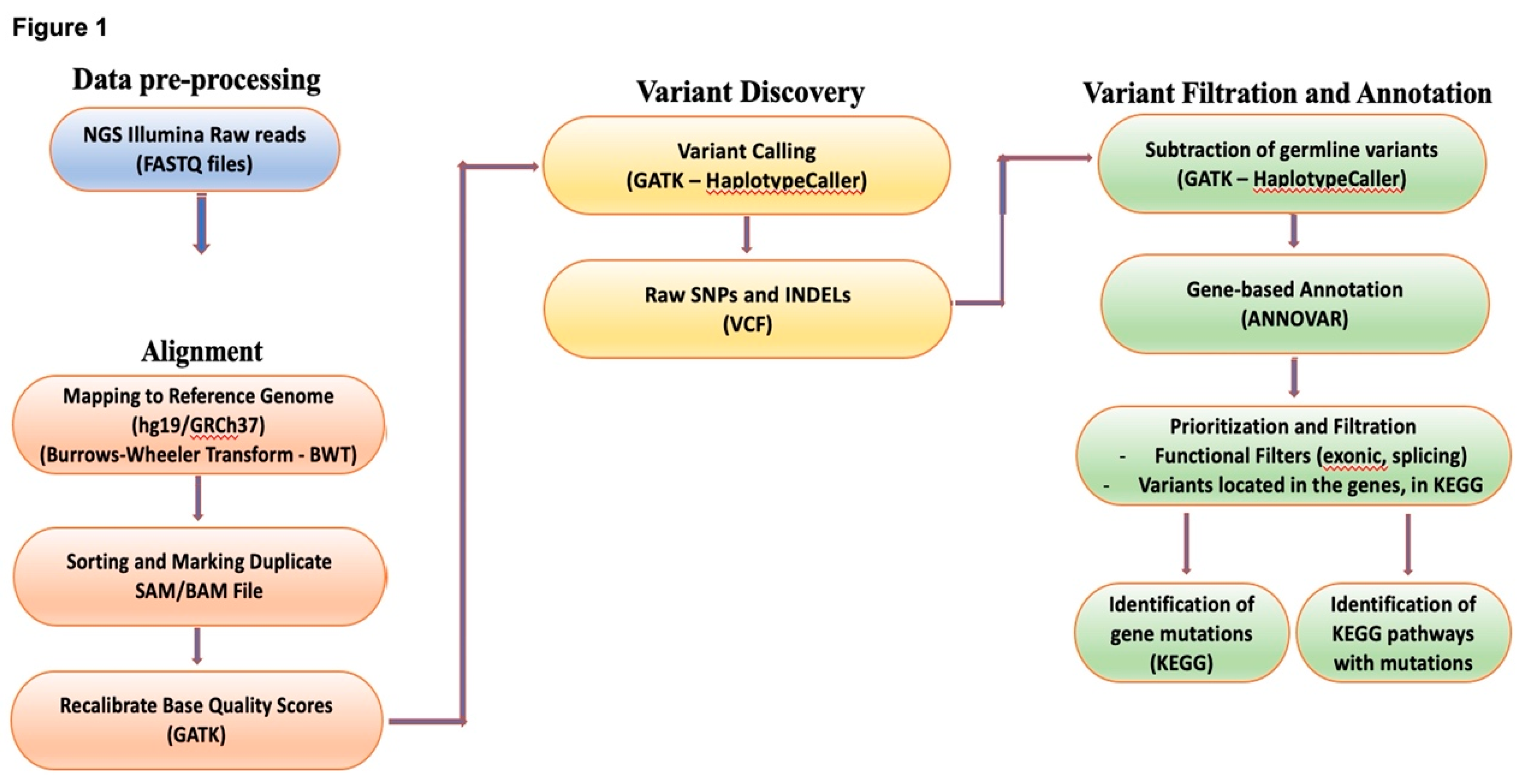
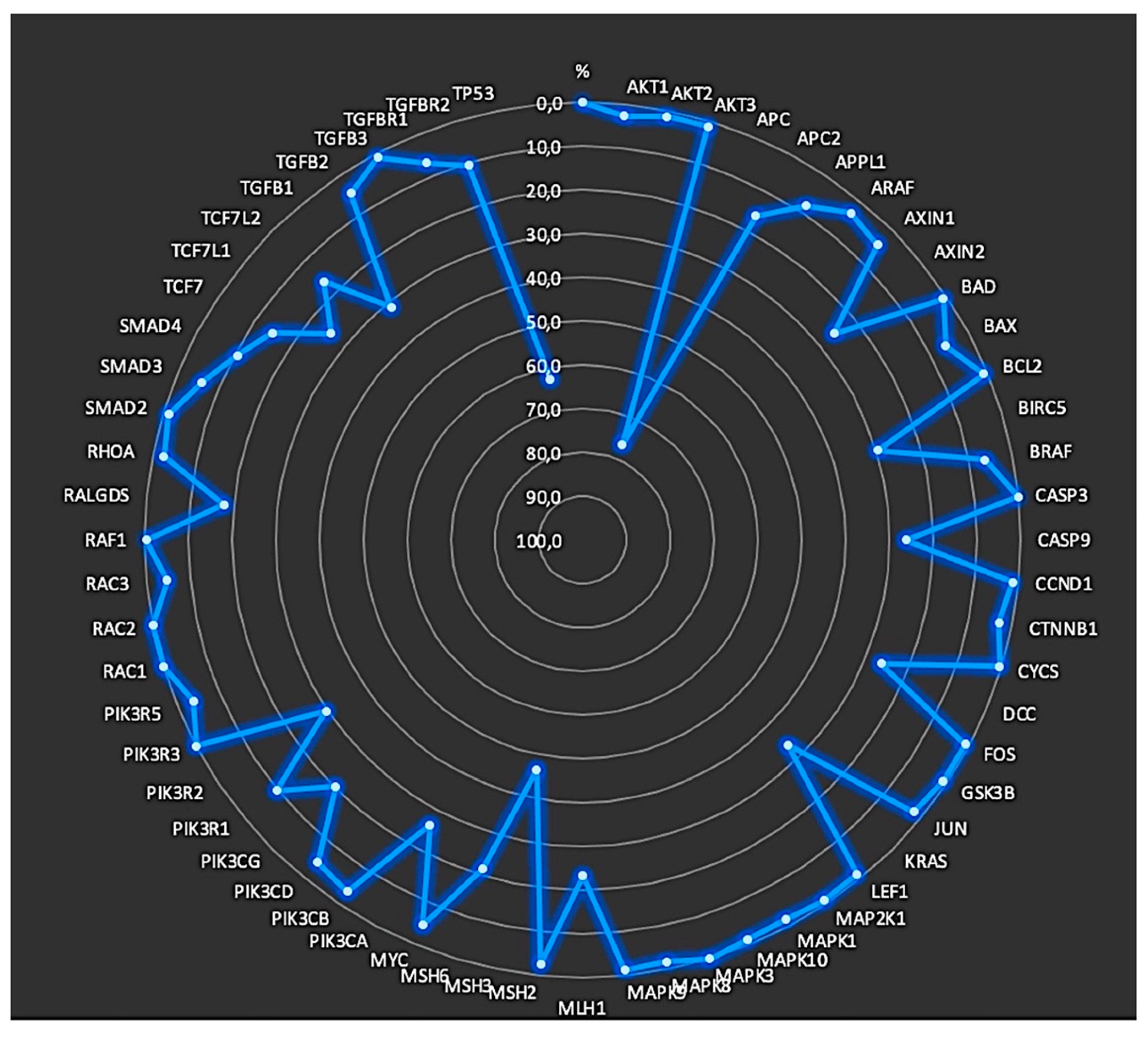
| Gene | Chromosome | Location based on GRCh37.p13 assembly | Source[https://www.ensembl.org/index.html] | |
|---|---|---|---|---|
| Start | End | |||
| AKT1 | 14 | 105235686 | 105262085 | Ensembl:ENSG00000142208 |
| AKT2 | 19 | 40736224 | 40791252 | Ensembl:ENSG00000105221 |
| AKT3 | 1 | 243651535 | 244014381 | Ensembl:ENSG00000117020 |
| APC | 5 | 112043195 | 112181936 | Ensembl:ENSG00000134982 |
| APC2 | 19 | 1450120 | 1473243 | Ensembl:ENSG00000115266 |
| APPL1 | 3 | 57261757 | 57307499 | Ensembl:ENSG00000157500 |
| ARAF | X | 47420604 | 47431307 | Ensembl:ENSG00000078061 |
| AXIN1 | 16 | 337440 | 402723 | Ensembl:ENSG00000103126 |
| AXIN2 | 17 | 63524681 | 63557766 | Ensembl:ENSG00000168646 |
| BAD | 11 | 64037300 | 64052176 | Ensembl:ENSG00000002330 |
| BAX | 19 | 49458132 | 49465055 | Ensembl:ENSG00000087088 |
| BCL2 | 18 | 60790579 | 60987002 | Ensembl:ENSG00000171791 |
| BIRC5 | 17 | 76210334 | 76221716 | Ensembl:ENSG00000089685 |
| BRAF | 7 | 140413128 | 140624729 | Ensembl:ENSG00000157764 |
| CASP3 | 4 | 185548850 | 185570601 | Ensembl:ENSG00000164305 |
| CASP9 | 1 | 15817896 | 15851285 | Ensembl:ENSG00000132906 |
| CCND1 | 11 | 69455924 | 69469242 | Ensembl:ENSG00000110092 |
| CTNNB1 | 3 | 41240996 | 41281934 | Ensembl:ENSG00000168036 |
| CYCS | 7 | 25158275 | 25164879 | Ensembl:ENSG00000172115 |
| DCC | 18 | 49866567 | 51062269 | Ensembl:ENSG00000187323 |
| FOS | 14 | 75745531 | 75748933 | Ensembl:ENSG00000170345 |
| GSK3B | 3 | 119540168 | 119813294 | Ensembl:ENSG00000082701 |
| JUN | 1 | 59246463 | 59249719 | Ensembl:ENSG00000177606 |
| KRAS | 12 | 25358180 | 25403863 | Ensembl:ENSG00000133703 |
| LEF1 | 4 | 108968704 | 109090088 | Ensembl:ENSG00000138795 |
| MAP2K1 | 15 | 66679250 | 66783882 | Ensembl:ENSG00000169032 |
| MAPK1 | 22 | 22113946 | 22221970 | Ensembl:ENSG00000100030 |
| MAPK10 | 4 | 86931558 | 87374348 | Ensembl:ENSG00000109339 |
| MAPK3 | 16 | 30125426 | 30134541 | Ensembl:ENSG00000102882 |
| MAPK8 | 10 | 49514720 | 49647403 | Ensembl:ENSG00000107643 |
| MAPK9 | 5 | 179660143 | 179719083 | Ensembl:ENSG00000050748 |
| MLH1 | 3 | 3703500 | 37092337 | Ensembl:ENSG00000076242 |
| MSH2 | 2 | 47630206 | 47710367 | Ensembl:ENSG00000095002 |
| MSH3 | 5 | 79950471 | 80172634 | Ensembl:ENSG00000113318 |
| MSH6 | 2 | 48010284 | 48034092 | Ensembl:ENSG00000116062 |
| MYC | 8 | 128747680 | 128755197 | Ensembl:ENSG00000136997 |
| PIK3CA | 3 | 178866145 | 178957881 | Ensembl:ENSG00000121879 |
| PIK3CB | 3 | 138371540 | 138553770 | Ensembl:ENSG00000051382 |
| PIK3CD | 1 | 9711789 | 9789172 | Ensembl:ENSG00000171608 |
| PIK3CG | 7 | 106505727 | 106549425 | Ensembl:ENSG00000105851 |
| PIK3R1 | 5 | 67511584 | 67597649 | Ensembl:ENSG00000145675 |
| PIK3R2 | 19 | 18263973 | 18281342 | Ensembl:ENSG00000105647 |
| PIK3R3 | 1 | 46505812 | 46640573 | Ensembl:ENSG00000117461 |
| PIK3R5 | 17 | 8782233 | 8869024 | Ensembl:ENSG00000141506 |
| RAC1 | 7 | 6414158 | 6443598 | Ensembl:ENSG00000136238 |
| RAC2 | 22 | 37621310 | 37640309 | Ensembl:ENSG00000128340 |
| RAC3 | 17 | 79989554 | 79992080 | Ensembl:ENSG00000169750 |
| RAF1 | 3 | 12625100 | 12705616 | Ensembl:ENSG00000132155 |
| RALGDS | 9 | 135973109 | 136024597 | Ensembl:ENSG00000160271 |
| RHOA | 3 | 49396578 | 49449409 | Ensembl:ENSG00000067560 |
| SMAD2 | 18 | 45335328 | 45457243 | Ensembl:ENSG00000175387 |
| SMAD3 | 15 | 67357940 | 67487507 | Ensembl:ENSG00000166949 |
| SMAD4 | 18 | 48556583 | 48611412 | Ensembl:ENSG00000141646 |
| TCF7 | 5 | 133450372 | 133483901 | Ensembl:ENSG00000081059 |
| TCF7L1 | 2 | 85360515 | 85537510 | Ensembl:ENSG00000152284 |
| TCF7L2 | 10 | 114710006 | 114927437 | Ensembl:ENSG00000148737 |
| TGFB1 | 19 | 41836228 | 41859827 | Ensembl:ENSG00000105329 |
| TGFB2 | 1 | 218518678 | 218617961 | Ensembl:ENSG00000092969 |
| TGFB3 | 14 | 76424440 | 76449354 | Ensembl:ENSG00000119699 |
| TGFBR1 | 9 | 101867395 | 101916474 | Ensembl:ENSG00000106799 |
| TGFBR2 | 3 | 30647994 | 30735634 | Ensembl:ENSG00000163513 |
| TP53 | 17 | 7571739 | 7590808 | Ensembl:ENSG00000141510 |
| Pathway | KEGG CRC genes | References [https://www.genome.jp] |
|---|---|---|
| Cell cycle | CCND1, GSK3B, MYC, SMAD2, SMAD3, SMAD4, TGFB1, TGFB2, TGFB3, TP53 | https://www.genome.jp/pathway/hsa04110 |
| p53 signaling pathway | BAX, BCL2, CASP3, CASP9, CCND1, CYCS, TP53 | https://www.genome.jp/pathway/hsa04115 |
| Apoptosis | AKT1, AKT2, AKT3, BAD, BAX, BCL2, BIRC5, CASP3, CASP9, CYCS, FOS, JUN, KRAS, MAP2K1, MAPK1, MAPK10, MAPK3, MAPK8, MAPK9, PIK3CA, PIK3CB, PIK3CD, PIK3R1, PIK3R2, PIK3R3, RAF1, TP53, DCC, APPL1 | https://www.genome.jp/pathway/hsa04210 |
| mTOR signaling pathway | BRAF, GSK3B, KRAS, MAP2K1, MAPK1, PIK3CA, PIK3CB, PIK3CD, PIK3R1, PIK3R2, PIK3R3, RAF1, RHOA | https://www.genome.jp/pathway/hsa04150 |
| PI3K-Akt signaling pathway | AKT1, AKT2, AKT3, BAD, BCL2, CASP9, GSK3B, KRAS, MAP2K1, MAPK1, MAPK3, MYC, PIK3CA, PIK3CB, PIK3CD, PIK3CG, PIK3R1, PIK3R2, PIK3R3, PIK3R5, RAC1, RAF1, TP53 | https://www.genome.jp/pathway/hsa04151 |
| Wnt signaling pathway | APC, APC2, AXIN1, AXIN2, CCND1,CTNNB1, GSK3B, JUN, LEF1, MAPK10, MAPK8, MAPK9, MYC, RAC1, RAC2, RAC3, RHOA, SMAD3, SMAD4, TCF7, TCF7L1, TCF7L2, TP53 | https://www.genome.jp/pathway/hsa04310 |
| TGF-beta signaling pathway | MAPK1, MAPK3, MYC, RHOA, SMAD2, SMAD3, SMAD4, TGFB1, TGFB2, TGFB3, TGFBR1, TGFBR2, | https://www.genome.jp/pathway/hsa04350 |
| MAPK signaling pathway | AKT1, AKT2, AKT3, ARAF, BRAF, CASP3, FOS, JUN, KRAS, LEF1, MAP2K1, MAPK1, MAPK10, MAPK3, MAPK8, MAPK9, MYC, RAC1, RAC2, RAC3, RAF1, TGFB1, TGFB2, TGFB3, TGFBR1, TGFBR2, TP53 | https://www.genome.jp/pathway/hsa04010 |
| ErbB signaling pathway | AKT1, AKT2, AKT3, ARAF, BAD, BRAF, GSK3B, JUN, KRAS, MAP2K1, MAPK1, MAPK10, MAPK3, MAPK8, MAPK9, MYC, PIK3CA, PIK3CB, PIK3CD, PIK3R1, PIK3R2, PIK3R3, RAF | https://www.genome.jp/pathway/hsa04012 |
| MSI pathway | APC, AXIN1, AXIN2, BAD, BAX, BCL2, GSK3B, MLH1, MSH2, MSH3, MSH6, TGFBR2 | https://www.genome.jp/dbget-bin/www_bget?path:map05210 |
| Ras signaling pathway | AKT1, AKT2, AKT3, BAD, KRAS, MAP2K1, MAPK1, MAPK10, MAPK3, MAPK8, MAPK9, PIK3CA, PIK3CB, PIK3CD, PIK3R1, PIK3R2, PIK3R3, RAC1, RAC2, RAC3, RAF1, RALGDS, RHOA | https://www.genome.jp/pathway/hsa04014 |
| Characteristics | Number of patients (Ν=237) | % |
|---|---|---|
| Median age (range) | 64 (18–84) | |
| <70 | 159 | 67.1 |
| ≥70 | 78 | 32.9 |
| Gender | ||
| Males | 151 | 63.7 |
| Females | 86 | 36.3 |
| Tumor location | ||
| Cecum | 38 | 16.0 |
| Ascending | 42 | 17.7 |
| Transverse | 22 | 9.3 |
| Descending | 24 | 10.1 |
| Sigmoid | 111 | 46.8 |
| Sidedness | ||
| Left | 158 | 66.7 |
| Right | 79 | 33.3 |
| Performance status | ||
| 0-1 | 236 | 99.6 |
| >2 | 1 | .4 |
| Regimen | ||
| Folfox | 105 | 44.3 |
| Capox | 132 | 55.7 |
| Treatment duration | ||
| 3 months | 116 | 48.9 |
| 6 months | 121 | 51.1 |
| Gene | Mutant patients |
|---|---|
| AKT1 | 6 |
| AKT2 | 3 |
| AKT3 | 3 |
| APC | 181 |
| APC2 | 38 |
| APPL1 | 19 |
| ARAF | 8 |
| AXIN1 | 11 |
| AXIN2 | 61 |
| BAD | 2 |
| BAX | 14 |
| BCL2 | 2 |
| BIRC5 | 70 |
| BRAF | 15 |
| CASP3 | 0 |
| CASP9 | 62 |
| CCND1 | 3 |
| CTNNB1 | 7 |
| CYCS | 1 |
| DCC | 62 |
| FOS | 2 |
| GSK3B | 2 |
| JUN | 5 |
| KRAS | 80 |
| LEF1 | 3 |
| MAP2K1 | 2 |
| MAPK1 | 4 |
| MAPK10 | 3 |
| MAPK3 | 0 |
| MAPK8 | 4 |
| MAPK9 | 3 |
| MLH1 | 55 |
| MSH2 | 6 |
| MSH3 | 110 |
| MSH6 | 51 |
| MYC | 11 |
| PIK3CA | 62 |
| PIK3CB | 8 |
| PIK3CD | 11 |
| PIK3CG | 48 |
| PIK3R1 | 23 |
| PIK3R2 | 70 |
| PIK3R3 | 0 |
| PIK3R5 | 9 |
| RAC1 | 0 |
| RAC2 | 0 |
| RAC3 | 11 |
| RAF1 | 1 |
| RALGDS | 42 |
| RHOA | 6 |
| SMAD2 | 3 |
| SMAD3 | 14 |
| SMAD4 | 25 |
| TCF7 | 35 |
| TCF7L1 | 61 |
| TCF7L2 | 39 |
| TGFB1 | 74 |
| TGFB2 | 11 |
| TGFB3 | 2 |
| TGFBR1 | 16 |
| TGFBR2 | 25 |
| TP53 | 148 |
| Characteristics | Gene | No % (p value) |
|---|---|---|
| Gender | ||
| Male vs Female | JUN | 43.4% vs 25.0% (0.05) |
| MAPK3 | 57.5% vs 31.1% (0.05) | |
| Sideness | ||
| Left vs Right | MLH1 | 80% vs 20% (0.007) |
| MSH6 | 84.3% vs 15.7% (0.001) | |
| TCF7L1 | 77% vs 33% (0.019) | |
| Location | ||
| Colon vs Sigmoid | DCC | 90.3% vs 9.7% (0.04) |
| KRAS | 88.8% vs 11.2% (0.048) | |
| TGFBR2 | 96% vs 4% (0.043) | |
| Age | ||
| <70 vs ≥70 | TGFBR1 | 57.5% vs 33% (0.012) |
| 51-70 vs ≥70 vs <50 | BAD | 56.1% vs 30.7% vs 12.7% (<0.001) |
| 51-70 vs ≥70 vs <50 | RAC | 56.1% vs 30.7% vs 12.7% (0.016) |
| 51-70 vs ≥70 vs <50 | AKT, AKT2, AKT3, APC, APPL1, AXIN1, AXIN2, BIRC5, DCC, GSK3B, KRAS, MAPK1, MAPK8, MAPK9, MAPK10, MLH1, MSH6, PIK3CA, PIK3R1, PIK3R2, PIK3R3, PIK3R5, RAF1, RALGD5, SMAD2, SMAD3, TCF7L2, TGFB1, TGFB2 | 56.1% vs 30.7% vs 12.7% (0.037) |
| Relapse post Adj Chemotherapy | ||
| Yes vs No | ARAF, MAPK10 | 71.7% vs 20.5% (0.027) |
| CASP3 | 75.5% vs 22.6% (0.044) | |
| TCF7 | 75% vs 21.2% (0.003) | |
| TGFΒ3 | 74.5% vs 22.2% (0.037) |
| Cell cycle | Apoptosis | PI3K-Akt | Wnt | MAPK | ErbB | MSI | RAS | |
|---|---|---|---|---|---|---|---|---|
| Transverse Colon | X | X | X | X | ||||
| TaqI Homozygous | X | X | X | X | X | X | X | |
| ApaI Homozygous | X | X | X | X | ||||
| FokI Homozygous | X | X | X | |||||
| BsmI Homozygous | X | X | X | |||||
| TLR9 -T1237C Homozygous | X | X | X | |||||
| TLR9–T1486C Homozygous | X | X | X |
| Univariate | Multivariate | |||||||
|---|---|---|---|---|---|---|---|---|
| DFS | OS | DFS | OS | |||||
| Feature | HR (95%CI) | p-value | HR (95%CI) | p-value | HR (95%CI) | p-value | HR (95%CI) | p-value |
| Sidedness (Right vs Left) | 1.9 (1.2–3.2) | 0.012 | 2.1 (1.0–4.0) | 0.043 | 2.1 (1.1-4.0) | 0.019 | 2.2 (1.0-4.5) | 0.043 |
| AKT1 (Mutant vs Wild type) | 2.9 (1.1-8.2) | 0.039 | ||||||
| APC2 (Mutant vs Wild type) | 2.5 (1.4-4.5) | 0.002 | ||||||
| ARAF (Mutant vs Wild type) | 3.8 (1.2-12.1) | 0.027 | 5.0 (2.0-12.7) | 0.001 | 3.9 (1.2-13.1) | 0.027 | ||
| BAD (Mutant vs Wild type) | 8.6 (1.1-64.4) | 0.036 | ||||||
| MAPK10 (Mutant vs Wild type) | 43.0 (9.1-203.7) | <0.001 | 15.1 (4.6-50.3) | <0.001 | 49.0 (9.8-244.1) | <0.001 | ||
| MSH6 (Wild type vs Mutant) | 2.3 (1.0-5.1) | 0.041 | ||||||
| RAC3 (Mutant vs Wild type) | 4.7 (1.8-11.9) | 0.001 | 3.5 (1.1-10.7) | 0.029 | ||||
| RHOA (Mutant vs Wild type) | 4.8 (1.5-15.6) | 0.009 | 9.5 (1.9-47.7) | 0.006 | ||||
| TGFB2 (Mutant vs Wild type) | 2.6 (1.1-6.7) | 0.040 | ||||||
| TGFB3 (Mutant vs Wild type) | 9.0 (2.1-37.8) | 0.003 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





