Submitted:
14 August 2023
Posted:
16 August 2023
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Results
2.1. Sequence Analysis of DREB Proteins
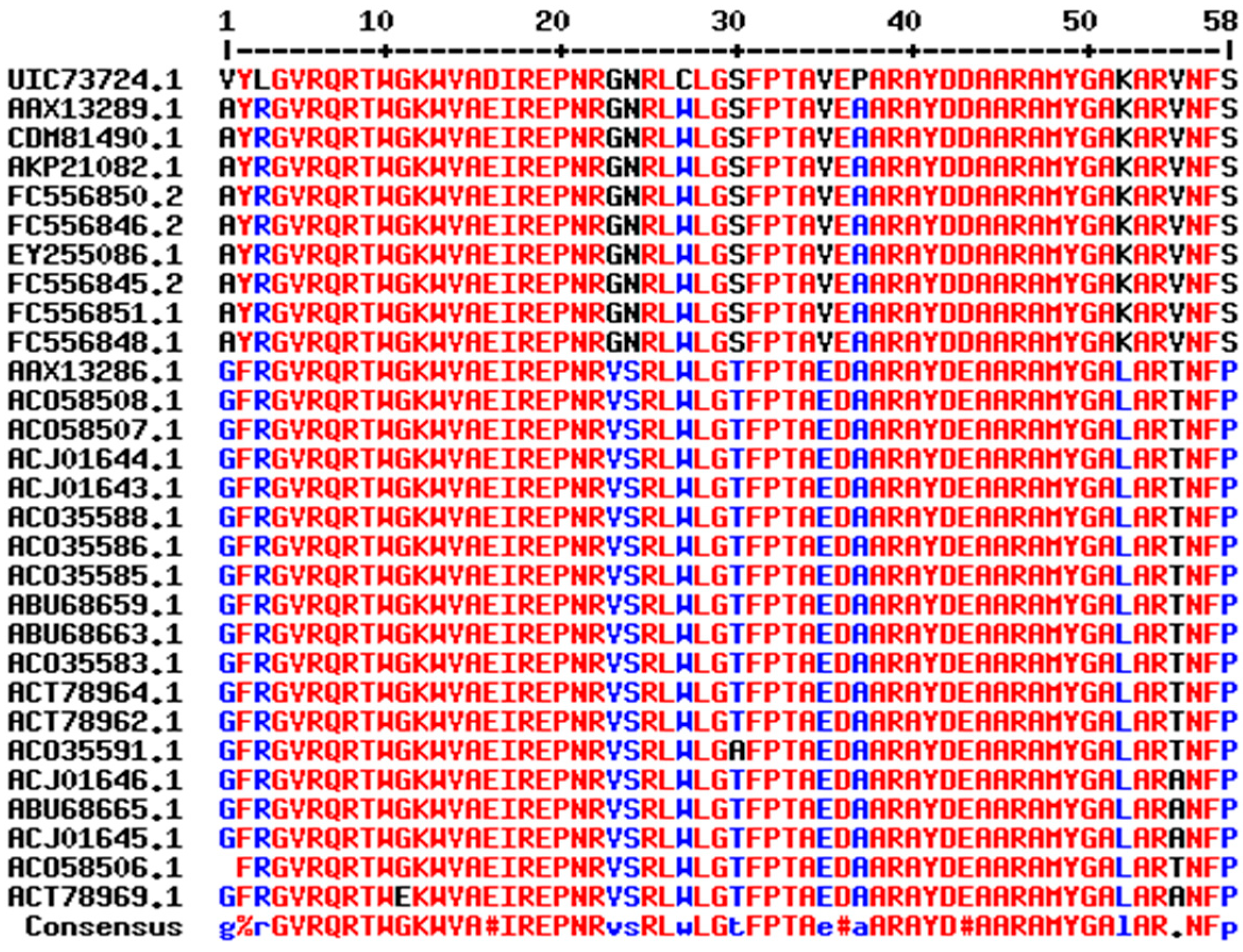
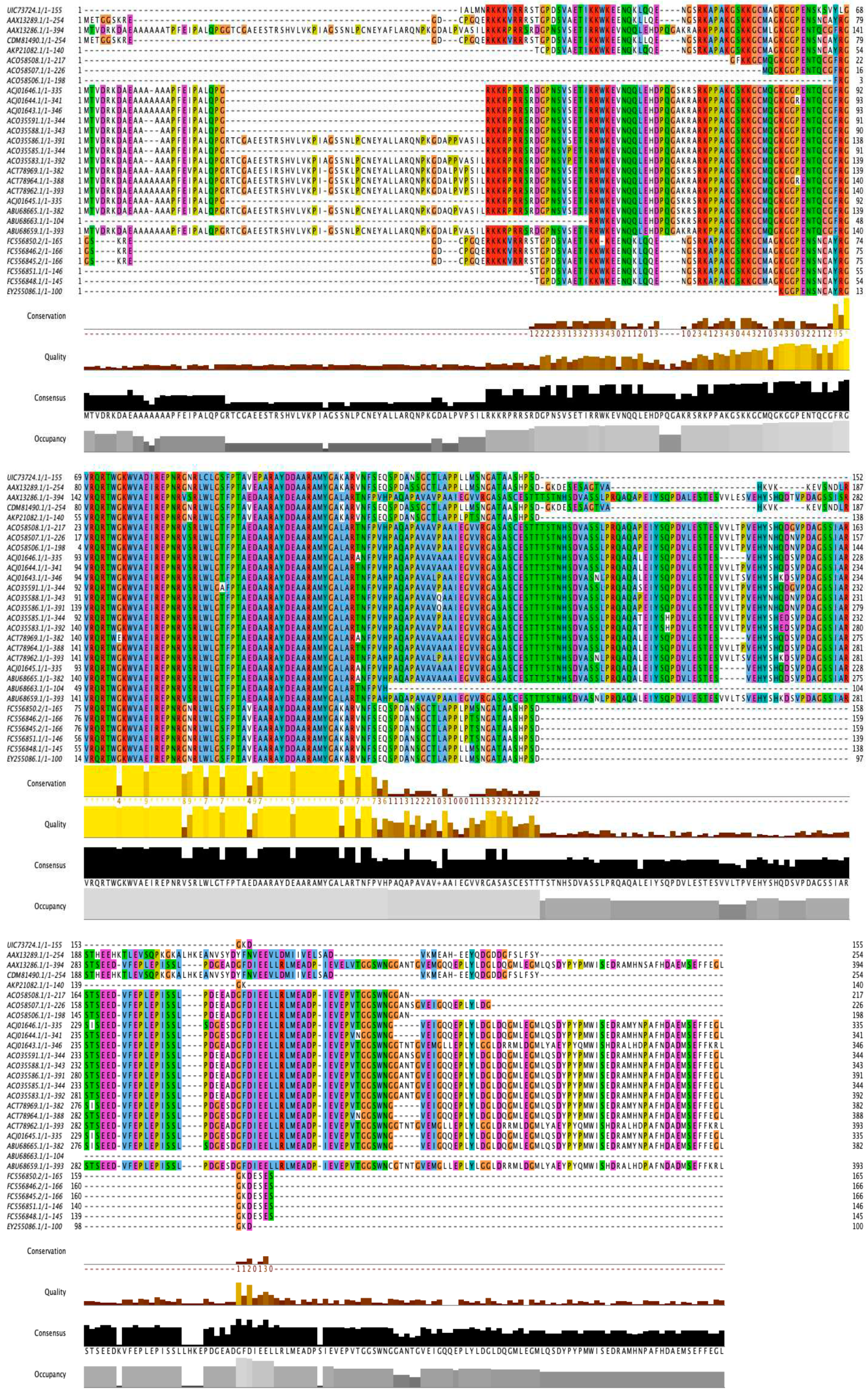
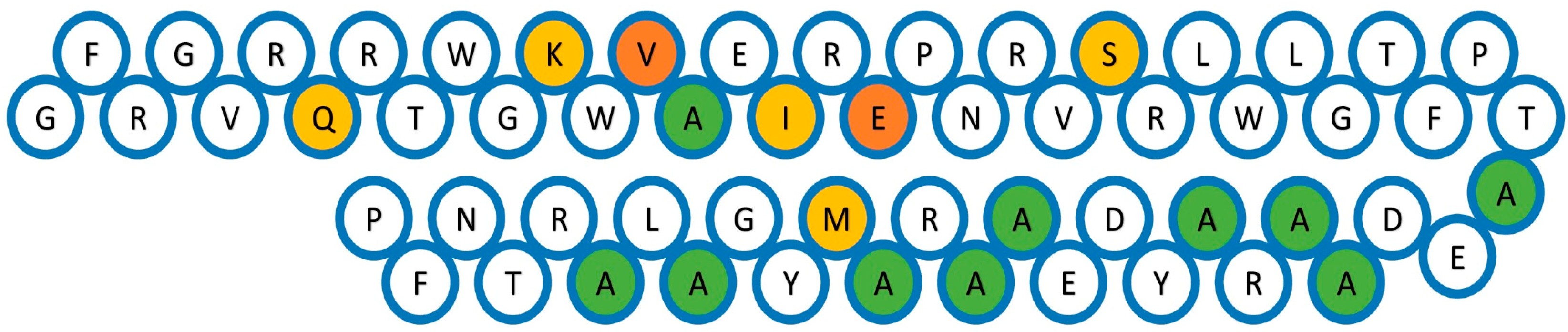
2.2. Conserved Motif Analysis
| Motif Number | Amino Acid Sequence |
|---|---|
| 1 | FRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGA |
| 2 | IEGVVRGASASCESTTTSTNHSDVASSLPRQAQALEIYSQPDVLESTESV |
| 3 | STSEEDVFEPLEPISSLPDGEADGFDIEELLRLMEADPIEVEPVTGGSWN |

| Amino acid | Motif 1 | Motif 2 | Motif 3 | Amino acid | Motif 1 | Motif 2 | Motif 3 | |
|---|---|---|---|---|---|---|---|---|
| Ala (A) | 18.0% | 10.0% | 4.0% | Lys (K) | 2.0% | 0.0% | 0.0% | |
| Arg (R) | 16.0% | 4.0% | 2.0% | Met (M) | 2.0% | 0.0% | 2.0% | |
| Asn (N) | 2.0% | 2.0% | 2.0% | Phe (F) | 4.0% | 0.0% | 4.0% | |
| Asp (D) | 4.0% | 4.0% | 10.0% | Pro (P) | 4.0% | 4.0% | 10.0% | |
| Cys (C) | 0.0% | 2.0% | 0.0% | Ser (S) | 2.0% | 20.0% | 10.0% | |
| Gln (Q) | 2.0% | 6.0% | 0.0% | Thr (T) | 6.0% | 10.0% | 4.0% | |
| Glu (E) | 8.0% | 10.0% | 20.0% | Trp (W) | 6.0% | 0.0% | 2.0% | |
| Gly (G) | 8.0% | 4.0% | 8.0% | Tyr (Y) | 4.0% | 2.0% | 0.0% | |
| His (H) | 0.0% | 2.0% | 0.0% | Val (V) | 6.0% | 10.0% | 6.0% | |
| Ile (I) | 2.0% | 4.0% | 6.0% | Pyl (O) | 0.0% | 0.0% | 0.0% | |
| Leu (L) | 4.0% | 6.0% | 10.0% | Sec (U) | 0.0% | 0.0% | 0.0% |
2.3. Phylogenetic Analysis of AP2 Domains
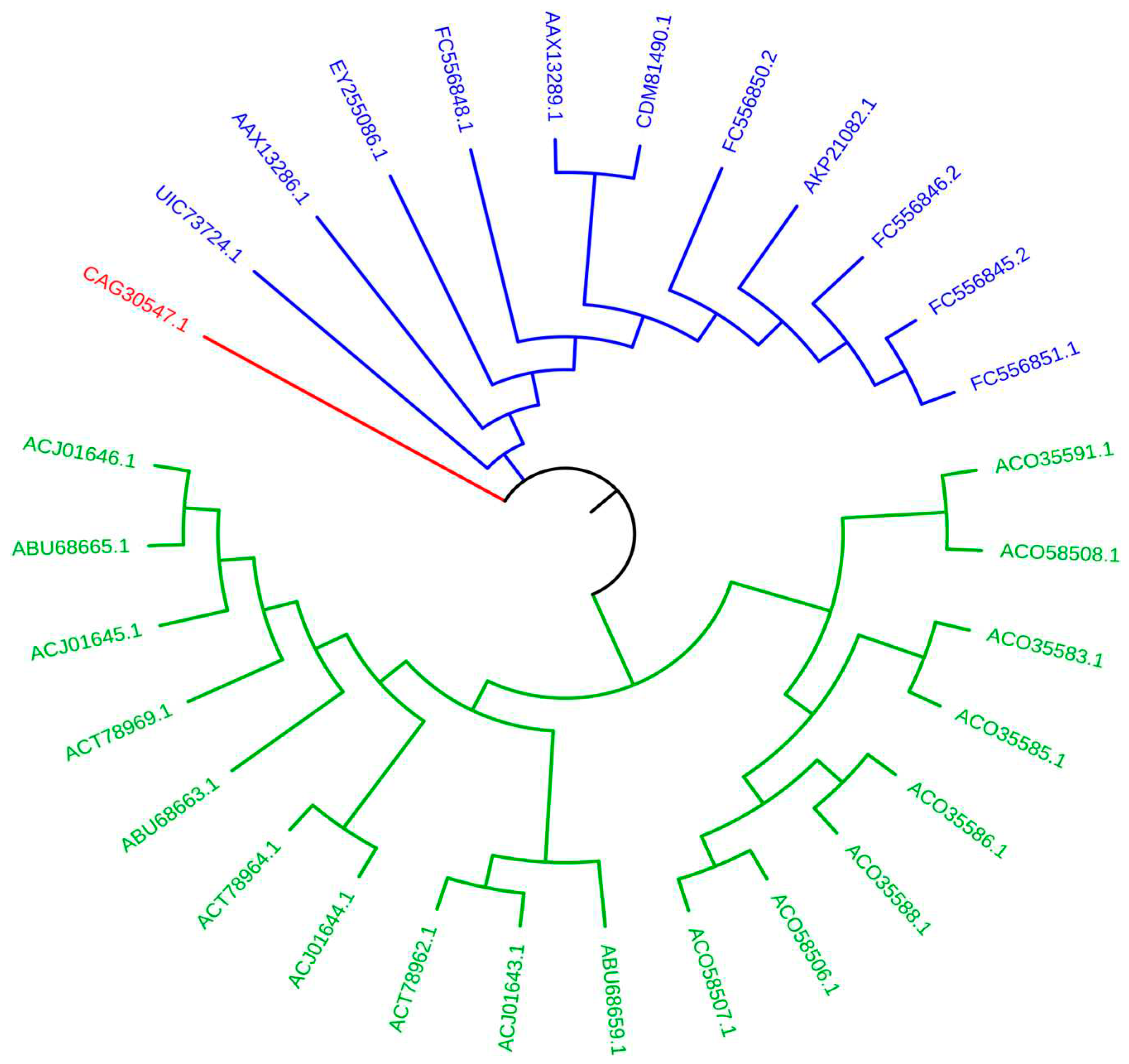
2.4. Structural Analysis of AP2 Domain
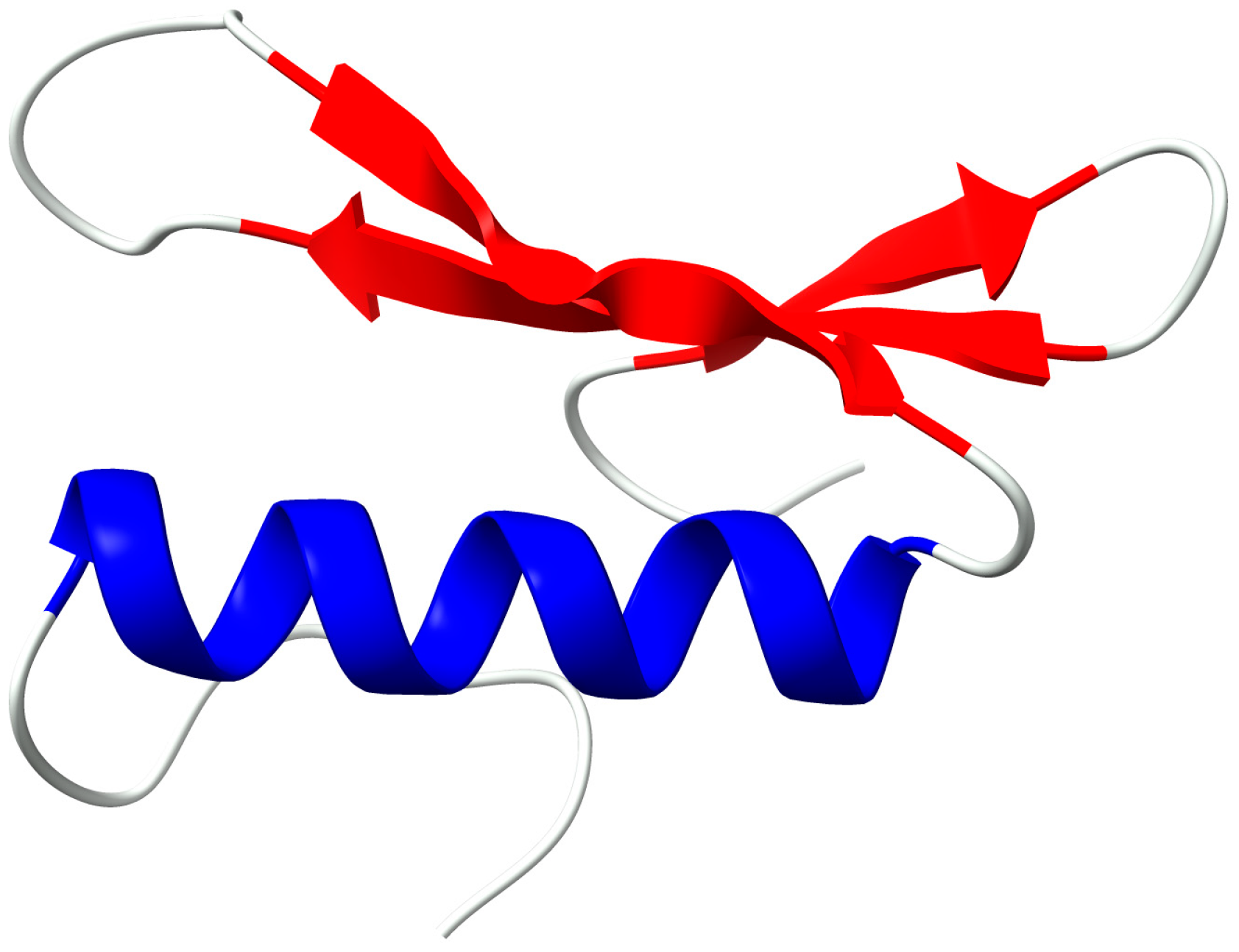
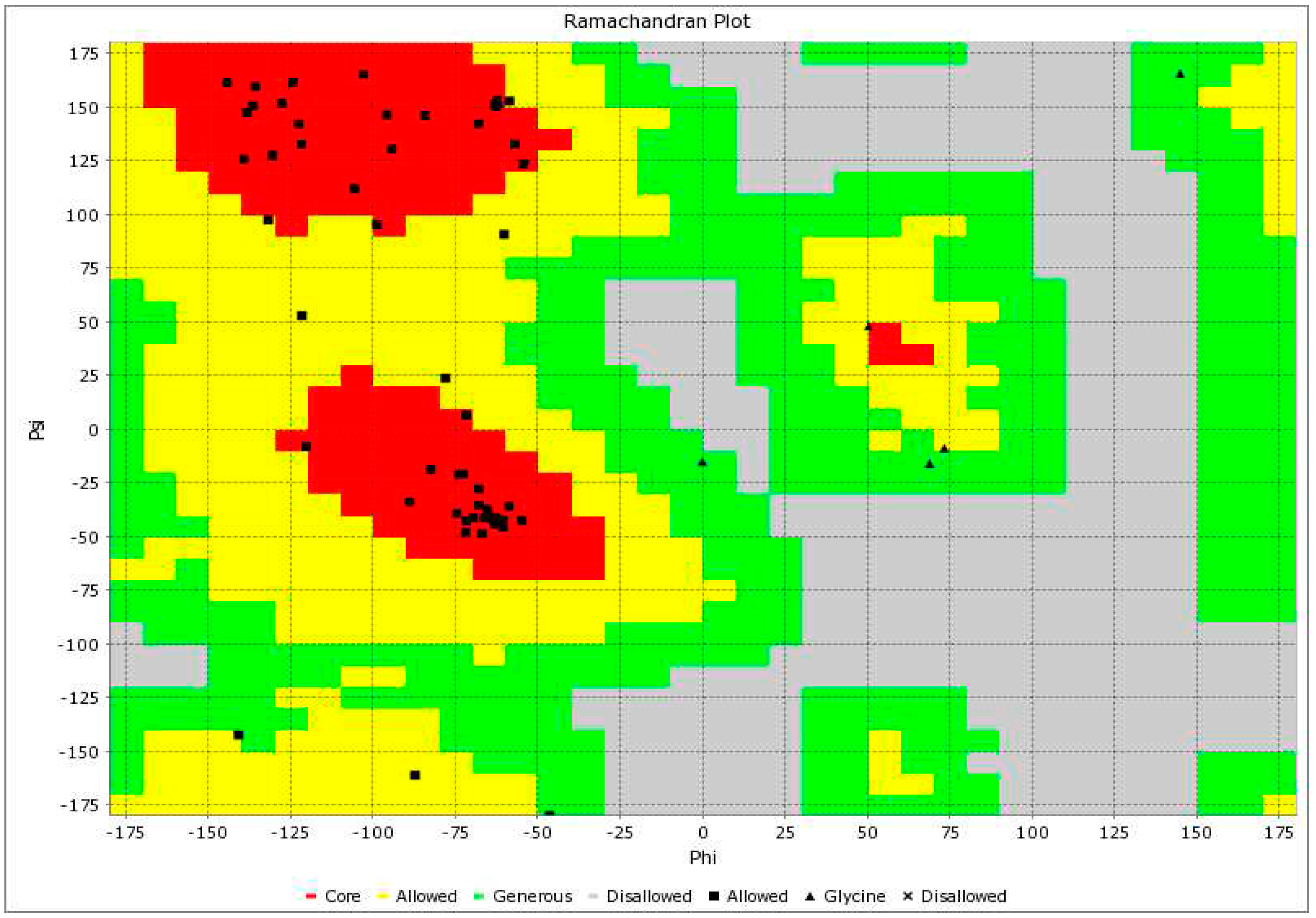
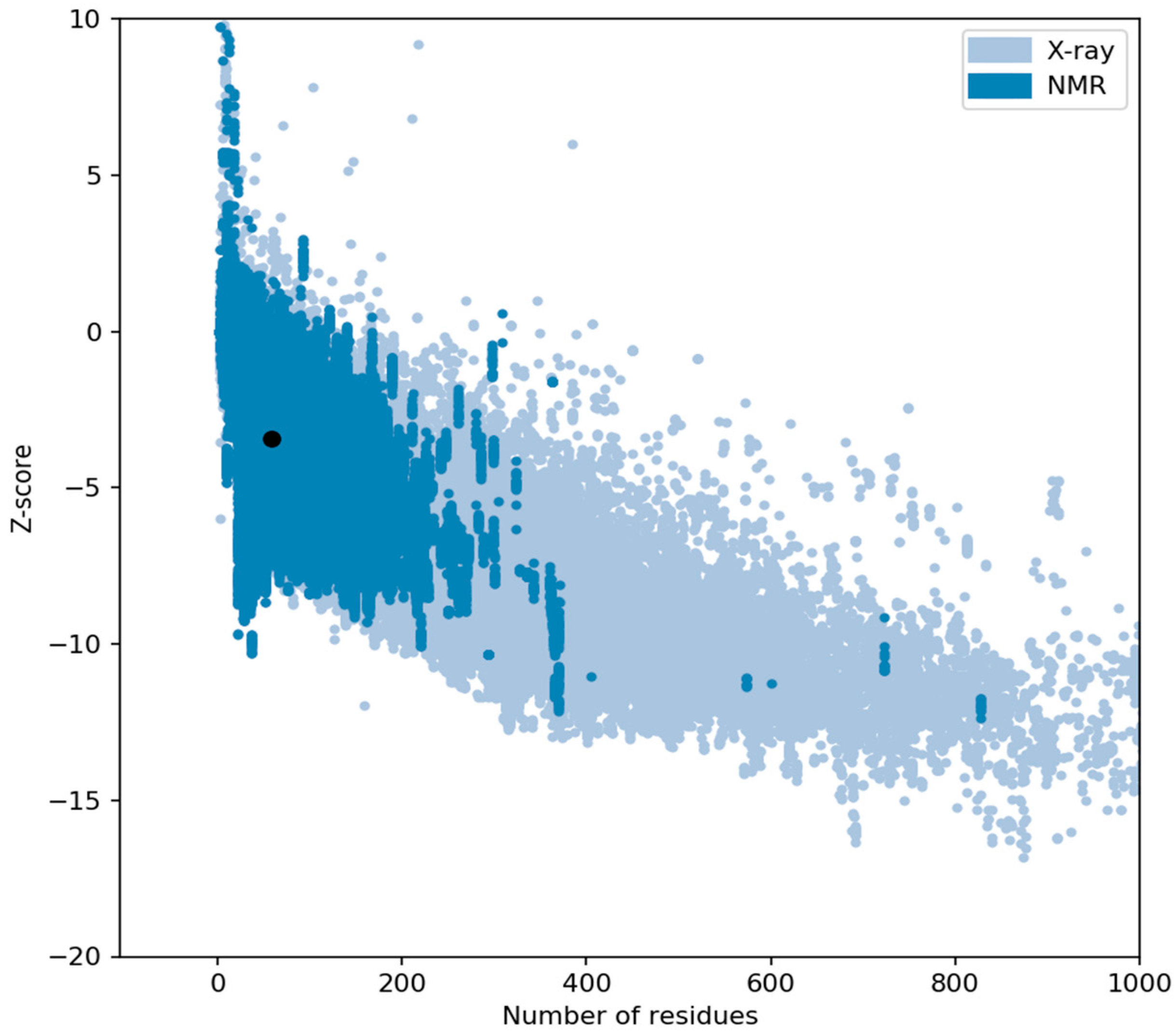
3. Discussion
4. Materials and Methods
4.1. Database Search of DREB and AP2s in Wheat
4.2. Multiple Sequence Alignment and Biochemical Characteristics
| Accession number of TS sequence | AP2 Domain Sequence | Country | Species |
|---|---|---|---|
| EY255086.1 | AYRGVRQRTWGKWVAEIREPNRGNRLWLGSFPTAVEAARAYDDAARAMYGAKARVNFSEQSPDA | Iran |
Triticum aestivum cv. Shahi1 |
| ABU68663.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPVH | Italy | Triticum turgidum subsp. Durum1 |
| AKP21082.1 | AYRGVRQRTWGKWVAEIREPNRGNRLWLGSFPTAVEAARAYDDAARAMYGAKARVNFSEQSPDA |
Afghanistan |
Triticum aestivum cv. Kalak Afghani |
| FC556848.1 | AYRGVRQRTWGKWVAEIREPNRGNRLWLGSFPTAVEAARAYDDAARAMYGAKARVNFSEQSPDA | Iran |
Triticum aestivum cv. Omid |
| FC556851.1 | AYRGVRQRTWGKWVAEIREPNRGNRLWLGSFPTAVEAARAYDDAARAMYGAKARVNFSEQSPDA | Iran |
Triticum aestivum cv. Zarrin |
| UIC73724.1 | VYLGVRQRTWGKWVADIREPNRGNRLCLGSFPTAVEPARAYDDAARAMYGAKARVNFSEQSPDA | Azerbaijan |
Triticum aestivum cv. Azerbaijan |
| FC556850.2 | AYRGVRQRTWGKWVAEIREPNRGNRLWLGSFPTAVEAARAYDDAARAMYGAKARVNFSEQSPDA |
Iran |
Triticum aestivum cv. Shahi2 |
| FC556846.2 | AYRGVRQRTWGKWVAEIREPNRGNRLWLGSFPTAVEAARAYDDAARAMYGAKARVNFSEQSPDA | Iran |
Triticum aestivum cv. Bayat |
| FC556845.2 | AYRGVRQRTWGKWVAEIREPNRGNRLWLGSFPTAVEAARAYDDAARAMYGAKARVNFSEQSPDA | Iran |
Triticum aestivum cv. Alvand |
| ACO58506.1 | FRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPVHPAQA | Italy |
Aegilops speltoides var. speltoides |
| ACO58508.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPVHPAQA | Italy |
Aegilops speltoides var. speltoides |
| ACO58507.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPVHPAQA | Italy |
Aegilops speltoides var. speltoides |
| AAX13289.1 | AYRGVRQRTWGKWVAEIREPNRGNRLWLGSFPTAVEAARAYDDAARAMYGAKARVNFSEQSPDA | China |
Triticum aestivum cv. China |
| CDM81490.1 | AYRGVRQRTWGKWVAEIREPNRGNRLWLGSFPTAVEAARAYDDAARAMYGAKARVNFSEQSPDA | France |
Triticum aestivum cv. France |
| ACJ01646.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARANFPVHPAQA | Italy | Triticum turgidum subsp. durum2 |
| ACJ01645.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARANFPVHPAQA | Italy | Triticum turgidum subsp. durum3 |
| ACJ01644.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPVHPAQA | Italy |
Triticum aestivum cv. Italy1 |
| ACO35588.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPVHPAQA | Italy |
Aegilops speltoides var. speltoides |
| ACO35591.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGAFPTAEDAARAYDEAARAMYGALARTNFPVHPAQA | Italy | Aegilops speltoides var. speltoides |
| ACO35585.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPVHPAQA | Italy |
Aegilops speltoides var. speltoides |
| ACJ01643.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPAHPAQA | Italy |
Triticum aestivum cv. Italy2 |
| ACT78969.1 | GFRGVRQRTWEKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARANFPVHPAQA | Italy |
Triticum aestivum cv. Italy3 |
| ABU68665.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARANFPVHPAQA | Italy |
Triticum aestivum cv. Italy4 |
| ACT78964.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPVHPAQA | Italy |
Triticum aestivum cv. Italy5 |
| ACO35586.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPVHPAQA | Italy | Aegilops speltoides var. speltoides |
| ACO35583.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPVHPAQA | Italy | Aegilops speltoides var. speltoides |
| ACT78962.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPAHPAQA | Italy | Triticum turgidum subsp. Durum4 |
| ABU68659.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPAHPAQA | Italy | Triticum turgidum subsp. Durum5 |
| AAX13286.1 | GFRGVRQRTWGKWVAEIREPNRVSRLWLGTFPTAEDAARAYDEAARAMYGALARTNFPVHPAQA | China |
Triticum aestivum cv. China |
4.3. Composition and Motif Analysis
4.4. Phylogenetic Analysis
4.5. Structure and Modeling of AP2 Domain
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Okamuro, J.K., Caster, B., Villarroel, R., Van Montagu, M.; Jofuku, K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 94, 7076–7081. https://doi.org/10.1073/pnas.94.13.7076. [CrossRef]
- Ma, Q., Xia, Z., Cai, Z., Li, L., Cheng, Y., Liu, J.; Nian, H. (2019). GmWRKY16 Enhances Drought and Salt Tolerance Through an ABA-Mediated Pathway in Arabidopsis thaliana. Frontiers in plant science, 9, 1979. https://doi.org/10.3389/fpls.2018.01979. [CrossRef]
- Zhu, P., Chen, Y., Zhang, J., Wu, F., Wang, X., Pan, T., Wei, Q., Hao, Y., Chen, X., Jiang, C.; Ji, K. (2021). Identification, classification, and characterization of AP2/ERF superfamily genes in Masson pine (Pinus massoniana Lamb.). Scientific reports, 11, 5441. https://doi.org/10.1038/s41598-021-84855-w. [CrossRef]
- Nakano, T., Suzuki, K., Fujimura, T.; Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant physiology, 140, 411–432. https://doi.org/10.1104/pp.105.073783. [CrossRef]
- Sakuma, Y., Liu, Q., Dubouzet, J.G., Abe, H., Shinozaki, K.; Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemical and biophysical research communications, 290, 998–1009. https://doi.org/10.1006/bbrc.2001.6299. [CrossRef]
- Feng, K., Hou, X.L., Xing, G.M., Liu, J.X., Duan, A.Q., Xu, Z.S., Li, M.Y., Zhuang, J.; Xiong, A.S. (2020). Advances in AP2/ERF super-family transcription factors in plant. Critical reviews in biotechnology, 40, 750–776. https://doi.org/10.1080/07388551.2020.1768509. [CrossRef]
- Aukerman, M.J.; Sakai, H. (2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. The Plant cell, 15, 2730–2741. https://doi.org/10.1105/tpc.016238. [CrossRef]
- Jofuku, K.D., Omidyar, P.K., Gee, Z.; Okamuro, J.K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proceedings of the National Academy of Sciences of the United States of America, 102, 3117–3122. https://doi.org/10.1073/pnas.0409893102. [CrossRef]
- Taketa, S., Amano, S., Tsujino, Y., Sato, T., Saisho, D., Kakeda, K., Nomura, M., Suzuki, T., Matsumoto, T., Sato, K., Kanamori, H., Kawasaki, S.; Takeda, K. (2008). Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proceedings of the National Academy of Sciences of the United States of America, 105, 4062–4067. https://doi.org/10.1073/pnas.0711034105. [CrossRef]
- Licausi, F., Ohme-Takagi, M.; Perata, P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. The New phytologist, 199, 639–649. https://doi.org/10.1111/nph.12291. [CrossRef]
- Liu, S., Wang, X., Wang, H., Xin, H., Yang, X., Yan, J., Li, J., Tran, L.S., Shinozaki, K., Yamaguchi-Shinozaki, K.; Qin, F. (2013). Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS genetics, 9, e1003790. https://doi.org/10.1371/journal.pgen.1003790. [CrossRef]
- Rashid, M., Guangyuan, H., Guangxiao, Y., Hussain, J.; Xu, Y. (2012). AP2/ERF Transcription Factor in Rice: Genome-Wide Canvas and Syntenic Relationships between Monocots and Eudicots. Evolutionary bioinformatics online, 8, 321–355. https://doi.org/10.4137/EBO.S9369. [CrossRef]
- Gil-Humanes, J., Pistón, F., Martín, A.; Barro, F. (2009). Comparative genomic analysis and expression of the APETALA2-like genes from barley, wheat, and barley-wheat amphiploids. BMC plant biology, 9, 66. https://doi.org/10.1186/1471-2229-9-66. [CrossRef]
- Greenwood, J.R., Finnegan, E.J., Watanabe, N., Trevaskis, B.; Swain, S.M. (2017). New alleles of the wheat domestication gene Qreveal multiple roles in growth and reproductive development. Development (Cambridge, England), 144, 1959–1965. https://doi.org/10.1242/dev.146407. [CrossRef]
- Simons, K.J., Fellers, J.P., Trick, H.N., Zhang, Z., Tai, Y.S., Gill, B.S.; Faris, J.D. (2006). Molecular characterization of the major wheat domestication gene Q. Genetics, 172, 547–555. https://doi.org/10.1534/genetics.105.044727. [CrossRef]
- Yu, Y., Yu, M., Zhang, S., Song, T., Zhang, M., Zhou, H., Wang, Y., Xiang, J.; Zhang, X. (2022). Transcriptomic Identification of Wheat AP2/ERF Transcription Factors and Functional Characterization of TaERF-6-3A in Response to Drought and Salinity Stresses. International journal of molecular sciences, 23, 3272. https://doi.org/10.3390/ijms23063272. [CrossRef]
- R. DePauw, and L. O’Brien, Wheat Breeding: Exploiting and Fixing Genetic Variation by Selection and Evaluation. In Encyclopedia of Food Grains, 2nd ed.; Colin Wrigley, Harold Corke, Koushik Seetharaman, Jon Faubion; Publisher: Academic Press, Cambridge, Massachusetts, US, 2016; Volume 4, pp. 279-286, doi.org/10.1016/B978-0-12-394437-5.00216-3.
- Yang, J.; Zhang, Y. (2015). I-TASSER server: new development for protein structure and function predictions. Nucleic acids research, 43(W1), W174–W181. https://doi.org/10.1093/nar/gkv342. [CrossRef]
- Zheng, W., Zhang, C., Li, Y., Pearce, R., Bell, E.W.; Zhang, Y. (2021). Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell reports methods, 1, 100014. https://doi.org/10.1016/j.crmeth.2021.100014. [CrossRef]
- Zhou, X., Zheng, W., Li, Y., Pearce, R., Zhang, C., Bell, E.W., Zhang, G.; Zhang, Y. (2022). I-TASSER-MTD: a deep-learning-based platform for multi-domain protein structure and function prediction. Nature protocols, 17, 2326–2353. https://doi.org/10.1038/s41596-022-00728-0. [CrossRef]
- Sippl M. J. (1993). Recognition of errors in three-dimensional structures of proteins. Proteins, 17, 355–362. https://doi.org/10.1002/prot.340170404. [CrossRef]
- Wiederstein, M.; Sippl, M.J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic acids research, 35(Web Server issue), W407–W410. https://doi.org/10.1093/nar/gkm290. [CrossRef]
- Hassan, S., Berk, K.; Aronsson, H. (2022). Evolution and identification of DREB transcription factors in the wheat genome: modeling, docking and simulation of DREB proteins associated with salt stress. Journal of biomolecular structure & dynamics, 40, 7191–7204. https://doi.org/10.1080/07391102.2021.1894980. [CrossRef]
- Karimi, J., Mohsenzadeh, S., Mohabatkar, H. (2009). Isolation and characterization of partial DREB gene from four Iranian Triticum aestivum cultivars. World Journal of Agricultural Sciences, 5, 561-566.
- Mohsenzadeh, S., Karimi, J., Mohabatkar, H. (2011). Study of dehydration-responsive element binding-factor gene in some Iranian bread wheat cultivars. Iranian Journal of Plant Biology. 3: 29-40.
- Mohsenzadeh, S., Sherafat, A., Karimi, J. (2021). Alignment and phylogenetic tree analysis of some Iranian and Chinese wheat cultivars based on DREB partial gene sequences. The Fourth International Biotechnology Congress of Iran. Paper code (biotech12-05990388).
- Sharma, M.K., Kumar, R., Solanke, A.U., Sharma, R., Tyagi, A.K.; Sharma, A.K. (2010). Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Molecular genetics and genomics : MGG, 284, 455–475. https://doi.org/10.1007/s00438-010-0580-1. [CrossRef]
- Filiz, E.; Tombuloğlu, H. (2014). In silico analysis of DREB transcription factor genes and proteins in grasses. Applied biochemistry and biotechnology, 174, 1272–1285. https://doi.org/10.1007/s12010-014-1093-x. [CrossRef]
- Trevino, S.R., Scholtz, J.M.; Pace, C.N. (2007). Amino acid contribution to protein solubility: Asp, Glu, and Ser contribute more favorably than the other hydrophilic amino acids in RNase Sa. Journal of molecular biology, 366, 449–460. https://doi.org/10.1016/j.jmb.2006.10.026. [CrossRef]
- Liu, Y., Zhao, T.J., Liu, J.M., Liu, W.Q., Liu, Q., Yan, Y.B.; Zhou, H.M. (2006). The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS letters, 580, 1303–1308. https://doi.org/10.1016/j.febslet.2006.01.048. [CrossRef]
- Jofuku, K.D., den Boer, B.G., Van Montagu, M.; Okamuro, J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. The Plant cell, 6, 1211–1225. https://doi.org/10.1105/tpc.6.9.1211. [CrossRef]
- Büttner, M.; Singh, K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proceedings of the National Academy of Sciences of the United States of America, 94, 5961–5966. https://doi.org/10.1073/pnas.94.11.5961. [CrossRef]
- "Multiple sequence alignment with hierarchical clustering" F. CORPET, 1988, Nucl. Acids Res., 16 (22), 10881-10890.
- Waterhouse, A.M., Procter, J.B., Martin, D.M.A, Clamp, M. and Barton, G.J. (2009) "Jalview Version 2 - a multiple sequence alignment editor and analysis workbench", Bioinformatics 25 (9) 1189-1191 doi: 10.1093/bioinformatics/btp033. [CrossRef]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A.; Protein Identification and Analysis Tools on the Expasy Server; (In) John M. Walker (ed): The Proteomics Protocols Handbook, Humana Press (2005), pp. 571-607.
- Tamura, K., Stecher, G.; Kumar, S. (2021). MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular biology and evolution, 38, 3022–3027. https://doi.org/10.1093/molbev/msab120. [CrossRef]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542.
- Eslami-Farouji, A.,Khodayari, H., Assadi, M. et al. (2021). Phylogeny and biogeography of the genus Hesperis (Brassicaceae, tribe, Hesperideae) inferred from nuclear ribosomal DNA sequence data. Plant Syst Evol. 307, 17-35.
- Leigh Willard, Anuj Ranjan, Haiyan Zhang, Hassan Monzavi, Robert F. Boyko, Brian D. Sykes, and David S. Wishart "VADAR: a web server for quantitative evaluation of protein structure quality" Nucleic Acids Res. 2003 July 1; 31 (13): 3316–3319.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





