Introduction
Alfalfa, a polyploid legume forage, is an important crop with strong resistance to stress, and both of its yield and quality are excellent [
1]. In addition, alfalfa owns biological nitrogen-fixing capacity, which can improve the soil structure and the soil fertility, making it a crop for sustainable agriculture[
2]. Previouslly, researchers has made several remarkable achievements in alfalfa gene expression atlas[
3] and genome assembly[
4,
5,
6]. Studies about alfalfa lncRNAs were rarely reported. Although Chao et al.[
7] and Wan et al.[
8] predicted alfalfa lncRNAs using bioinfomatic methods based on long read or short read data, respectively, these lncRNAs are merely associated with leaf development and drought response. Systematic identification and characterization of alfalfa lncRNA have not been reported.
Long non-coding RNAs (lncRNAs) are a class of RNAs that are greater than 200 nt in length and have no ro low proteincoding potential[
9]. Several studies reported that they expressed differently among different tissues[
10,
11,
12], and they present low conservation on nucleic acid sequences among species[
13]. LncRNA plays important roles in important life processes such as gene expression regulation, chromatin remodeling, and epigenetics[
14,
15,
16]. lncRNA could act as a competitive endogenous RNA to regulate gene function at post-translation level. For example, transcription of an antisense lncRNA suppresses PHO84 mRNA transcription[
17]. lncRNA can also acts as a miRNA sponge or target mimics, binding a large number of miRNAs based on complementary base pairing, thereby positively regulating the expression of target genes[
18]. Additionally, lncRNAs encode short peptides with biological functions have been found in animals and plants, such as Toddler[
19], Myoregulin (MLN)[
20], DWORF[
21] and ENOD40[
22]. And all these lncRNAs encode small peptides with 11 to 58 amino acid[
19,
20,
21,
22]. The discovery of these small peptides indicates that there is the possibility of small open read frames (sORFs) encoding short peptides in ncRNA, and the short peptides encoded by them may play some important roles in the growth and development of organisms. Therefore, it is important to systematically study the distribution and coding potential of sORFs in lncRNA. At the current stage, the research on the coding region in lncRNA is still in its infancy, many sORFs translated from lncRNA need to be discovered.
Full-length transcriptome sequencing is a newly developed nucleic acid sequencing technology. Compared to the second-generation sequencing technology, the full-length transcriptome sequencing obtained longer reads and achievable, and the full-length transcript can be directly obtained, and the obtained transcript sequence was more accurate. In order to identify the lncRNA of alfalfa in genome-wide, we analyzed the third-generation full-length transcriptome sequencing data generated by PacBio sequencing technology, and predicted a large number of lncRNA using our prediction pipeline. We also detected the expression of some sequence conserved lncRNAs and the interaction between lncRNA and miRNA. In this research, we obtained the omics information of alfalfa lncRNAs for the first time, and these data sets about lncRNA and miRNA provided abundant sources to develop the research field of alfalfa lncRNA function.
Results
PacBio sequencing and data analysis
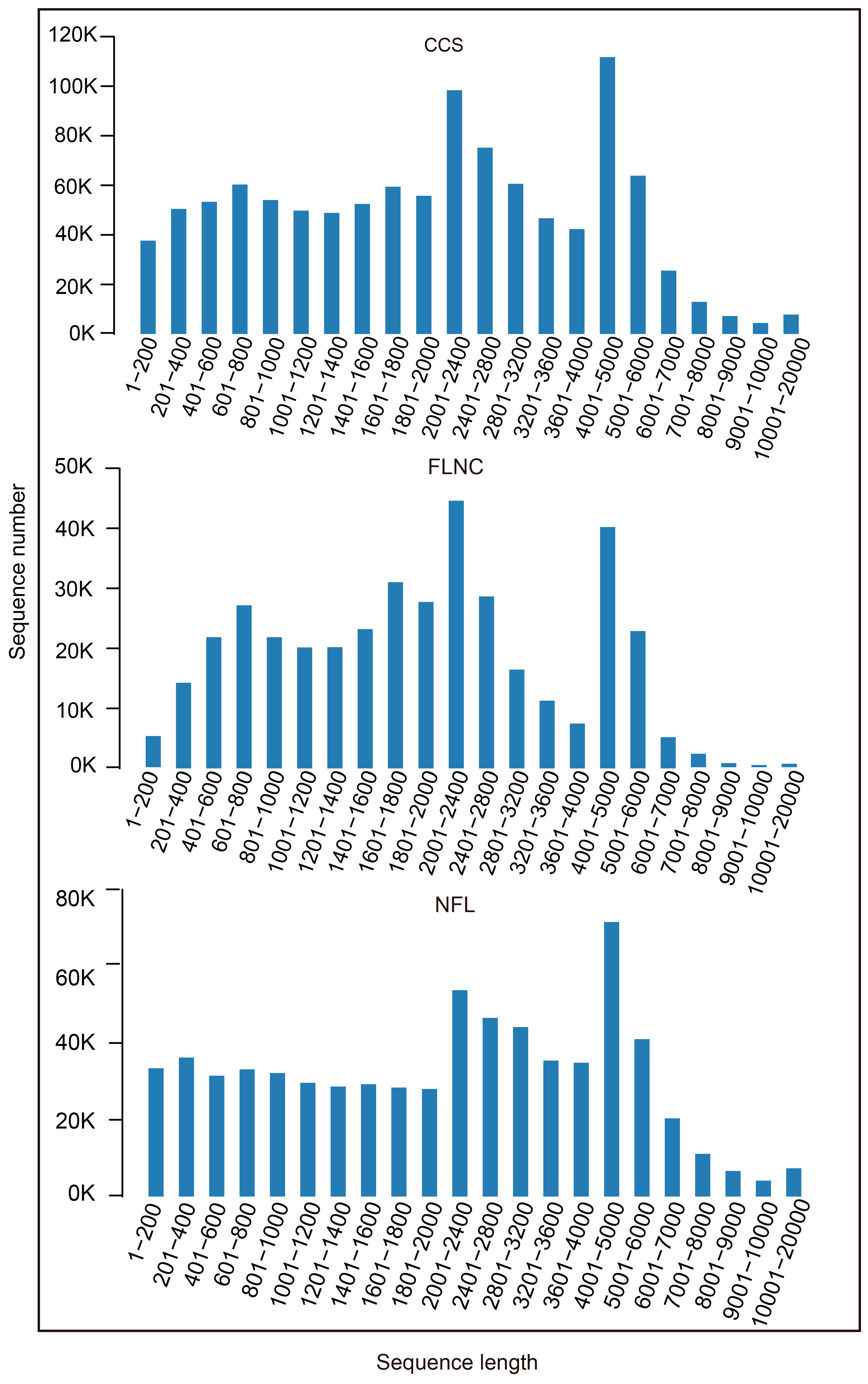
After cutting off low quality reads, we got 1,089,299 circular consensus sequences (CCS) by PACBIO sequencing. 5′ prime reads, 3′ prime reads and polyA reads were counted and the result was listed in
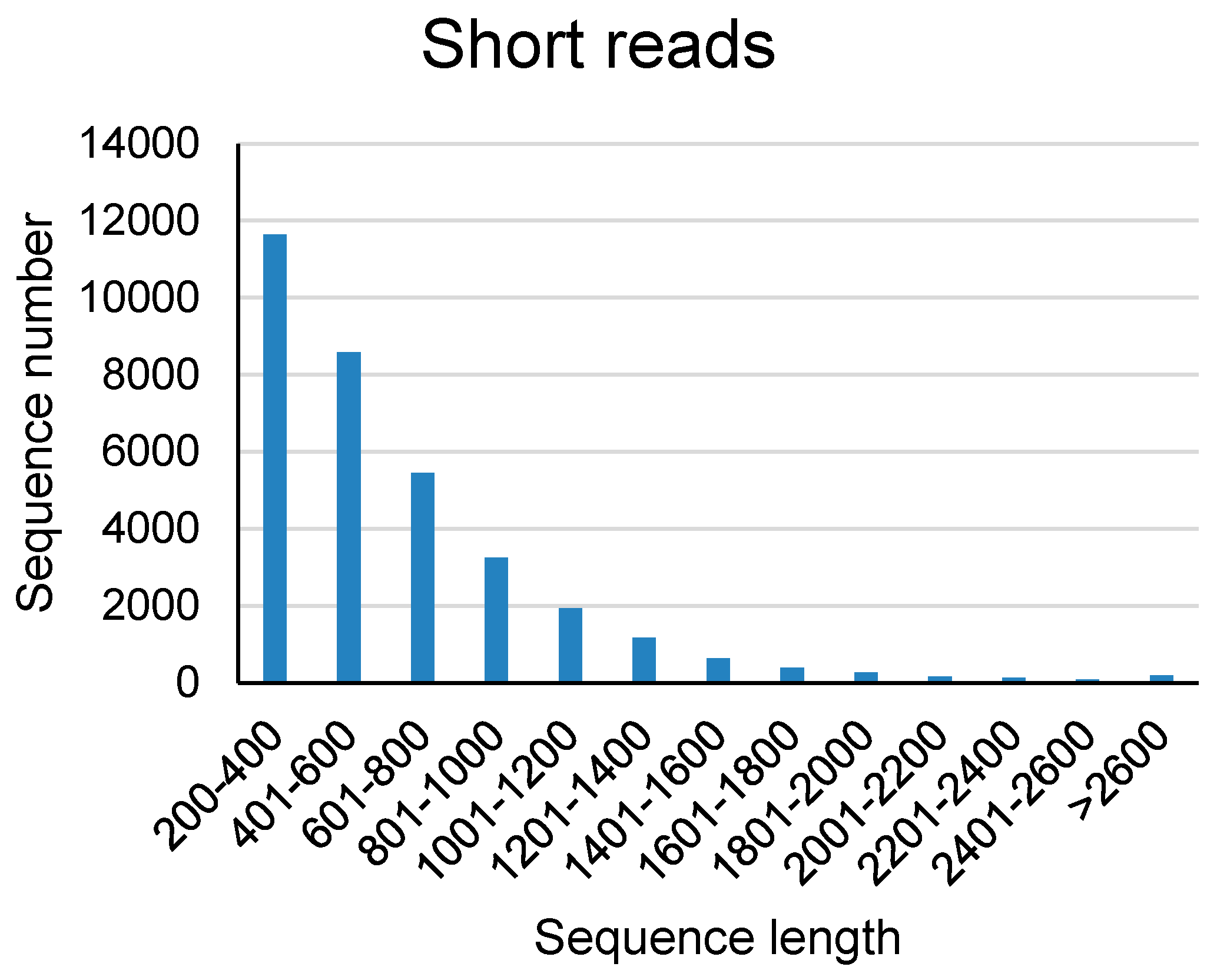
Table 1. In this research, we got 391, 677 full length non-chimeric reads (FLNCs) and 687, 477 non-full length reads (NFLs), which were 35.96% and 63.11% of CCS, respectively. Average FLNCs length is 2300.8 nt. Length distribution of CCS, FLNC and NFL was showed in
Figure 1. All the FLNCs and NFLs were clustered using CD-Hit software, then 539, 260 unigenes were obtained.
Identification of lncRNAs from alfalfa long and short read RNA-seq data
The assembled transcripts of alfalfa based on PacBio sequencing were used for lncRNA identification. This file contains 539,260 reliably expressed transcripts. We dumped transcripts with length <200 nt. Then CPC2 and PLEK software were introduced to screen out transcripts with low or without coding potential, and 174345 transcripts were filtered out. Further, we filtered out the transcripts with known protein-coding genes by mapping transcripts to pfam, Nr, Swissprot, KEGG, GO and COG databae, and 45,116 transcripts as expressed putative lncRNAs were left.
The length distribution of lncRNAs was shown in
Figure 2, and three obvious peaks were consistent with the fraction size of CCS and FLNC. The length distribution analysis showed that more than 38.87 % of the lncRNAs were in the range of 200 to 2000 bp, and about 61.14 % of the lncRNAs were in the range of 2001 to 4000 bp.
We also identified lncRNAs from transcripts based on short read RNA-seq data released by AGED database [
3] using the same identification pipline. And we got 37733 lncRNA transcripts from the short read RNA-seq data. The details of these short read based lncRNAs were list in
Table S1. The IDs and gene expression data in
Table S1 were retrived from AGED database.
Length distribution of lncRNAs identified from short read RNA-seq was statisticed. The result showed that more than 50% of the identified lncRNAs were in the range of 200 to 2000 bp, and about 30% of the lncRNAs were more than 2000 bp in length.
Sequence conservation of lncRNAs between species
We tried to find out lncRNAs highly conserved among species by aligning the alfalfa lncRNA sequences with
Medicago truncatula [
23] and
Arabidopsis thaliana [
24] lncRNAs sequences, respectively. The alignment result showed that only five lncRNAs were homologous with
Medicago truncatula and
Arabidopsis thaliana lncRNAs (
Table S2). And we noticed that two of the five lncRNAs, fl11.68878518.31_2627_CCS and fl8.47251612.31_2377_CCS, aligned their targets with high identity and great hit length. Then we blast their sequences in NCBI using blastn, and found that the two lncRNAs may drived form alfalfa chloroplast or mitochondrial genomes, since fl11.68878518.31_2627_CCS contains a fraction of 18S ribosomal RNA in alfalfa chloroplast genome, and fl8.47251612.31_2377_CCS is a part of large subunit ribosomal RNA in alfalfa mitochondrial genome. The alignment results of the novel lncRNAs (fl11.68878518.31_2627_CCS) of alfalfa with its homologies in
A. thaliana and
M. truncatula was shown in
Figure S1.
Figure S1. Alignment of the nucleotide sequences of fl11.68878518.31_2627_CCS and its two homologies in in A. thaliana (NONATHT003850.1) and M. truncatula (chr4_490072_490302). Red and white backgrounds indicate conserved and non-conserved residues, respectively.
Table S2. Blast result of the five lncRNAs which were homologous with Medicago truncatula and Arabidopsis thaliana lncRNAs.
Identification and characterizition of plastid lncRNAs
Considering the above results and the fact that plastid genome is more conservative than nuclear genome, we hypothesized that there may be highly conserved lncRNAs in the plastid genome among species. In order to more comprehensively analyze the sequence characteristics of alfalfa lncRNA, we fused three set of alfalfa lncRNA transcripts, including the long read sequencing lncRNA transcripts obtained from the leaves of Zhongmu 1 at different developmental stages, the short read sequencing lncRNA transcripts from different tissues, and the long read sequencing lncRNA transcripts from different tissues of alfalfa (cv. Aohan) obtained in this study. An lncRNA sequence set for subsequent analysis was obtained by transcript clustering.
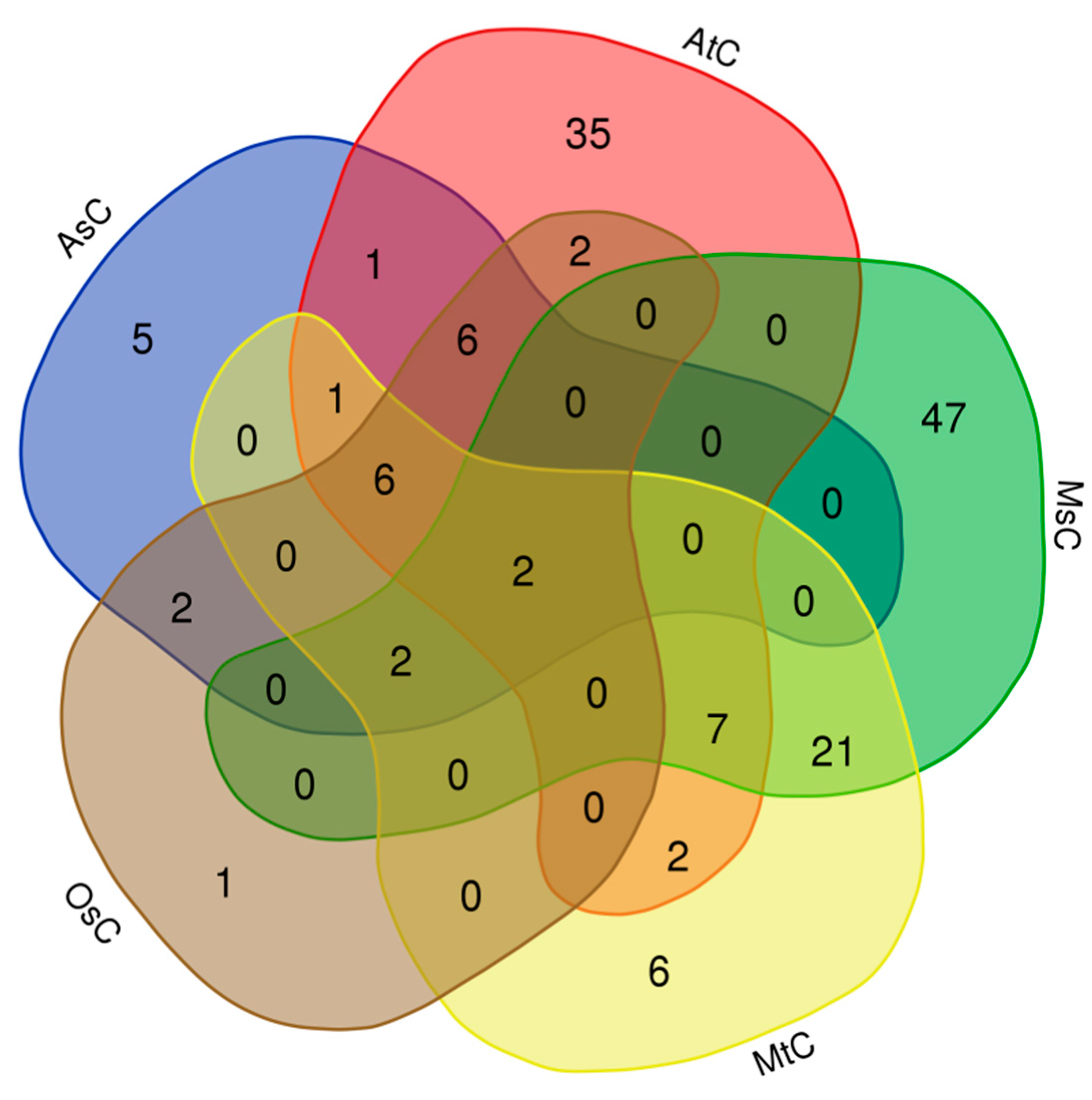
Then we the carried out the alignments between the fused lncRNA sequence set and chloroplast/mitochondrial genomes of different species, and found some lncRNAs had homologies in the chloroplast/mitochondrial genome. Firstly, we found that 62, 21, 25, 47 and 79 lncRNAs had homologies in the chloroplast genomes of Arabidopsis (AtC), rice (OsC), oats (AsC),
Medicago truncatula (MtC) and alfalfa (MsC), respectively (
Table S3,
Figure 4). This result implies that some of the identified lncRNAs may be plastid lncRNAs. interestingly, it was found that sequences of conting86830 and conting82120 lncRNAs were highly conserved in the chloroplast genomes of the above five species, which implys these lncRNAs may play important roles in plant growth and development.
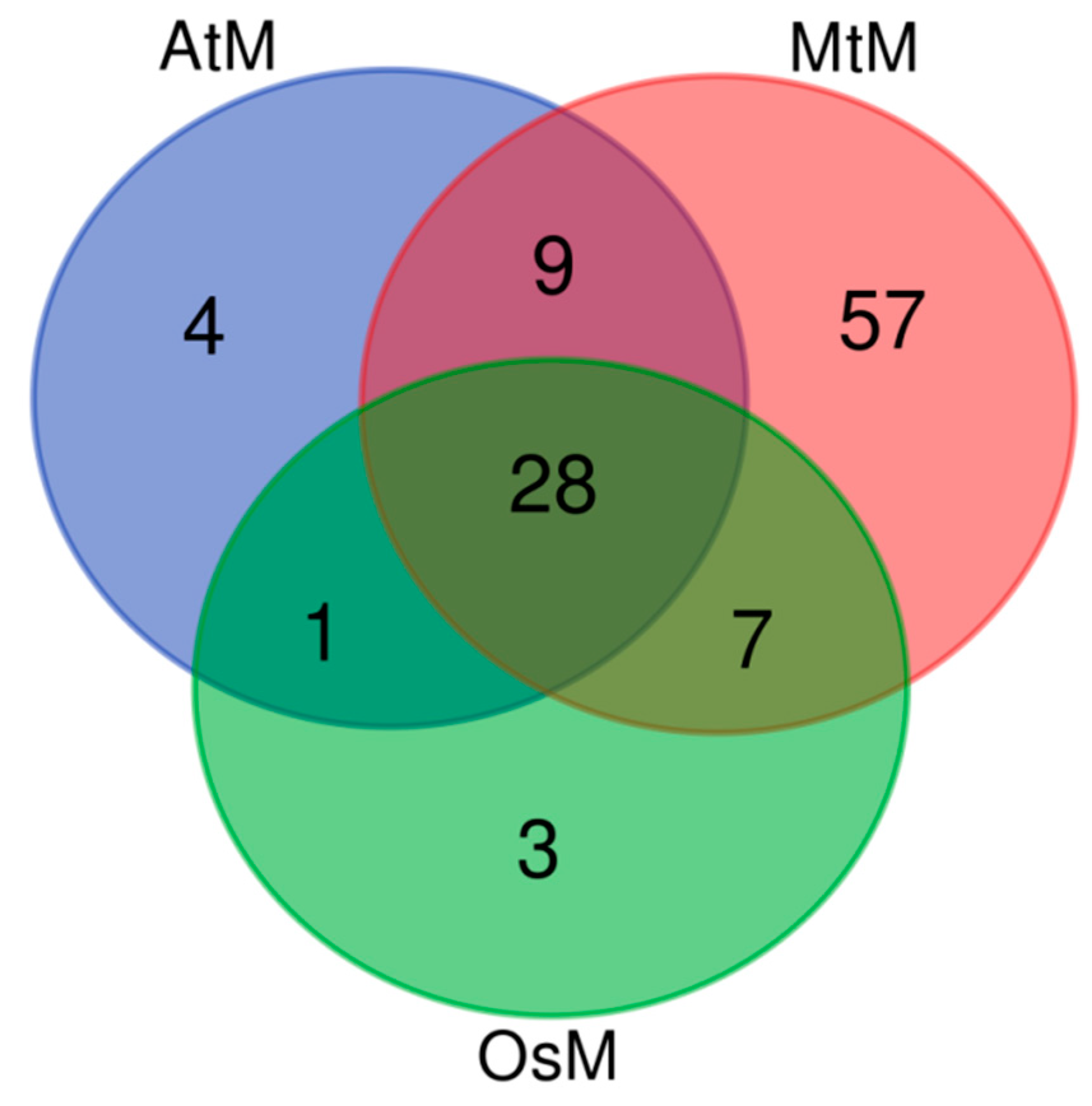
Secondly, we found that 109 lncRNAs had homologies in the mitochondrial genomes of Arabidopsis (AtM), rice (OsM),
M. truncatula (MtM) (
Table S4). And the venn diagram showed that it had the most homologies in the
M. truncatula mitochondrial genome as expected, and 28 lncRNAs own homologies in all the three mitochondrial genomes (
Figure S2).
Figure S2.
Venn diagram of alfalfa lncRNA homologies in mitochondrial genomes of three species.
Figure S2.
Venn diagram of alfalfa lncRNA homologies in mitochondrial genomes of three species.
lncRNAs associated miRNAs
In order to explore the lncRNAs associated with miRNAs, all lncRNA sequences were submitted to TargetFinder and mapped to miRNAs of
Medicago truncatula. Then we found that 85 miRNAs could be mapped to 34 lncRNAs without mismatch, which implys these lncRNAs may be precursors or targets of the 85 miRNAs (
Table S5). The relationship between these lncRNAs and miRNAs was shown in
Figure 5. For convenience of presentation, the miRNAs belongs to the same family were collapsed into one node. The details of this network were list in
Table S5. To further investigate relationship between these lncRNAs and miRNAs, we submitted the sequences of 34 lncRNAs into RNAfold and analysed secondary structure of the lncRNAs. We found sequences of 16 miRNAs were located at hairpin area in secondary structures of 19 lncRNAs (
Table S5).
Figure 5.
Network of miRNAs and their target lncRNAs.
Figure 5.
Network of miRNAs and their target lncRNAs.
Small ORFs analysis of alfalfa lncRNAs
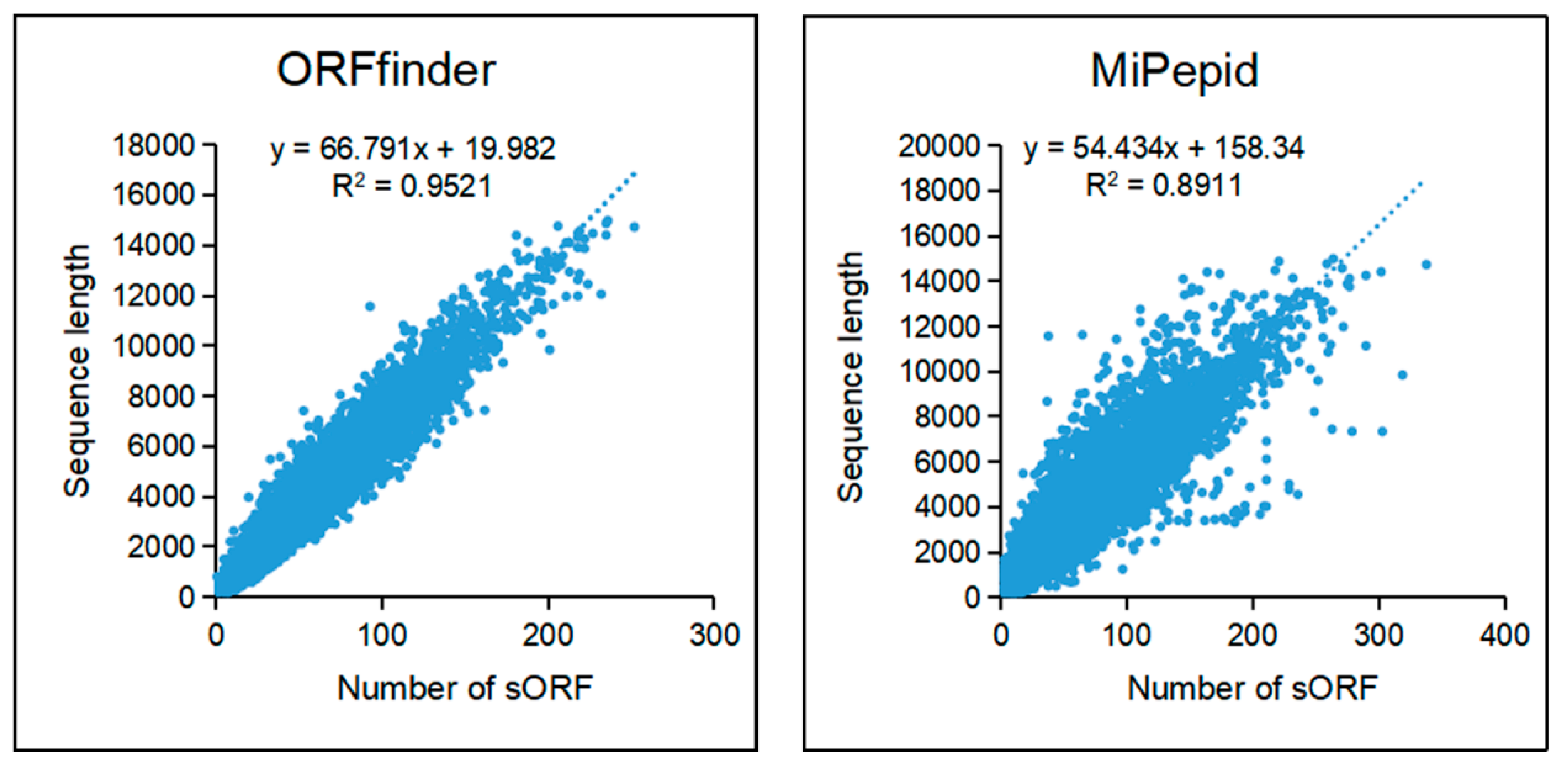
Small ORFs were predicted with ORFfinder and MiPepid. A total of 2,334,873 sORFs was predicted by ORFfinder from 88,558 lncRNAs and 2,617,979 sORFs were predicted by MiPepid from 85,710 lncRNAs (
Figure S3). Sequences of sORFs predicted by the two methods were list in Supplemetary file S1 and Supplementary file S2. Further, we investigated relationships between sequence length of lncRNA and number of sORF which it contains.
Figure 6 shows that there is a positive correlation between the length of the transcripts and the number of sORFs predicted in the transcripts, that is, the longer the transcript, the more candidate sORFs it contains.
Figure S3.
Venn diagram of sORF numbers predicted by ORFfinder and MiPepid, respectively.
Figure S3.
Venn diagram of sORF numbers predicted by ORFfinder and MiPepid, respectively.
Figure 6.
The correlation between the length of lncRNA and the quantity of sORFs.
Figure 6.
The correlation between the length of lncRNA and the quantity of sORFs.
Discussion
In this research, we sequenced RNA samples isolated from 4 different alfalfa tissues with PacBio sequencing technology and finally got 391,677 FLNCs from the sequencing data. The number of FLNC is about 2.6 times that of the previous report[
7], which prefigures more sequence information containing in our dataset. Based on this long read sequencing data and the other two published alfalfa transcripts, the genome-wide lncRNAs were predicted by using the pepline developed by oursevles, and the sequence conservation and small peptide coding of these lncRNAs were analyzed. It is found that quantity of lncRNAs predicted from transcripts of different experimental materials various greatly. We identified 45,116 lncRNAs from our full-length RNA-seq data derived from 4 different alfalfa tissues. However, Chao et al. idetified 20,915 lncRNAs from alfalfa leaf[
7]. The difference between the two studies may be caused by prediction methods, or more likely caused by factors such as genotype, physiological state, development stage, tissue type, since expression of lncRNA is tissue specific and stage specific[
10,
11,
12].
The sequence conservation analysis revealed that homology of lncRNAs was extremely low between alfalfa and other species. The result surpported the current point that lncRNAs present low conservation on nucleic acid sequences among species[
10,
13]. Although most of the lncRNAs present low sequence conserdvation, we still found some lncRNAs annotated as chloroplast genomic sequence showed high sequence homology among species. So we blast the predicted lncRNA sequences with the chloroplast genomes of several species to systematically identify sequence conserved plastid lncRNAs. The results showed that the number of lncRNAs aligned to the chloroplast genome of alfalfa was the largest, followed by
M. truncatula and Arabidopsis, and finally oats and rice. We also blasted the lncRNA sequences with 3 mitochondrial genomes, the results also showed the same characteristics. The number of lncRNA aligned to the mitochondrial genome of
M. truncatula was the largest, followed by Arabidopsis and rice. The alignment results showed that the sequence conservation of some lncRNAs from mitochondria or chloroplasts is very high, such as fl11.68878518.31_2627_CCS (
Figure 4). LncRNAs in mitochondrial DNA have been found previously in animals[
25], and also present high sequence conservation among species, which is agreed with our result.
In addition, we also found that some interesting lncRNAs of which only dozens of bp nucleotide showed high homology with the mitochondrial or chloroplast genomic sequences. These homologous sequences may be some conserved motifs or the result of the exchange between nuclear genome and plastid genome. These short sequences are likely to have some biological functions, since these kind of lncRNAs may play roles of miRNA sponage or transcription suppressor.
To discover lncRNAs function as miRNA sponage, we mapped the lncRNAs to miRNAs of M. truncatula, try to figure out how the lncRNAs work with miRNAs. As expected, it was found that 34 lncRNAs could bind with one or more miRNAs including miR167, miR171, miR393 and miR398, through completely complementary base pairing. Considering that miRNA play pvital roles in the processes of plant growth, development and stress response, these lncRNAs, such as potential biologically function in alfalfa. According to the result from secondary structure analysis, 16 miRNAs were located at hairpin area in secondary structures of 19 lncRNAs, which implys these lncRNAs maybe pri-miRNAs of the 16 alfalfa miRNAs.
More and more studies proved that sORF-encoded micropeptides play important roles in regulating various biological activities [
19,
20,
21,
22]. Using bioinformatic methods, we found more than 96% lncRNAs identified in this study could encode small peptides, which suggests that lncRNA has great potential to regulate some life processes through the synthesis of small peptides, although the existence of these small peptides needs further experimental verification. To date, there isn’t a bioinformatic way developed to annotated biological function of sORFs yet, and functional characterization of sORFs for plants is far behind that of other species. So, except for experimental methods, bioinformatic methods for investigating sORF function should be developed as soon as possible.
In this study, we identified alfalfa lncRNAs from combined long and short read sequencing data, resulting a tremendous number of putative lncRNAs. And we reported a set of plastid lncRNAs in plant and predicted sORFs of alfalfa lncRNAs for the first time. Our research not only provided abundant sequence infromation of alfalfa lncRNA, but also omrs a fresh perspective to study them.
Material and methods
Plant materials and sampling
Alfalfa plants were grown in plastic pots (20 cm×20 cm×30 cm), and cultured under natural light condition for 3 months. And the alfalfa plants were watered with MS solution (PH7.0) every 3 days. Root, node, stem, leaf and shoot apex tissue were collected respectively and frozen with liquid nitrogen immediately. Each of these tissues was collected up to 3 g for total RNA isolation.
Total RNA isolation and PACBIO library construction
Total RNA of each sample was isolated with RNA purification reagent (Invitrogen) according the instruction. The concentration and purity of total RNA was detected with Nanodrop2000, the integrity of total RNA was checked by agarose gel electrophoresis, and RIN was quantified by Agilent2100. Then, using Clontech-SMARTer™ PCR cDNA Synthesis Kit reverse transcribed total RNA into cDNA. Finally, the library was constructed with evrogen-Trimmer-2 Kit and SMRTbell Template Prep Kit 1.0.
Analysis of PacBio sequencing data
Analysis of PacBio sequencing data was performed by the transcriptome analysis software of PacificBiosciences[
26]. Sequences from raw data were combined into circular consensus sequence (CCS), then 5 primer, 3 primer and polyA sequence were checked for each read. After filtering out short reads and chimeric reads, full-length non-chimeric reads (FLNCs) and non-full-length reads (NFLs) were obtained respectively. Further, in order to get unigenes, FLNCs and NFLs were clustered using cdhit software[
27]. The raw data was already uploaded to the National Genomics Data Center (
https://www.cncb.ac.cn/) and the accession number is CRA009238.
The pipeline to identify lncRNA from transcriptome data
Sequence conservation analysis of lncRNAs
To reveal the sequence conservation features of alfalfa lncRNAs predicted from the PACBIO long read sequencing data, and the sequences of these putative lncRNAs were searched for homologs from the lncRNA sequences data sets of
Arabidopsis thaliana and
Medicago truncatula using TBtools[
30] with default parameters. The homologies were screened with the cutoff of identity >=90%. The lncRNA sequences of
Arabidopsis thaliana were downloaded from NONCODE database[
24], and the lncRNA sequences of
Medicago truncatula was extracted from
Medicago truncatula genome files according to the chromosome location published by Wang et al.[
23].
Prediction of microRNA target mimics
The target mimics mechanism of lncRNA–microRNA and their potential roles in gene expression were reported in plants [
18]. To explore the possibility of putative lncRNAs as microRNA targets, all lncRNA sequences were submitted to Targetfinder[
31] with default parameter. Then the alignment result was screened with the cutoff of score = 0.
Small ORFs analysis of alfalfa lncRNAs
Small ORFs were predicted with ORFfinder [
32] and MiPepid [
33], respectively. The parameter of S was set as “0” when using ORFfinder to predicted sORFs. And it should be noted that the sequences of transcript containing N were removed before we predicted sORFs using MiPepid with default parameters, since the software could not recognize those sequences.
Supplementary Materials
Supplementary Figure S1. Alignment of the nucleotide sequences of fl11.68878518.31_2627_CCS and its two homologies in in A. thaliana (NONATHT003850.1) and M. truncatula (chr4_490072_490302). Red and white backgrounds indicate conserved and non-conserved residues, respectively. Supplementary Figure S2. Venn diagram of alfalfa lncRNA homologies in mitochondrial genomes of three species. Supplementary Figure S3. Venn diagram of sORF numbers predicted by ORFfinder and MiPepid, respectively. Supplemental Table S1: Details of the lncRNAs predicted form short reads RNA-seq data. Supplemental Table S2: Blast result of the five lncRNAs which were homologous with Medicago truncatula and Arabidopsis thaliana lncRNAs. Supplemental Table S3: homologies in the chloroplast genomes of Arabidopsis (AtC), rice (OsC), oats (AsC), Medicago truncatula (MtC) and alfalfa (MsC), respectively. Supplemental Table S4: homologies in the mitochondrial genomes of Arabidopsis (AtM), rice (OsM), M. truncatula (MtM). Supplemental Table S5: Details of the miRNA-lncRNA interaction network. Supplemental File S1: Sequences of sORF candidates predicted by ORFfinder. Supplemental File S2: Sequences of sORF candidates predicted by MiPepid.
Author Contributions
Y. L., J. K. and Y. S. conceived the idea and structured the manuscript. Y. L. performed all the alignments and sORF analyses, Y. L. and C. W. interpreted the results. Overall data analyses were completed by Y. L., H. C., K. Z., F. J., C. M. and Y. L., C. W. and Y. S. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the National Natural Science Foundation of China (No. 32271763).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data of PacBio sequencing was already uploaded to the National Genomics Data Center (
https://www.cncb.ac.cn/) and the accession number is CRA009238.
Conflflicts of Interest
The authors declare no conflflict of interest.
References
- Wang, C.; Ma, B.L.; Yan, X.; Han, J.; Guo, Y.; Wang, Y.; Li, P. Yields of alfalfa varieties with different fall-dormancy levels in a temperate environment. Agronomy Journal 2009, 101, 1146–1152. [Google Scholar] [CrossRef]
- Li, Y.; Wan, L.; Bi, S.; Wan, X.; Li, Z.; Cao, J.; Tong, Z.; Xu, H.; He, F.; Li, X. Identification of drought-responsive microRNAs from roots and leaves of alfalfa by high-throughput sequencing, Genes 2017, 8.
- O’Rourke, J.A.; Fu, F.; Bucciarelli, B.; Yang, S.S.; Samac, D.A.; Lamb, J.F.S.; Monteros, M.J.; Gronwald, J.W.; Krom, N.; Li, J.; et al. The medicago sativa gene index 1. 2: a web-accessible gene expression atlas for investigating expression differences between medicago sativa subspecies. BMC Genomics 2015, 16, 1–17. [Google Scholar]
- Long, R.; Zhang, F.; Zhang, Z.; Li, M.; Chen, L.; Wang, X.; Liu, W.; Zhang, T.; Yu, L.X.; He, F.; et al. Genome assembly of alfalfa cultivar zhongmu-4 and identification of SNPs associated with agronomic traits. Genomics, Proteomics & Bioinformatics 2022. [CrossRef]
- Chen, H.; Zeng, Y.; Yang, Y.; Huang, L.; Tang, B.; Zhang, H.; Hao, F.; Liu, W.; Li, Y.; Liu, Y.; et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 2020, 11, 2494. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Du, H.; Chen, Z.; Lu, H.; Zhu, F.; Chen, H.; Meng, X.; Liu, Q.; Liu, P.; Zheng, L.; et al. The chromosome-level genome sequence of the autotetraploid alfalfa and resequencing of core germplasms provide genomic resources for alfalfa research. Molecular Plant 2020, 13, 1250–1261. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.; Yuan, J.; Guo, T.; Xu, L.; Mu, Z.; Han, L. Analysis of transcripts and splice isoforms in medicago sativa l. by single-molecule long-read sequencing. Plant Molecular Biology 2019, 99, 219–235. [Google Scholar]
- Wan, L.; Li, Y.; Li, S.; Li, X. Transcriptomic profling revealed genes involved in response to drought stress in alfalfa, Journal of plant growth regulation 2022, 41: 92-112.
- Ng, S.Y.; Lin, L.; Soh, B.S.; Stanton, L.W. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013, 29, 461–468. [Google Scholar] [CrossRef]
- Song, X.; Sun, L.; Luo, H.; Ma, Q.; Zhao, Y.; Pei, D. Genome-Wide Identification and Characterization of Long Non-Coding RNAs from Mulberry (Morus notabilis) RNA-seq Data. Genes (Basel) 2016, 7, 11. [Google Scholar] [CrossRef]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Wahrisch, S.; Beisaw, A.; Macura, K.; Blass, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. 2008, 105, 716–721. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012 22, 1775–1789. [CrossRef]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an arabidopsis polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Bi, X. Functions of chromatin remodeling factors in heterochromatin formation and maintenance. Sci. China Life Sci. 2012, 55, 89−96. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, Q.J.; Li, J.; Jiang, D.G.; Zhou, L.Y.; Wu, P.; Lu, S.; Li, F.; Zhu, L.Y.; Liu, Z.L.; et al. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 2012, 22, 649–60. [Google Scholar] [CrossRef]
- Camblong, J. ; Beyrouthy, N,; Guffanti, E. ; Schlaepfer, G.; Steinmetz, L.M.; Stutz, F. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009, 23, 1534–1545. [Google Scholar]
- Shin, H.; Shin, H.S.; Chen, R.; Harrison, M.J. Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J. 2006, 45, 712−726. [Google Scholar] [CrossRef]
- Pauli, A.; Norris, M.L.; Valen, E.; Chew, G.L.; Gagnon, J.A.; Zimmerman, S.; Mitchell, A.; Ma, J.; Dubrulle, J.; Reyon, D.; et al. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 2014, 343, 1248636. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.F.; Reese, A.L.; McAnally, J.R.; Chen, X.W.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science, 2016, 351, 271–275. [Google Scholar] [CrossRef]
- Crespi, M.D.; Jurkevitch, E.; Poiret, M.; d’Aubenton-Carafa, Y.; Petrovics, G.; Kondorosi, E.; Kondorosi, A. enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994, 13, 5099−5112. [Google Scholar] [CrossRef]
- Wang, T.Z.; Liu, M.; Zhao, M.G.; Chen, R.; Zhang, W.H. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 2015, 15, 131. [Google Scholar] [CrossRef]
- Liu, C.; Bai, B.; Geir, S.; Lun, C.; Deng, W.; Zhang, Y.; Bu, D.; Zhao, Y.; Chen, R. Noncode: an integrated knowledge database of non-coding RNAs. Nucleic Acids Research 2005, 33, D112–D115. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Tian, X.; Chang, H.; Sun, Y.; Wu, Z.; Cheng, Z.; Dong, P.; Zhao, Q.; Ruan, J.; Bu, W. Two novel lncrnas discovered in human mitochondrial dna using pacbio full-length transcriptome data. Mitochondrion 2018, 38, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Mccarthy, A. Third generation dna sequencing: pacific biosciences’ single molecule real time technology. Chemistry & Biology 2010, 17, 675–676. [Google Scholar]
- Li, W.; Godzik, A. CD-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences”, Weizhong Li & Adam Godzik Bioinformatics 2006, 22, 1658-1659.
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic acids research 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhang, J.; Zhou, Z. PLEK: a tool for predicting long non-coding rnas and messenger rnas based on an improved k-mer scheme. BMC Bioinformatics 2014, 15, 311. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H. R.; Frank, M. H.; He, Y.; Xia, R. Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lavorgna, G.; Guffanti, A.; Borsani, G.; Ballabio, A.; Boncinelli, E. Targetfinder: searching annotated sequence databases for target genes of transcription factors. Bioinformatics 1999, 15, 172–173. [Google Scholar] [CrossRef]
- Rombel, I.T.; Sykes, K.F.; Rayner, S.; Johnston, S. A. Orf-finder: a vector for high-throughput gene identification. Gene 2002, 282, 33–41. [Google Scholar] [CrossRef]
- Zhu, M.; Gribskov, M. MiPepid: MicroPeptide identification tool using machine learning. BMC Bioinformatics 2019, 20, 559. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).