1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most common type of pancreatic cancer, especially in Asian countries but also in Europe and the United States (1). With a slight male predominance (2), this cancer is the third leading cause of cancer-related death in Western countries (1,3). Only 20-30% of patients are diagnosed with resectable or borderline resectable tumors. In these patients, even with the adjuvant FOLFIRINOX, less than 30% will be cured (4). Patients who exhibit either a locally advanced or a metastatic disease at diagnosis cannot be treated with surgery. In locally advanced disease, a small number of patients can be treated by surgery after neoadjuvant treatment, including combined chemotherapy with or without chemoradiation, and a small percentage will be cured after these intense treatments. Nevertheless, most patients will die from their disease, especially in metastatic cases. The standard-of-care in first-line metastatic disease relies on a combination of 5-fluorouracil (5FU) plus irinotecan plus oxaliplatin (the FOLFIRINOX regimen) (4) or a combination of gemcitabine and nab-paclitaxel (5,6). With these regimens, median overall survival (OS) is close to 10 months (4,5). At progression, a second line of treatment is proposed to ~60% of patients. In patients receiving gemcitabine-based chemotherapy as the first line, oxaliplatin-based second-line treatment has shown conflicting results, therefore the most evidence-based second-line option is the use of nanoliposomal irinotecan combined with 5FU and folinic acid (NALIRI regimen)(7).
Molecular profiling (MP) of PDAC discovered several key gene alterations (GAs) involved in carcinogenesis mainly in four genes: KRAS, TP53, SMAD4 and CDKN2A (8). Except for the KRAS G12C mutation, which is found in less than 2% of PDAC cases, these GAs currently have no molecular target therapies (MTTs). Nevertheless, in around 25% of cases, actionable GAs have been found at low frequencies (9–12), including genes involved in the DNA damage response and repair (DDR), some specific pathways (BRCA1, BRCA2, PALB2), the AKT, PI3K, FGFR, NTRK, MYC, MET and NOTCH pathways, amplification of ERBB2, and mutations in RNF43, mTOR, PDGFR, ROS1, KIT, BRAF (8,13).
However, very few MTTs have been proven to be effective, and only olaparib (POLO trial) has been approved as treatment in patients who have responded to a first-line of platinum-based chemotherapy, with locally advanced or metastatic pancreatic cancer carrying germline mutation of BRCA1 or BRCA2 (14).
In recent years, several prospective studies have been developed to evaluate the feasibility and efficacy of precision medicine by next generation sequencing (NGS) in patients with various solid tumors (15–20). Of note, only 10-25% of patients in these studies received a specific therapy informed by MP (15–20), and fewer benefited from this approach. However, a recent prospective randomized precision-medicine trial in patients with breast cancer reported that the use of MTT improved progression-free survival (PFS), but only when GAs were classified as level I/II based on the ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT) (18). This was confirmed by data from the precision-medicine program at our institution (21).
Specifically in PDAC patients, results from several prospective and retrospective studies showed a benefit in terms of overall response rates, PFS, and median overall survival (OS) in patients with an actionable GA who received the corresponding MTT, compared with those who had an actionable GA but not treated by MTT (6,10,22,23). In their recently published retrospective study, Pishvaian and colleagues found a positive impact on OS of treatments targeting actionable mutations in PDAC. Their study showed better OS in patients harboring actionable Gas treated with MTT compared to those treated with standard therapies (10). Nevertheless, whether a MP approach could improve treatment decision making is yet to be demonstrated in patients with PDAC.
In this retrospective study, we analyzed the clinical outcomes of patients with PDAC from whom we had obtained their MP through our institutional precision-medicine programs. We aimed to report the clinical applicability of precision medicine in PDAC and assess the impact of MTT in patients with actionable GAs.
2. Materials and Methods
Study Design
We conducted a retrospective analysis of all >18 year-old patients with histologically confirmed PDAC who underwent tumor MP between 2011 and 2020 in our institution as part of personalized medicine trials. We excluded patients with non-contributive, or when was obtained by liquid biopsy only. Included patients had received at least one line of treatment at our institution.
Clinical characteristics, as well as treatment-related outcomes, were retrospectively collected by hospital chart review (demographics, clinical characteristics at diagnosis, primitive lesion treatments, systemic treatments and outcomes). The primary endpoint was OS, defined as time between diagnosis of metastatic disease and death or loss of follow-up. A minimal follow-up of 6 months was required after molecular profiling. Secondary endpoints were PFS for patients receiving MTTs.
This retrospective study complies with the French MR004 methodology regarding general data protection regulation for non-interventional retrospective health research (Délibération n° 2018-155 du 3 mai 2018) and was approved by our institutional review board (CSET N°2023-175), in compliance with Helsinki declaration. All patients gave signed informed consent for genomic analyses as part of precision-medicine trials.
Genomic Analyses
MOSCATO
The primary objective of both versions of the MOSCATO study was to evaluate the clinical benefit of with evaluation of outcomes following MTTs. This trial included patients from December 2011 until March 2016. The inclusion criteria (24) specified all patients with a histologically proven solid tumor (including PDAC), refractory or in locoregional or metastatic relapse considered as not curable with conventional treatments, were eligible. Patients then underwent a biopsy of an accessible tumor site (primary tumor or metastasis) according to the local recommendations of our institution.
A conventional histological evaluation was performed first to determine the percentage of tumor cells. Samples showing more than 30% tumor cells were selected for whole molecular analysis. In samples with 10-30% tumor cells, only targeted sequencing was performed.
DNA and RNA extractions were performed using the Qiagen method (RNeasy/ DNeasy) at the genomic functional unit (GFU). Array comparative genomic hybridization (aCGH) was performed on DNA for samples having a quantity of DNA greater than 1 g. High-level amplifications (log2ratio < 0.89) and genomic loss (log2ratio < -0.28) were determined by the bio informaticians of the GFU.
Separately, 700 ng of DNA were used for screening of somatic mutations, covering more than 5,000 mutations described in the COSMIC database. The Ion AmpliSeq Cancer Panel (CP1), covering 190 amplicons in 40 cancer genes (ThermoFisher Scientific), was used between May and November 2012. From December 2012 to September 2013, the panel was expanded (Ion AmpliSeq Cancer Hotspot Panel v2) to include 207 amplicons in 50 cancer genes (ThermoFisher Scientific). Finally, starting September 2013, the last panel (MOSC3) covered 75 cancer genes using Ion AmpliSeq custom design, combining 1,218 amplicons with the CHP2 panel.
RAGNAR Trial
As part of the RAGNAR phase II trial on the efficacy and toxicity profile of erdafitinib in patients with solid tumor with an alteration of the FGFR gene, eligible patients benefited first from a molecular screening for an alteration in the FGFR gene. Archived tumor tissue or fresh samples were sent to the Central Laboratory for testing. After DNA extraction, molecular analysis was carried out by NGS based on the FoundationOne®CDx platform, allowing the detection of mutations in 324 genes as well as the status of microsatellite instability and tumor mutational burden.
STARTRK Trial
As part of the STARTRK phase II basket trial, which evaluated entrectinib in the treatment of patients with locally advanced or metastatic solid tumors carrying the NTRK1/2/3, ROS1 or ALK gene rearrangements, eligible patients benefited from a first phase of molecular screening, based on the FoundationOne®CDx platform, allowing the detection of mutations in 324 genes (Table 4, Appendix), as well as the determination of and status.
Treatment Decision
For all patients, were discussed during a weekly multidisciplinary precision-medicine tumor board. Results were reported in the medical file of each patient. Actionability of GAs was classified according to ESMO Actionability for molecular Targets (ESCAT) guidelines (25). Considerations for MTT based on GAs relied on variant annotation databases (OncoKB, CIViC, My Cancer Genome, and literature) as well as European Medicines Agency (EMA) approval, temporary authorization of use (ATU) of the drug and phase I clinical trials available in our institution. In the absence of an actionable GA, the patient received the physician’s standard treatment of choice.
Hence, three groups of patients were defined: group A included patients with actionable GAs who received MTT; group B included patients with actionable GAs who did not receive MTT but were treated with standard treatment; and group C included patients without actionable GA,s who received the physician’s standard treatment choice.
Statistical Analysis
The groups were compared with the chi-squared test or Fisher’s exact test for binary variables and with the Mann Whitney nonparametric test for continuous variables. Survival was calculated from the date of onset of metastases. Multivariate analysis was performed with a Cox model. The effect of administering targeted therapy was studied, considering targeted therapy as a time-dependent variable. Other studied variables were the presence of hepatic or lung metastases, the number of metastatic sites and sex. All statistical analyses were performed using Rstudio software
3. Results
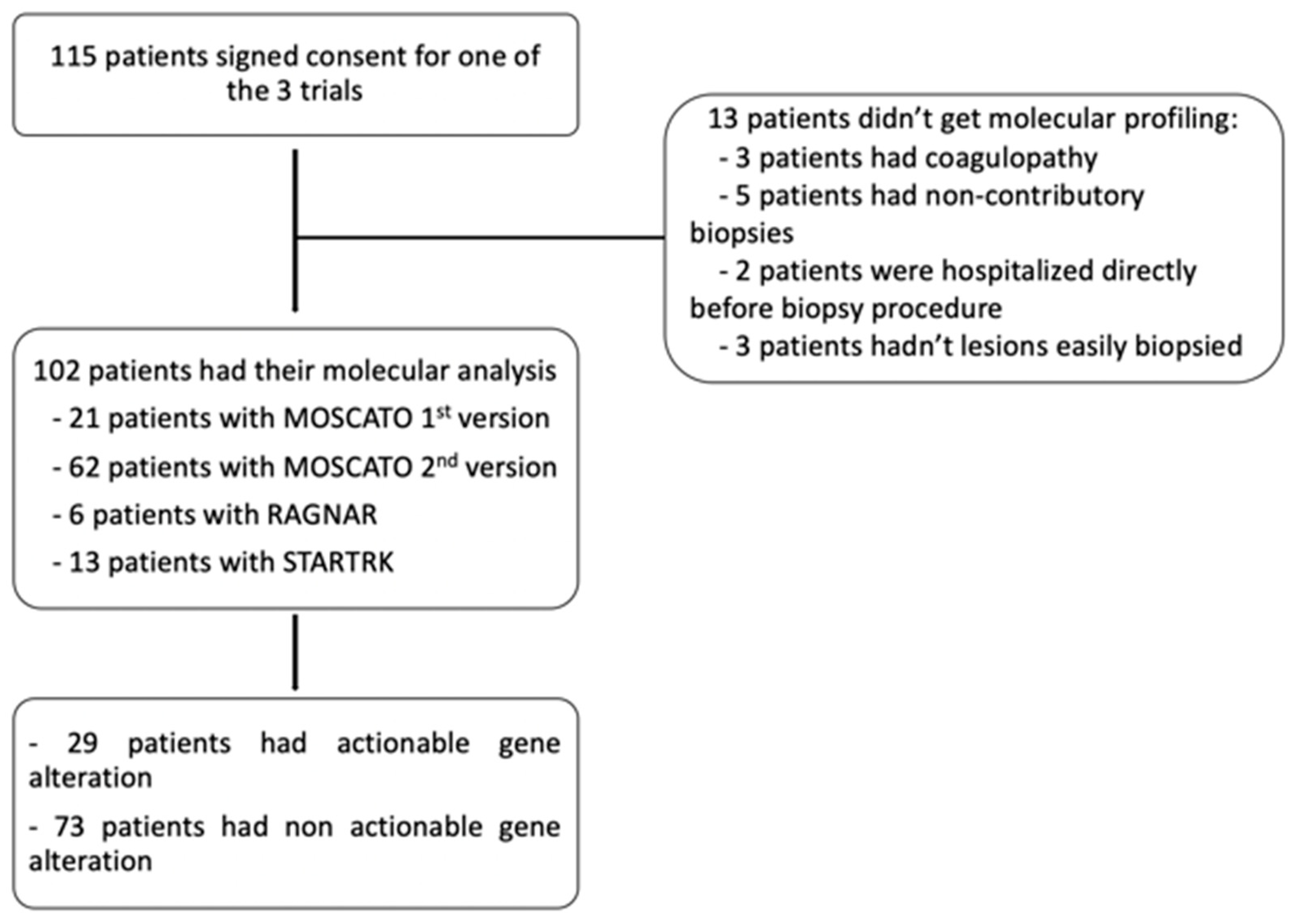
Out of 115 eligible patients, 102 patients got MP (exclusion causes were: biopsy contra-indication for coagulopathy (n=3), cancelled biopsy for inter-current disease (n=2), not-contributive sample (n=5) and no accessible lesion (n=3) in patients without available tumor tissue) (
Figure 1).
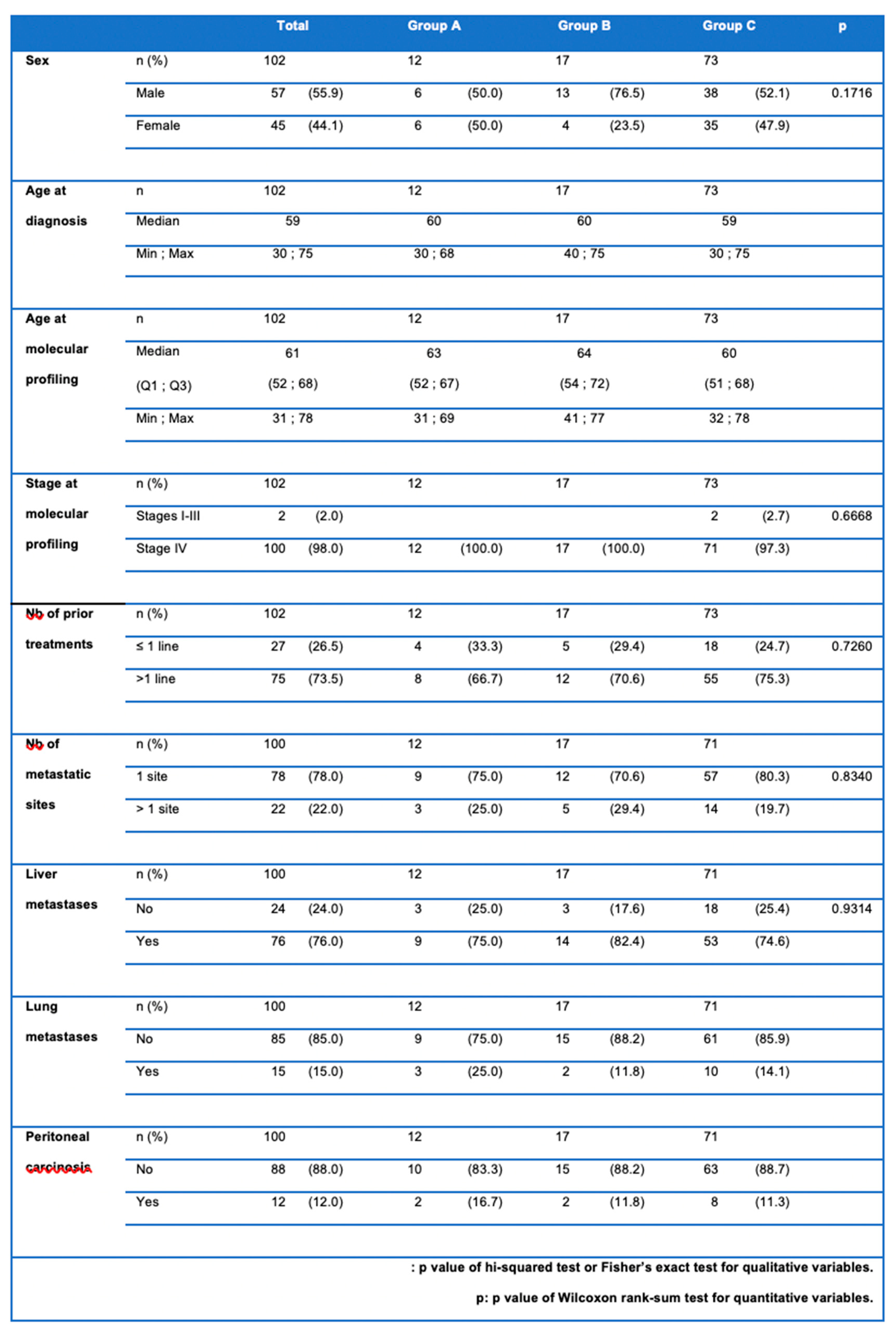
Patients’ characteristics are presented in
Table 1. With a slight male predominance (55.9%), the mean age at diagnosis was 59 years [37; 75]. More than half of the patients were metastatic at diagnosis (61.8%), with mainly hepatic metastases (76%). MP was performed 2 years after diagnosis most of the time. At that time, 98% of patients presented with metastatic disease, and 75% of them had already received two or more lines of treatment.
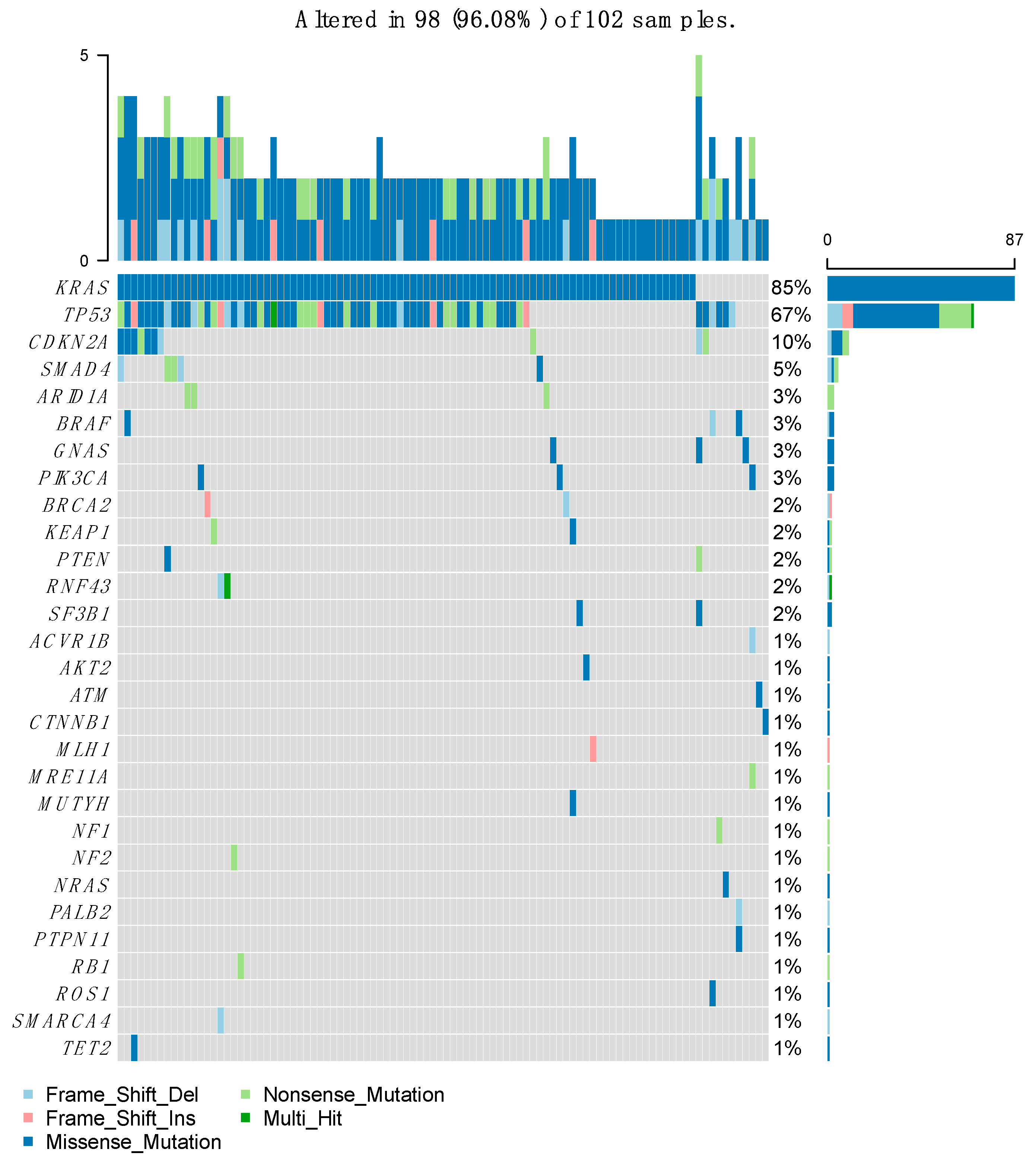
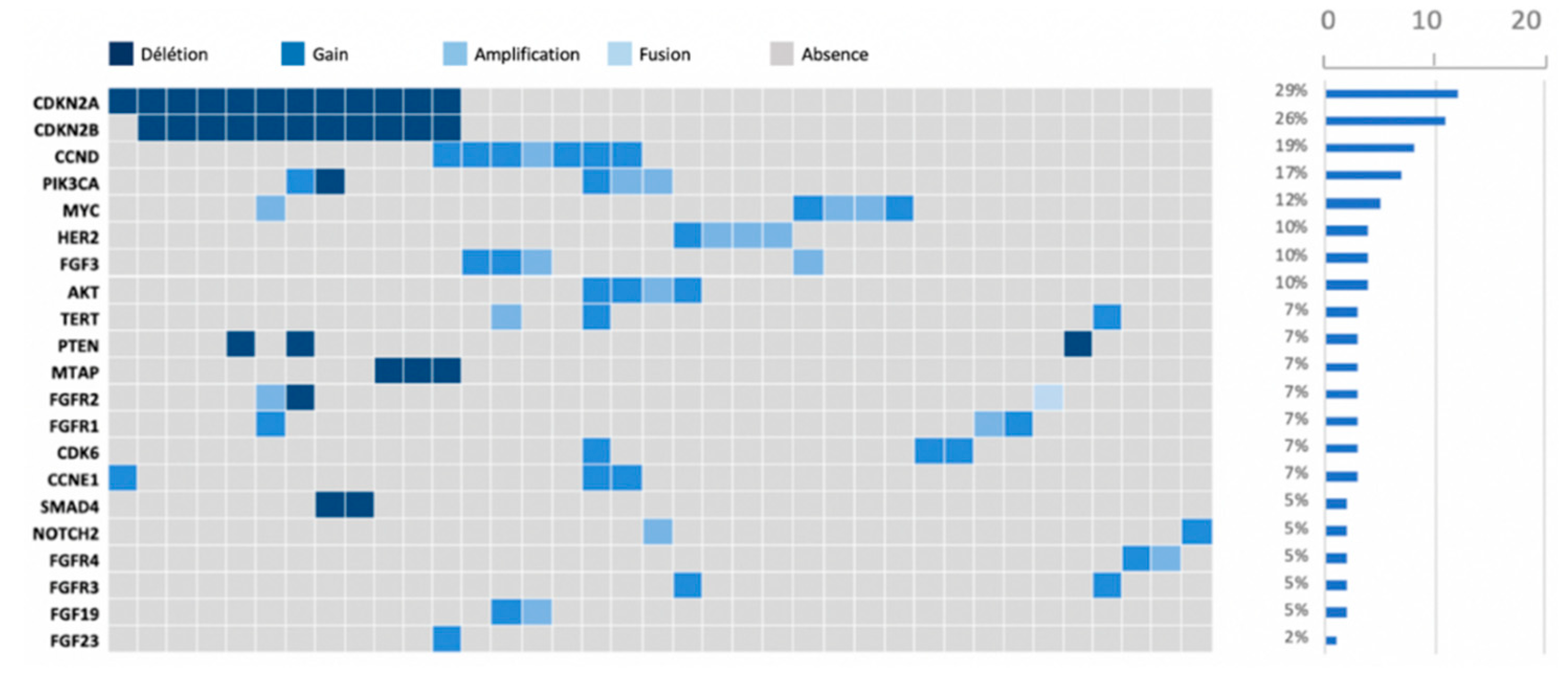
After performing the molecular sequencing, 359 abnormalities in 94 different genes were observed in the whole population (
Figure 2). Most of these abnormalities were mutations (n= 232, 64.6%), but there were also gene amplifications (n= 48, 13.4%), gains (n= 31, 8.6%), and deletions (n= 48, 13.4%) (
Figure 3). The most frequently altered gene was KRAS (found in 87 patients out of 102, 85.3%) followed by TP53 (in 77 patients, 75.5%) and CDKN2A (in 23 patients, 22.5%). Regarding KRAS, the most frequent mutation was G12D in 51% of cases, followed by G12V and G12R. The G12C mutation was found only in 4 patients. Regarding other types of GAs, loss of CDKN2A and CDKN2B were found in 12 and 11 patients, respectively (11.8% and 10.8%). HER2 was amplified in 4 patients (3.9%).
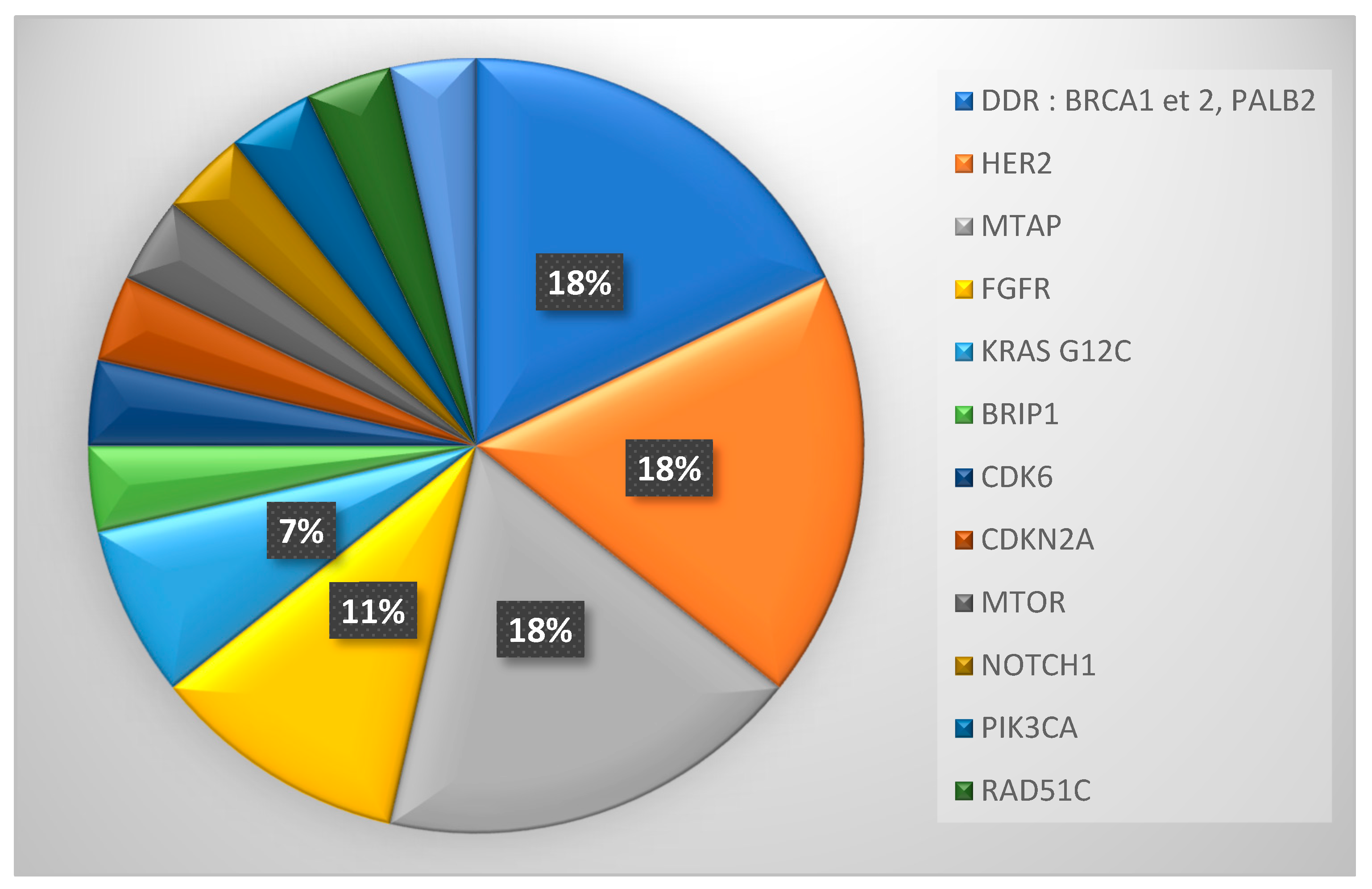
Twenty-nine patients (28.4%) exhibited a potentially actionable molecular alteration according to ESCAT classification at time of inclusion (ESCAT I-IV). The most common actionable mutations were amplification of HER2 and mutations in DNA repair genes (BRCA2 and PALB2), n=5 for both (
Figure 4).
Of these 29 patients, 12 patients received MTT. Reasons for not receiving MTT for the other 17 patients are detailed in
Table 2. MTT was usually received as the second line of treatment (median: 2; [1 4]).
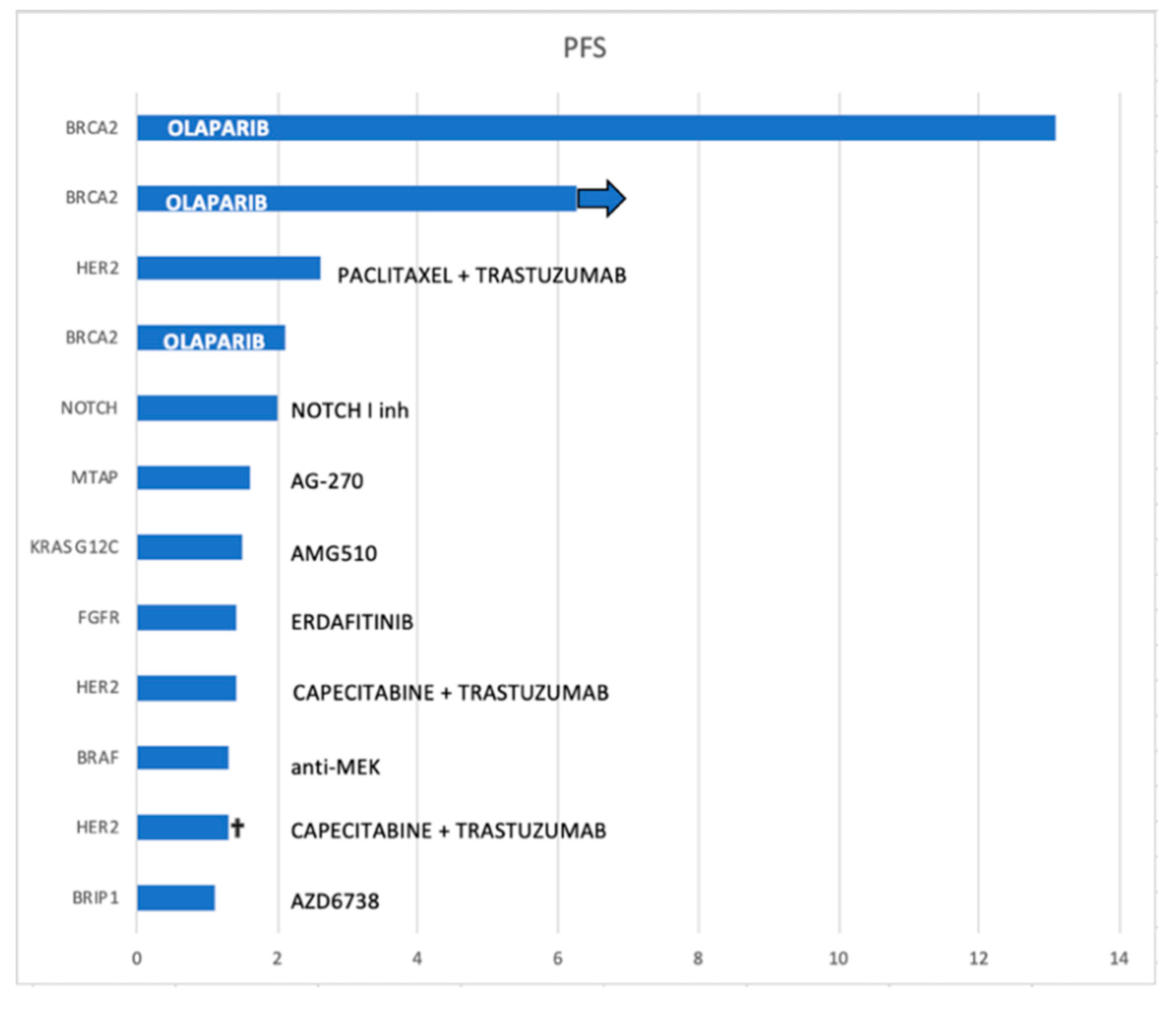
The MTTs were olaparib (in three patients with BRCA mutations), trastuzumab-based treatment regimen (in three patients with HER2 amplification), NOTCH 1 inhibitor (in one patient), anti-MEK (in one patient harboring a BRAF mutation), AMG510 (in one patient with a KRAS G12C mutation), erdafitinib (in one patient with an FGFR alteration), AG-270 (in one patient with an MTAP deletion) and AZD6738 (an ATR inhibitor used in one patient with a BRIP1 alteration).
The median duration of response to MTT was 1.55 months [95%CI: 1.1–not reached (NR)]. Despite the low sample size, patients with BRCA2 mutation seemed to have longer PFS following olaparib treatment, reaching 13.1 months in one patient, while a second patient was still on treatment at the time of the statistical analysis (follow-up of 7 months) (
Figure 5).
Better PFS is seen in patients with BRCA2 mutations that were treated with olaparib.
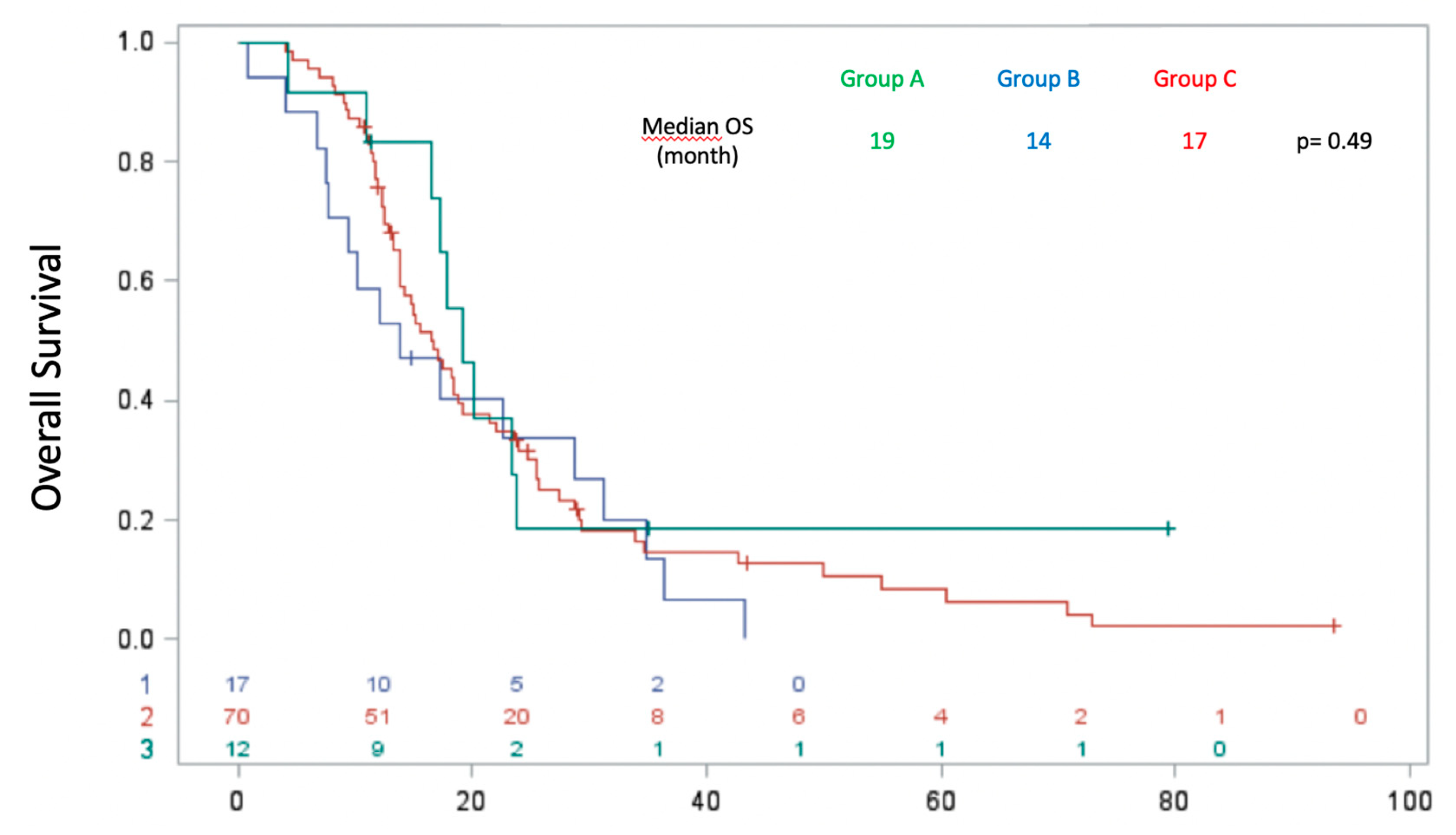
Regarding OS, there was no significant difference between the three groups, with a median OS of 19 months in group A, 14 months in group B and 17 months in group C (p = 0.4889) (
Figure 6). Multivariate analysis using the Cox model with time-dependent variables revealed better OS of patients without liver metastases compared with those with liver metastases at the time of molecular profiling with HR = 0.471 (p = 0.01).
4. Discussion
Here we report our real-life data of the clinical applicability of precision medicine in a tertiary center for the therapeutic management of PDAC in routine clinical practice. We show that is feasible in PDAC, but only 45% of patients benefitted from MTT when actionable GAs were found.
In our study, we found a comparable number of molecular profiles to previous studies (8–11) with the most common mutations in KRAS (85.3% of which 51% were G12D), followed by TP53 (67%) and CDKN2A (10%). With 28.4% of patients having a potentially actionable mutation based on ESCAT classification, our results are similar to that exhibited in Pishvaian et al (10), in which 26% of patients had potentially actionable mutations (10). Despite the limited number of patients actually treated with MTT, this is similar to that already described in large scale precision medicine trials for solid tumors (16,24,26). The proportion of patients receiving MTT was low due to several reasons. First, accessibility to treatment is difficult, especially accessibility to early phase I trials. Indeed, these trials are frequently restricted to a pre-selected population. Second, is generally only performed after several treatment lines have failed, when the general health status of the patient is poorer, which limits the potential clinical benefit from MTT. Performing at diagnosis or in early treatment might circumvent the rapid decrease in health status frequently observed in patients with PDAC. Third, the notion of mutation actionability is dynamic, and new drugs might allow previously non-actionable molecular alteration to become actionable. The key mutation in PDAC is KRAS, which is notoriously resistant to drugs; however, KRAS inhibitors are indeed emerging, with the recent FDA approvals in non-small cell lung cancers of two KRAS G12C-targeting drugs, sotorasib (27) and adagrasib (28). In the small subset of patients with PDAC (n=12), results of sotorasib were encouraging, with one partial response and nine stable diseases. Despite these encouraging results, their impact is limited given the small proportion of patients with PDAC associated with the G12C mutation in KRAS (1-2%) (27). With ~40% of PDAC cases harboring a KRAS G12D mutation (29), the pending trials of long-awaited KRAS G12D inhibitors should be more promising.
BRAF targeting therapy has changed over time and varies depending on the underlying pathology. Currently, anti-BRAF anti-MEK double blockade is the standard of care. In our study, the patient with a BRAF mutation was treated with only a MEK inhibitor, which can explain the poor PFS observed with this treatment and, consequently, the unimproved OS.
Therefore, unlike other studies highlighting a positive impact of therapies targeting actionable mutations on OS, our study shows no difference in survival curves in the three defined groups.
The absence of liver metastases appears as a good prognostic factor for overall survival, with a decrease in the risk of death of 53% (p = 0.01), independent of group. These results are concordant with the analysis of prognostic factors made in recent randomized studies of pancreatic cancer (4).
Our study has several limitations. First, this is a retrospective and monocentric study. Second, sample sizes were limited, with few patients actually treated with MTT. However, this is attributable to PDAC treatment recommendations as few MTT are approved. MTTs are available for very rare mutations, such as for high; however, the molecular landscape of PDAC is dominated by KRAS mutations, the inhibitors of which are pending. Third, was performed on both primary and metastatic, which might induce heterogeneity in the results. However, this heterogeneity reflects real-life clinical practice, and available tissues for NGS analyses (specific biopsy or archived tissue) were chosen as part of routine therapeutic management with the least risk for the patient. Finally, we did not address impact of liquid biopsy a simple, non-invasive, effective method that is rapidly expanding (30) and might be of interest for PDAC patients. Further exploration of the potential impact and the clinical utility of liquid biopsy will be needed for further PDAC treatment studies.
5. Conclusions
Our study reports real-life precision medicine with outcomes regarding routine in clinical practice. A potential actionable mutation was found in 28% of patients in advanced PDAC, of which 44% received a MMT; however, there was no improvement of OS.
References
- Rawla, P.; Sunkara, T.; Gaduputi, V. Eidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouché, O.; Guimbaud, R.; Bécouarn, Y.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Goldstein, D.; El-Maraghi, R.H.; Hammel, P.; Heinemann, V.; Kunzmann, V.; Sastre, J.; et al. nab-Paclitaxel plus gemcitabine for metastatic pancreatic cancer: long-term survival from a phase III trial. J Natl Cancer Inst. 2015, 107, dju413. [Google Scholar] [CrossRef]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013, 369, 1691–1703. [Google Scholar] [CrossRef]
- Kieler, M.; Unseld, M.; Bianconi, D.; Scheithauer, W.; Prager, G.W. A real-world analysis of second-line treatment options in pancreatic cancer: liposomal-irinotecan plus 5-fluorouracil and folinic acid. Ther Adv Med Oncol. 2019, 11, 1758835919853196. [Google Scholar] [CrossRef]
- Collisson, E.A.; Bailey, P.; Chang, D.K.; Biankin, A.V. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2019, 16, 207–220. [Google Scholar] [CrossRef]
- Aguirre, A.J.; Nowak, J.A.; Camarda, N.D.; Moffitt, R.A.; Ghazani, A.A.; Hazar-Rethinam, M.; et al. Real-time Genomic Characterization of Advanced Pancreatic Cancer to Enable Precision Medicine. Cancer Discov. 2018, 8, 1096–1111. [Google Scholar] [CrossRef]
- Pishvaian, M.J.; Blais, E.M.; Brody, J.R.; Lyons, E.; DeArbeloa, P.; Hendifar, A.; et al. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: a retrospective analysis of the Know Your Tumor registry trial. Lancet Oncol. 2020, 21, 508–518. [Google Scholar] [CrossRef]
- Singhi, A.D.; George, B.; Greenbowe, J.R.; Chung, J.; Suh, J.; Maitra, A.; et al. Real-Time Targeted Genome Profile Analysis of Pancreatic Ductal Adenocarcinomas Identifies Genetic Alterations That Might Be Targeted With Existing Drugs or Used as Biomarkers. Gastroenterology. 2019, 156, 2242–2253. [Google Scholar] [CrossRef] [PubMed]
- Lowery, M.A.; Jordan, E.J.; Basturk, O.; Ptashkin, R.N.; Zehir, A.; Berger, M.F.; et al. Real-Time Genomic Profiling of Pancreatic Ductal Adenocarcinoma: Potential Actionability and Correlation with Clinical Phenotype. Clin Cancer Res. 2017, 23, 6094–6100. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, S.B.; Chang, D.K.; Bailey, P.; Biankin, A.V. Pancreatic Cancer Genomes: Implications for Clinical Management and Therapeutic Development. Clin Cancer Res. 2017, 23, 1638–1646. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Hammel, P.; Reni, M.; Van Cutsem, E.; Macarulla, T.; Hall, M.J.; et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N Engl J Med. 2019, 381, 317–327. [Google Scholar] [CrossRef]
- Massard, C.; Michiels, S.; Ferté, C.; Le Deley, M.C.; Lacroix, L.; Hollebecque, A.; et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov. 2017, 7, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Le Tourneau, C.; Delord, J.P.; Gonçalves, A.; Gavoille, C.; Dubot, C.; Isambert, N.; et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015, 16, 1324–1334. [Google Scholar] [CrossRef]
- Trédan, O.; Wang, Q.; Pissaloux, D.; Cassier, P.; de la Fouchardière, A.; Fayette, J.; et al. Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: analysis from the ProfiLER trial. Ann Oncol. 2019, 30, 757–765. [Google Scholar] [CrossRef]
- Andre, F.; Filleron, T.; Kamal, M.; Mosele, F.; Arnedos, M.; Dalenc, F.; et al. Genomics to select treatment for patients with metastatic breast cancer. Nature. 2022, 610, 343–348. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- Tuxen, I.V.; Rohrberg, K.S.; Oestrup, O.; Ahlborn, L.B.; Schmidt, A.Y.; Spanggaard, I.; et al. Copenhagen Prospective Personalized Oncology (CoPPO)-Clinical Utility of Using Molecular Profiling to Select Patients to Phase I Trials. Clin Cancer Res. 2019, 25, 1239–1247. [Google Scholar] [CrossRef]
- Martin-Romano, P.; Mezquita, L.; Hollebecque, A.; Lacroix, L.; Rouleau, E.; Gazzah, A.; et al. Implementing the European Society for Medical Oncology Scale for Clinical Actionability of Molecular Targets in a Comprehensive Profiling Program: Impact on Precision Medicine Oncology. JCO Precis Oncol. 2100. [Google Scholar]
- Tsimberidou, A.M.; Hong, D.S.; Ye, Y.; Cartwright, C.; Wheler, J.J.; Falchook, G.S.; et al. Initiative for Molecular Profiling and Advanced Cancer Therapy (IMPACT): An MD Anderson Precision Medicine Study. JCO Precis Oncol. 2017, 2017. [Google Scholar]
- Schwaederle, M.; Zhao, M.; Lee, J.J.; Eggermont, A.M.; Schilsky, R.L.; Mendelsohn, J.; et al. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. J Clin Oncol. 2015, 33, 3817–3825. [Google Scholar] [CrossRef] [PubMed]
- Massard, C.; Michiels, S.; Ferté, C.; Le Deley, M.C.; Lacroix, L.; Hollebecque, A.; et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: Results of the MOSCATO 01 trial. Cancer Discovery. 2017, 7, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Chakravarty, D.; Dienstmann, R.; Jezdic, S.; Gonzalez-Perez, A.; Lopez-Bigas, N.; et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann Oncol. 2018, 29, 1895–1902. [Google Scholar] [CrossRef]
- Cobain, E.F.; Wu, Y.M.; Vats, P.; Chugh, R.; Worden, F.; Smith, D.C.; et al. Assessment of Clinical Benefit of Integrative Genomic Profiling in Advanced Solid Tumors. JAMA Oncol. 2021, 7, 525–533. [Google Scholar] [CrossRef]
- Hong, D.S.; Fakih, M.G.; Strickler, J.H.; Desai, J.; Durm, G.A.; Shapiro, G.I.; et al. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med. 2020, 383, 1207–1217. [Google Scholar] [CrossRef]
- Si, O.; Pa, J.; Ta, L.; Ii, R.; Jk, S.; Ma, B.; et al. First-in-Human Phase I/IB Dose-Finding Study of Adagrasib (MRTX849) in Patients With Advanced KRASG12C Solid Tumors (KRYSTAL-1). Journal of clinical oncology : official journal of the American Society of Clinical Oncology. Available online: https://pubmed.ncbi.nlm.nih.gov/35167329/ (accessed on 6 December 2022).
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb Perspect Med. 2018, 8, a031435. [Google Scholar] [CrossRef]
- Bayle, A.; Peyraud, F.; Belcaid, L.; Brunet, M.; Aldea, M.; Clodion, R.; et al. Liquid versus tissue biopsy for detecting actionable alterations according to ESCAT in patients with advanced cancer: A study from the French National Center for Precision Medicine (PRISM). Ann Oncol. 2022, 33, S0923–7534. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).