Submitted:
10 August 2023
Posted:
11 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
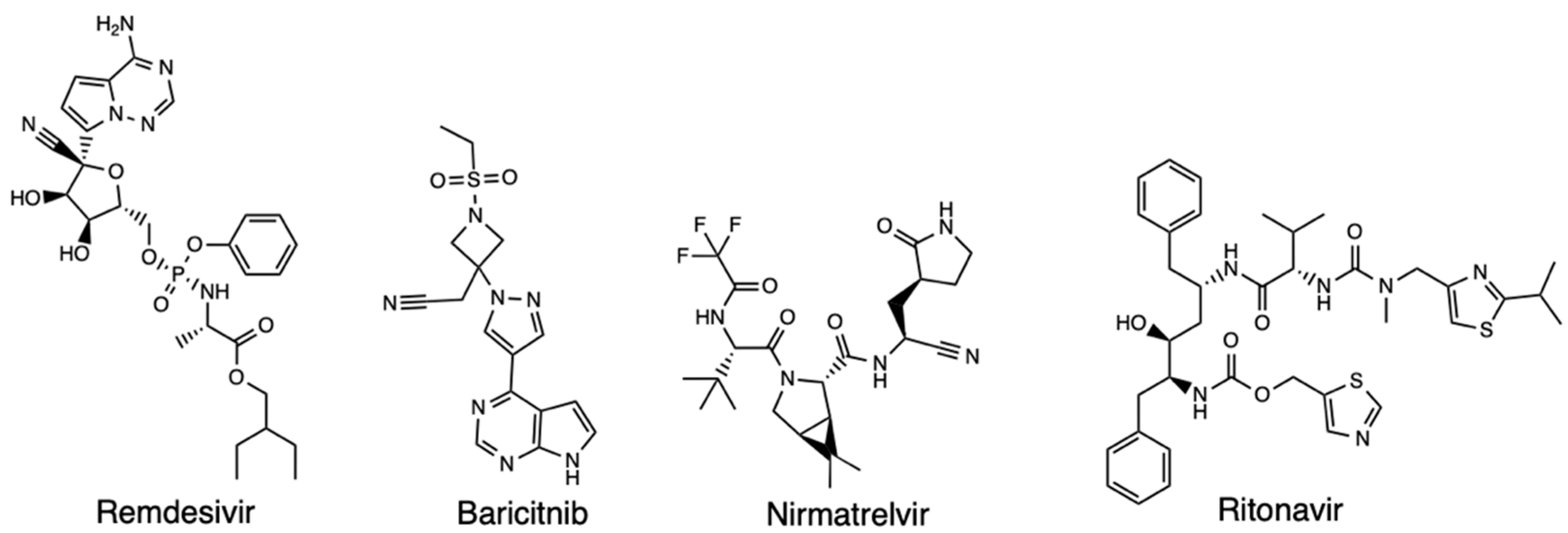
2. Computational Discovery of Covid-19 Therapeutics
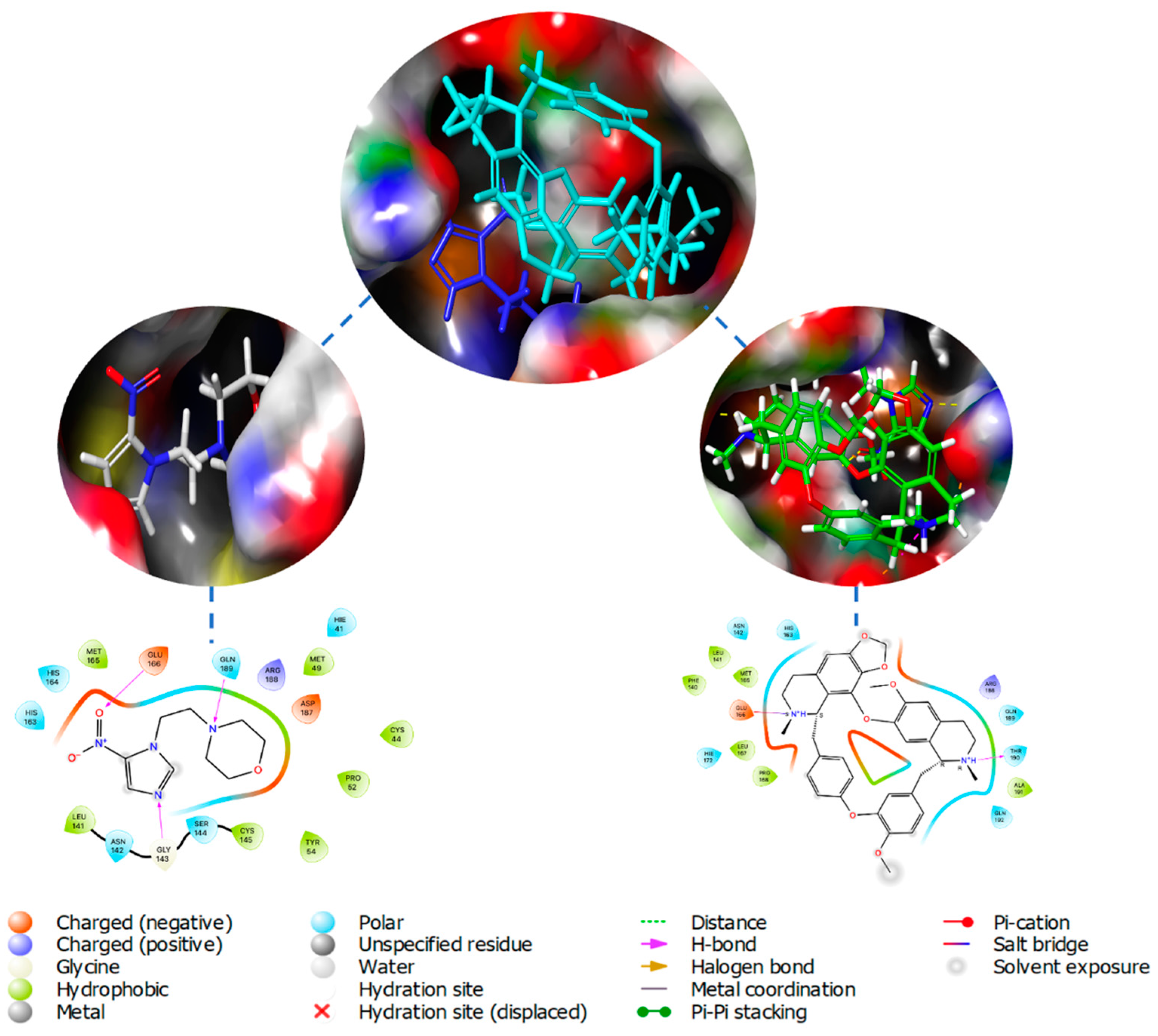
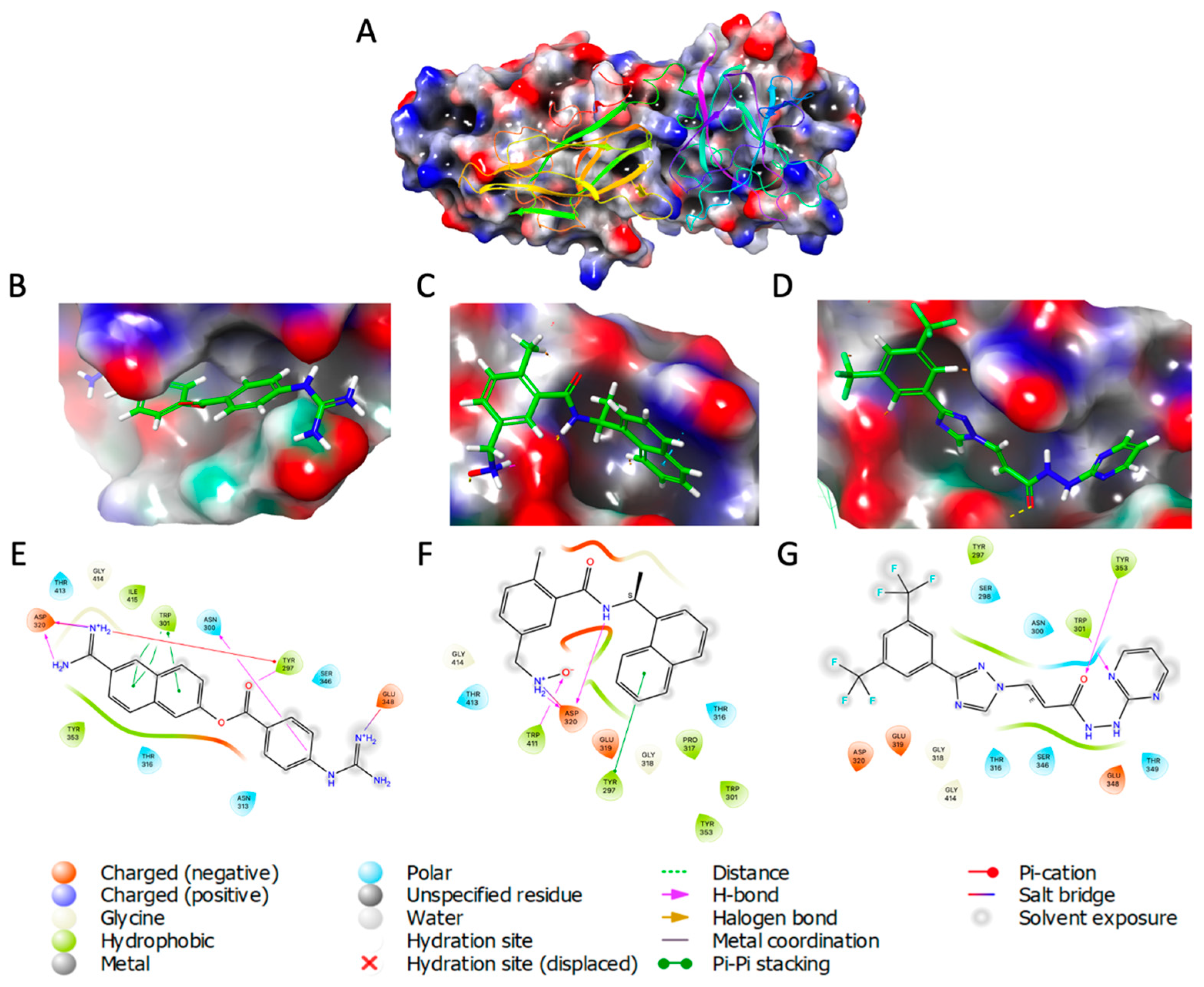
2.1. Molecular Docking
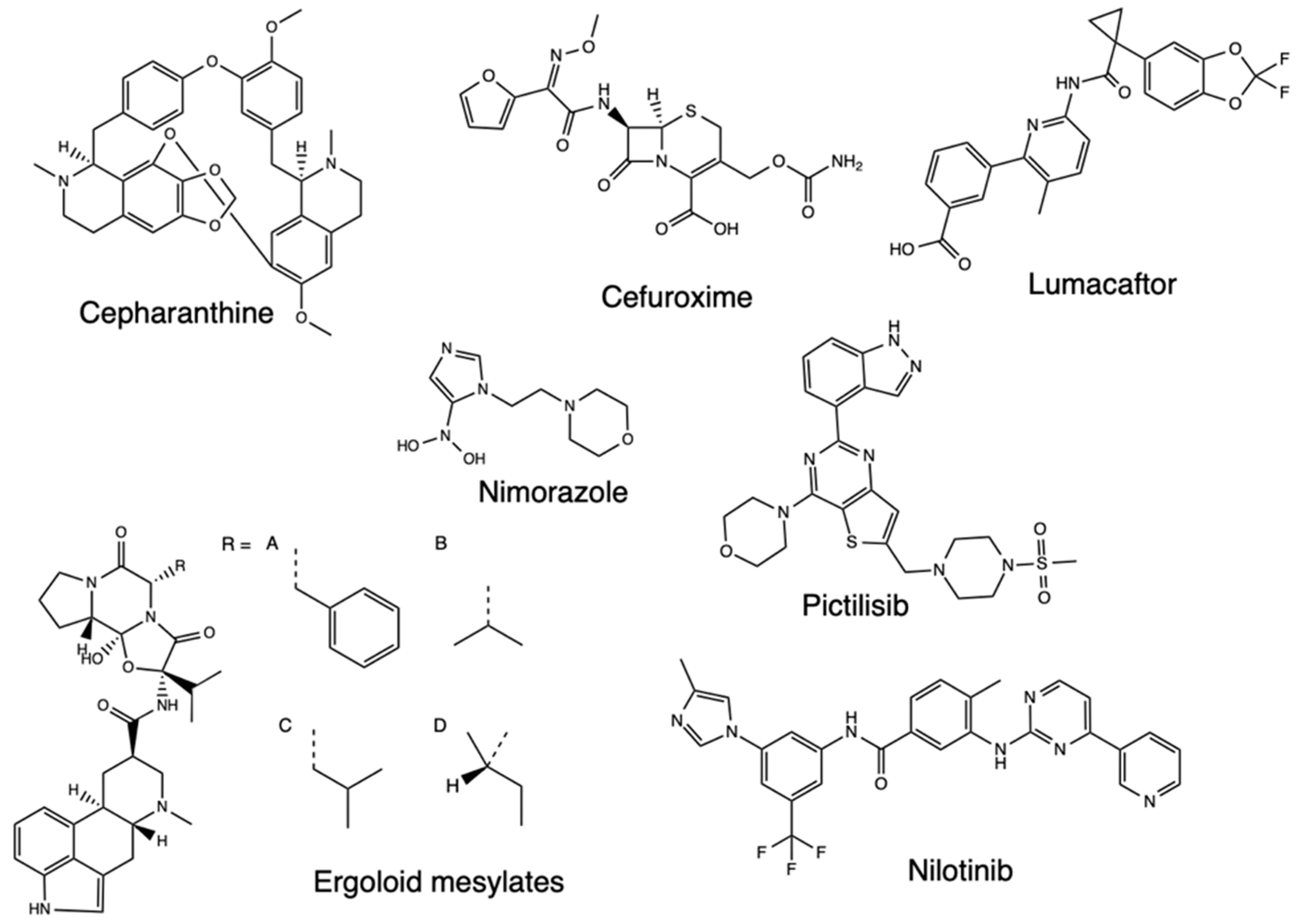
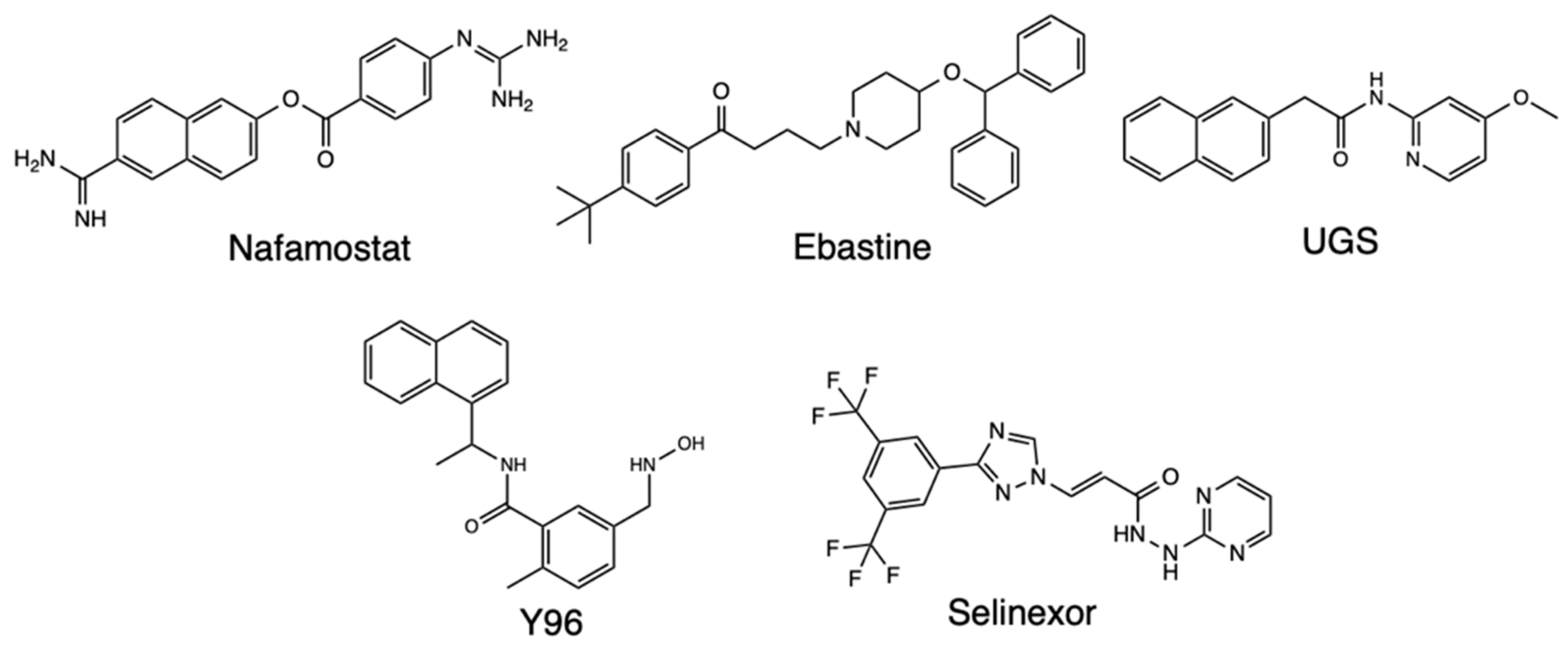
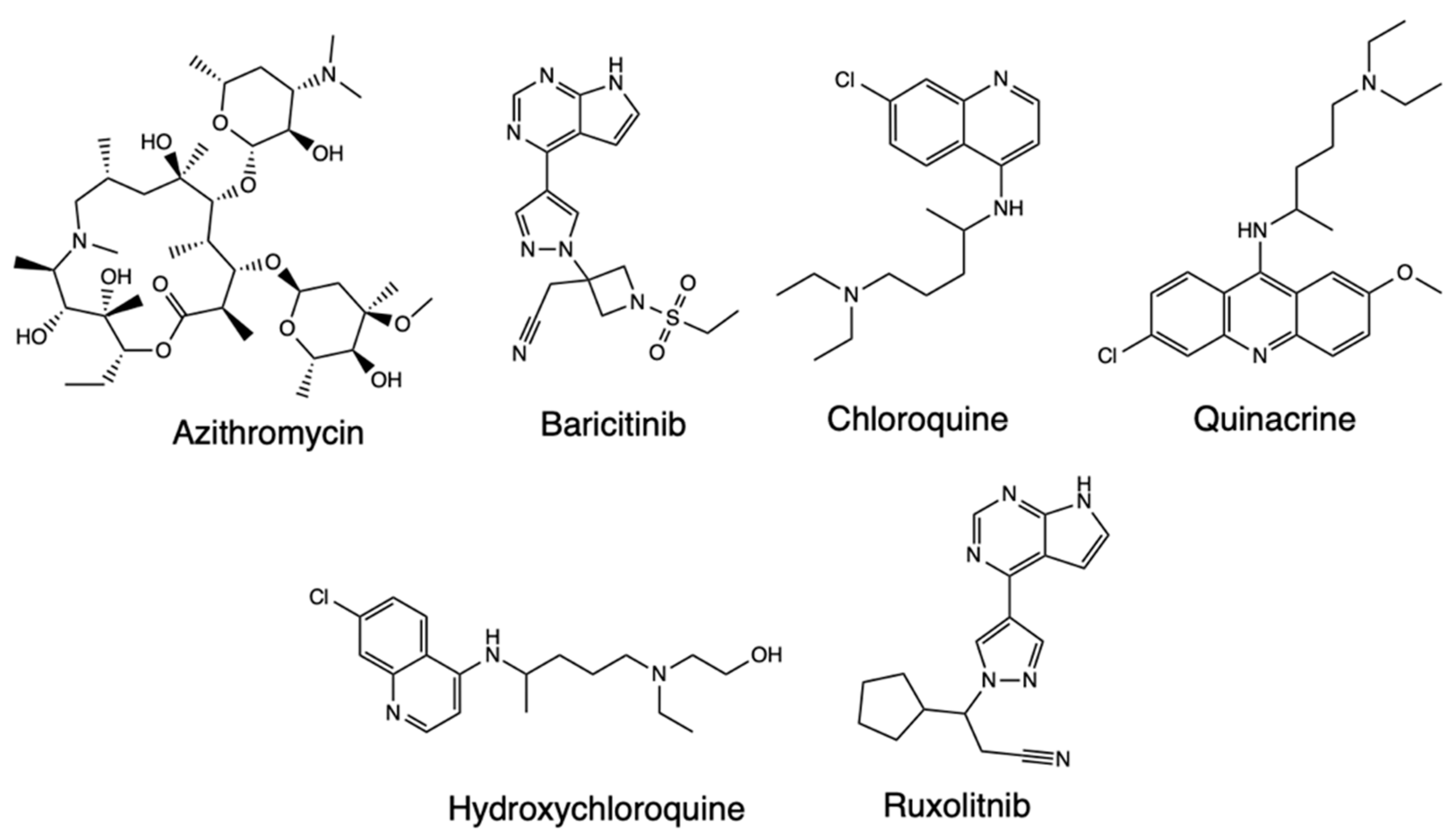
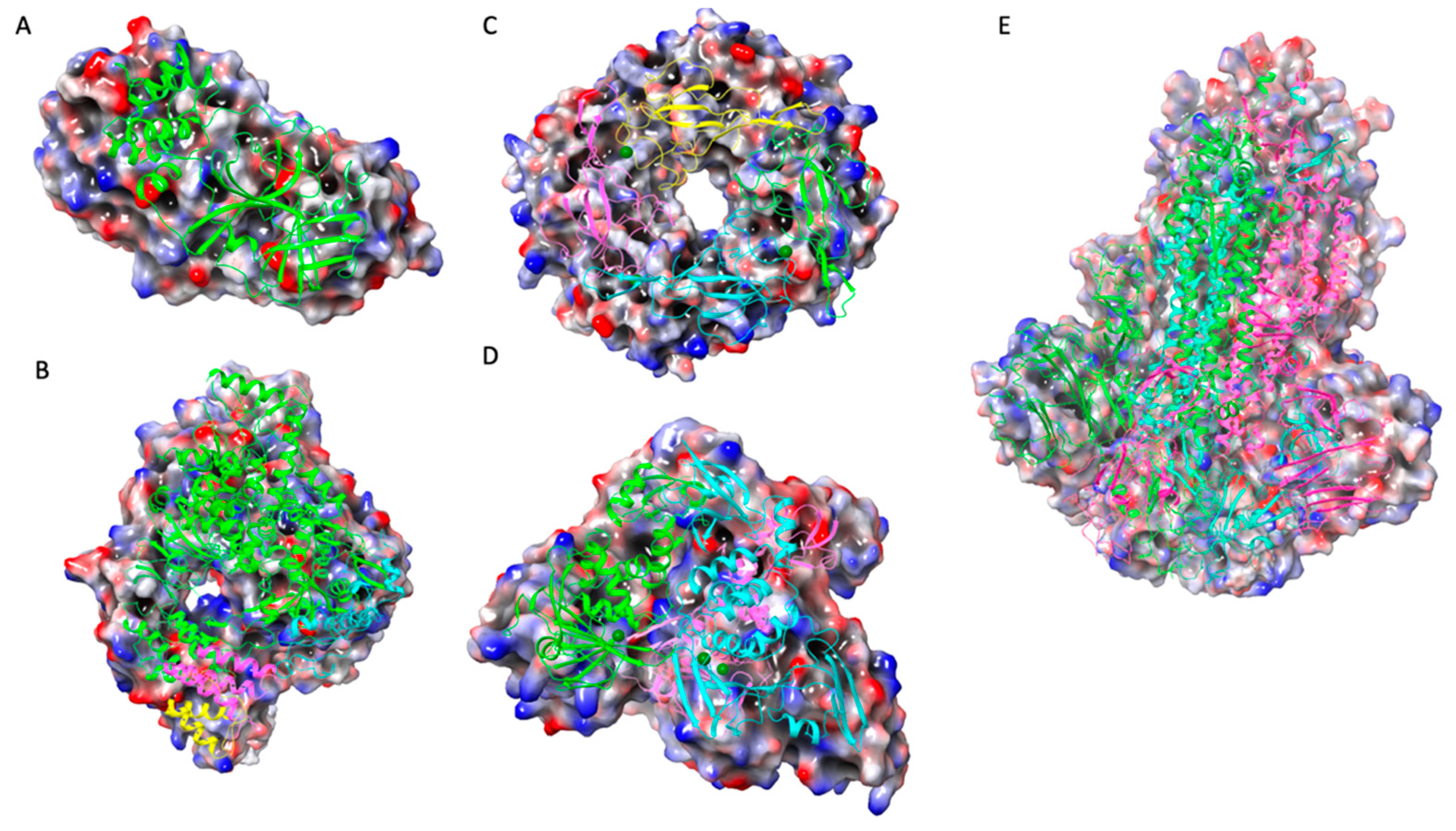
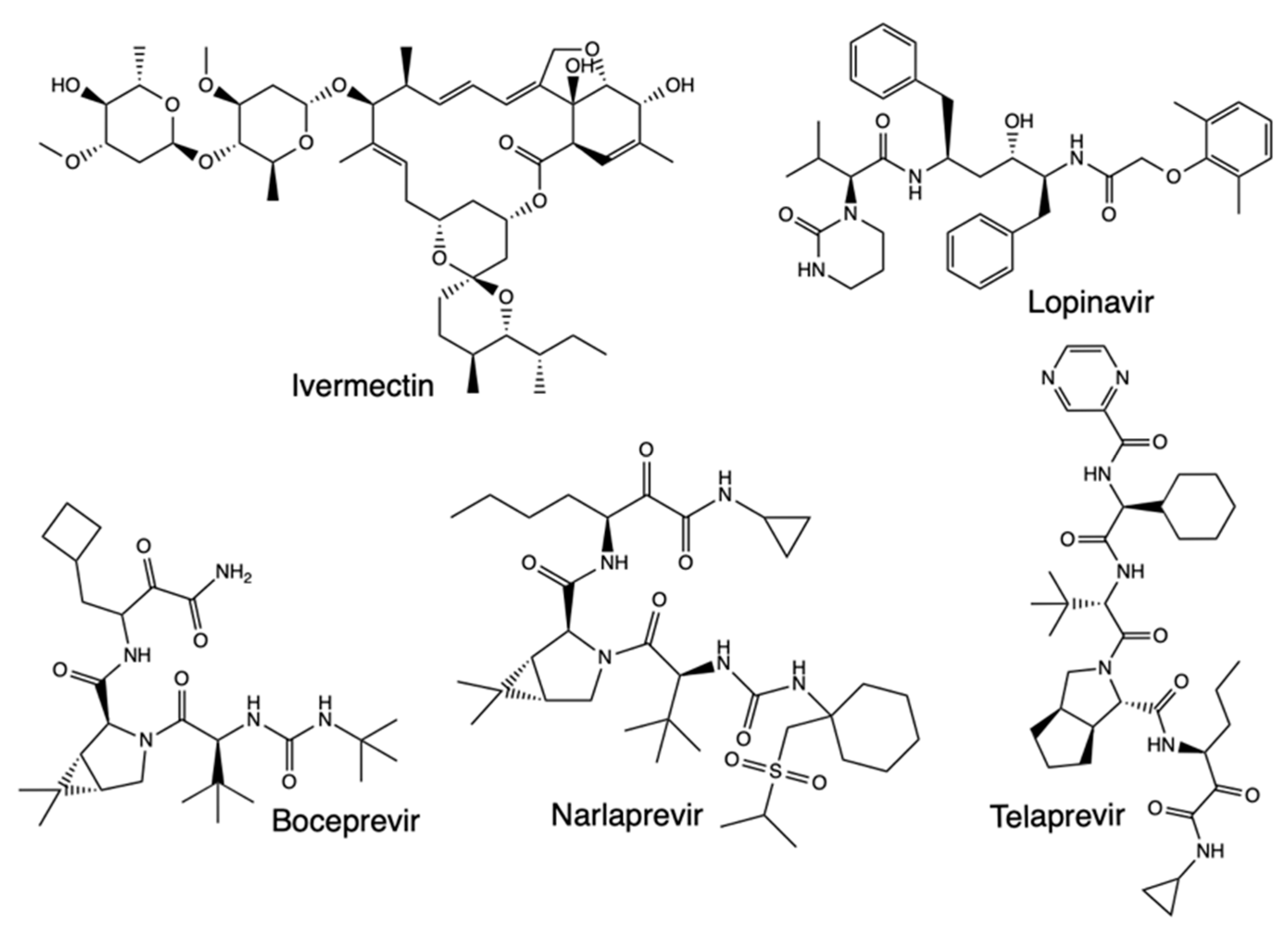
2.2. Quantitative Structure-Activity Relationships (QSAR)
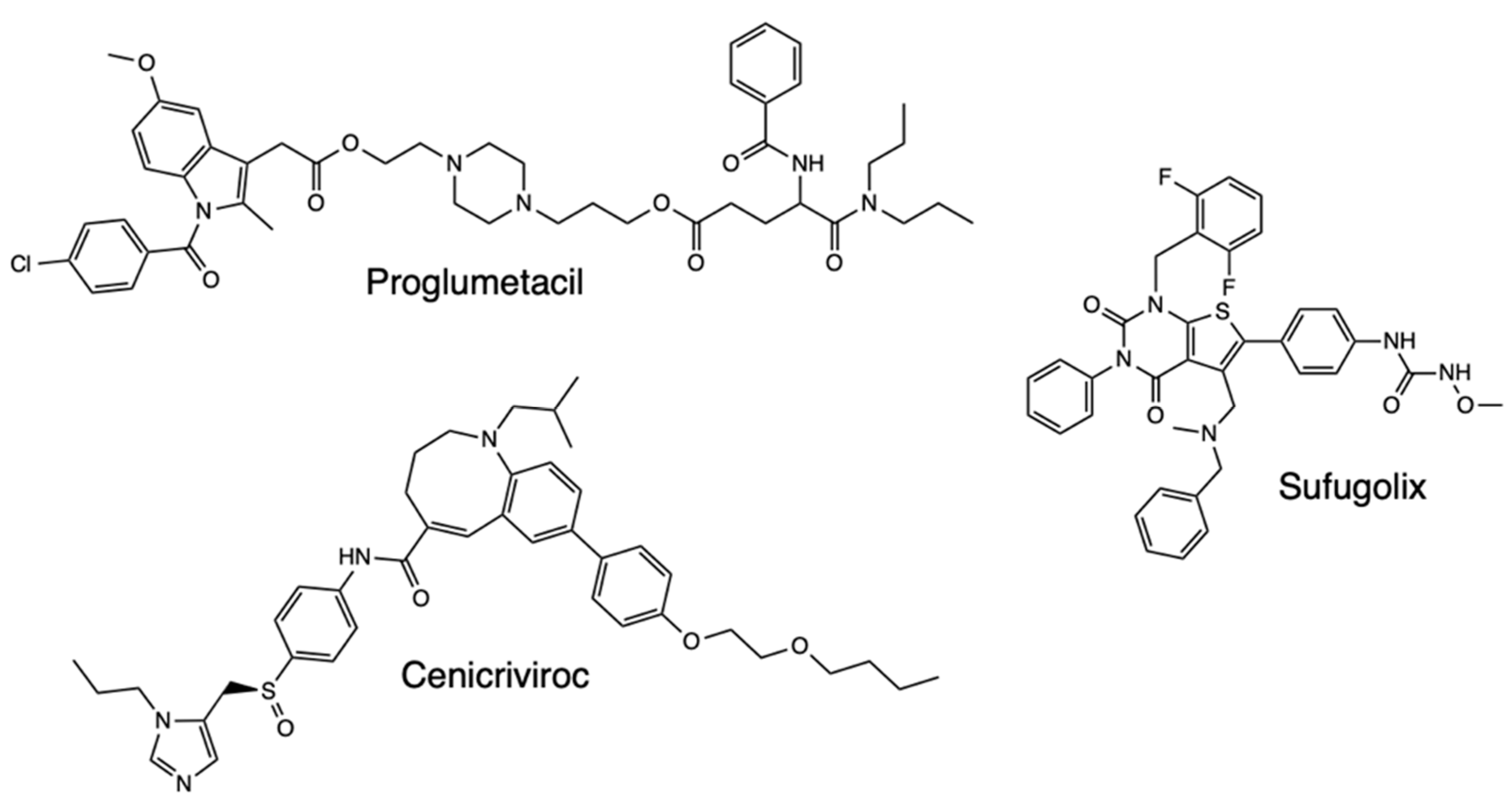
2.3. Molecular Dynamics (MD) Simulations
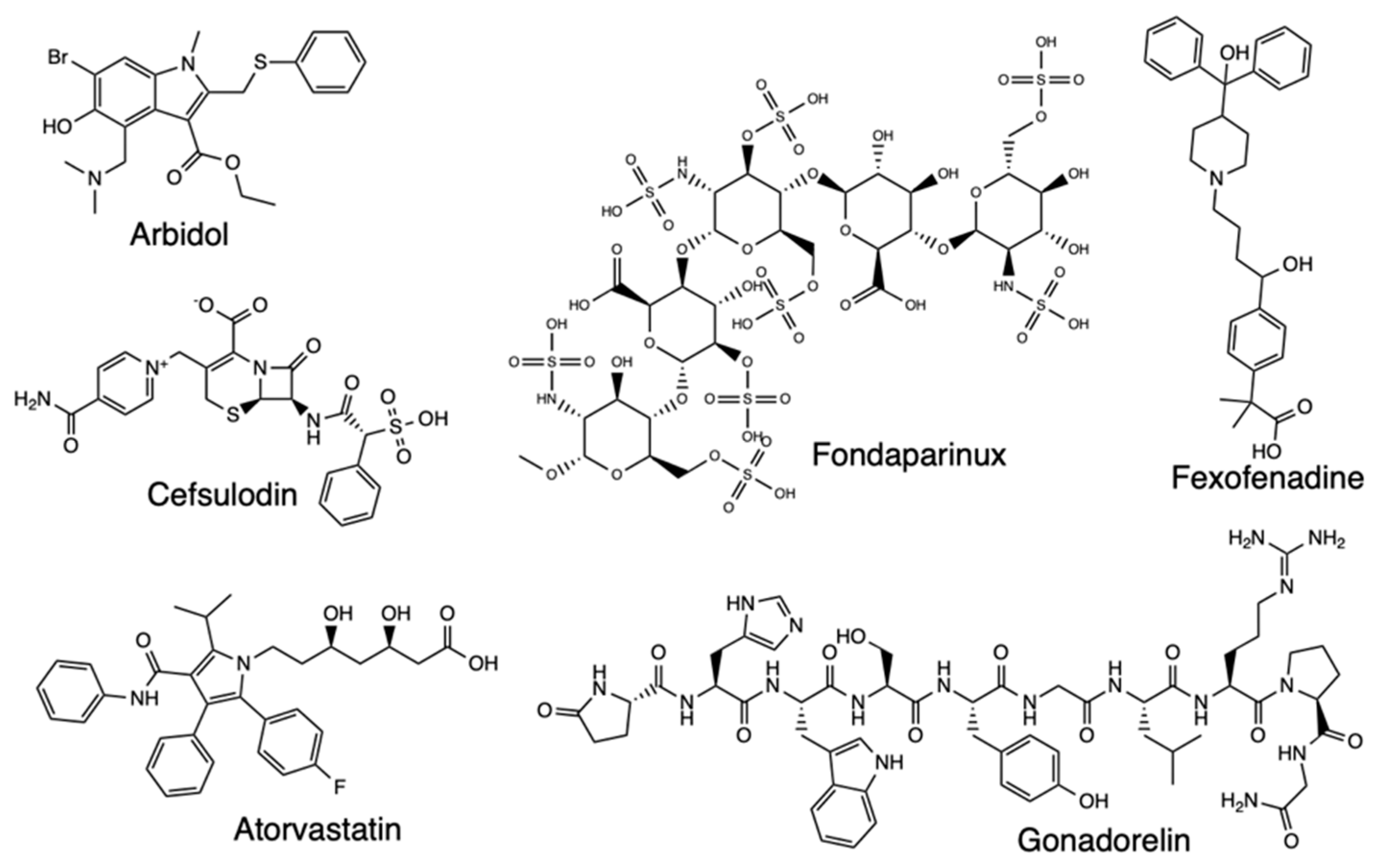
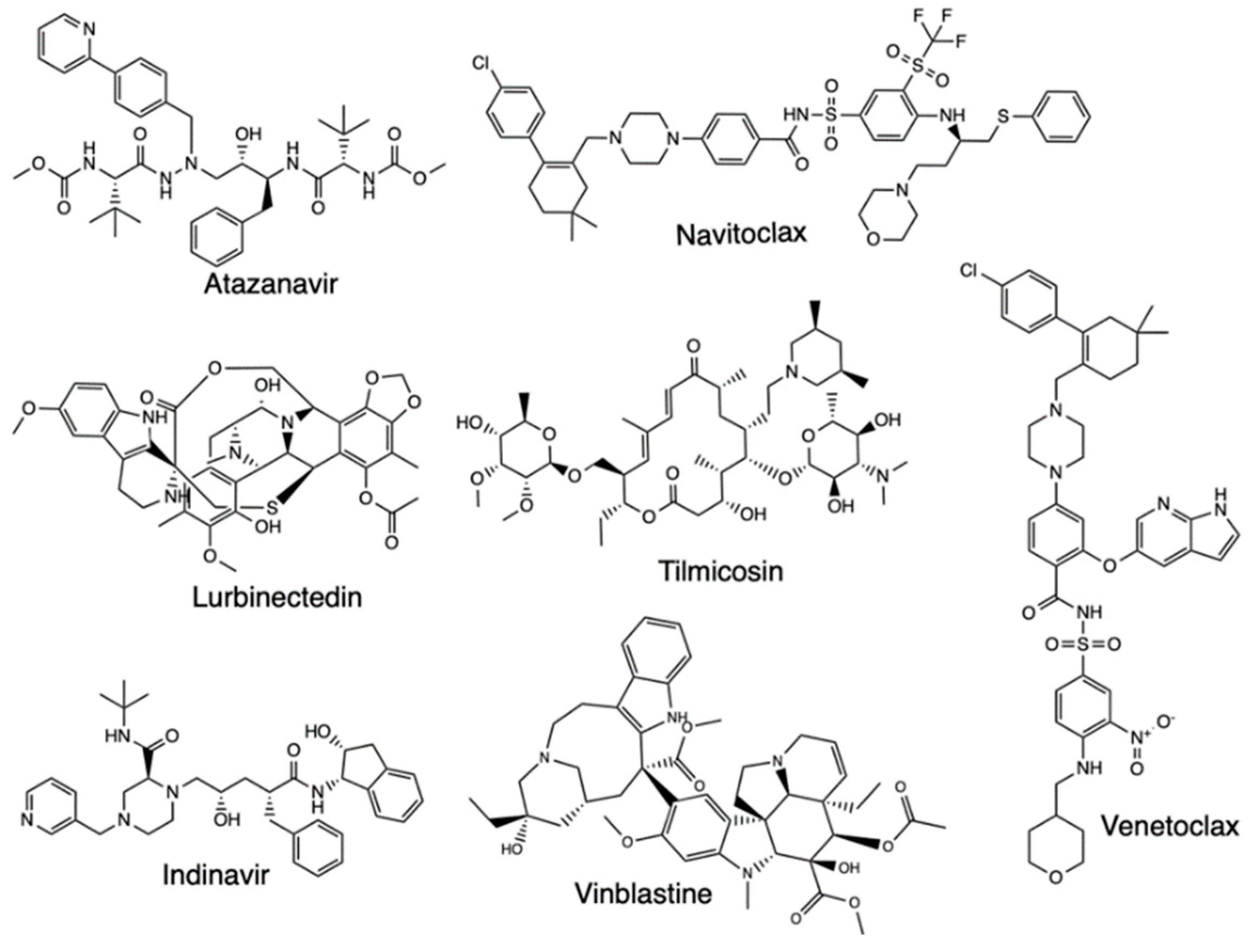
2.4. Pharmacophore Modeling
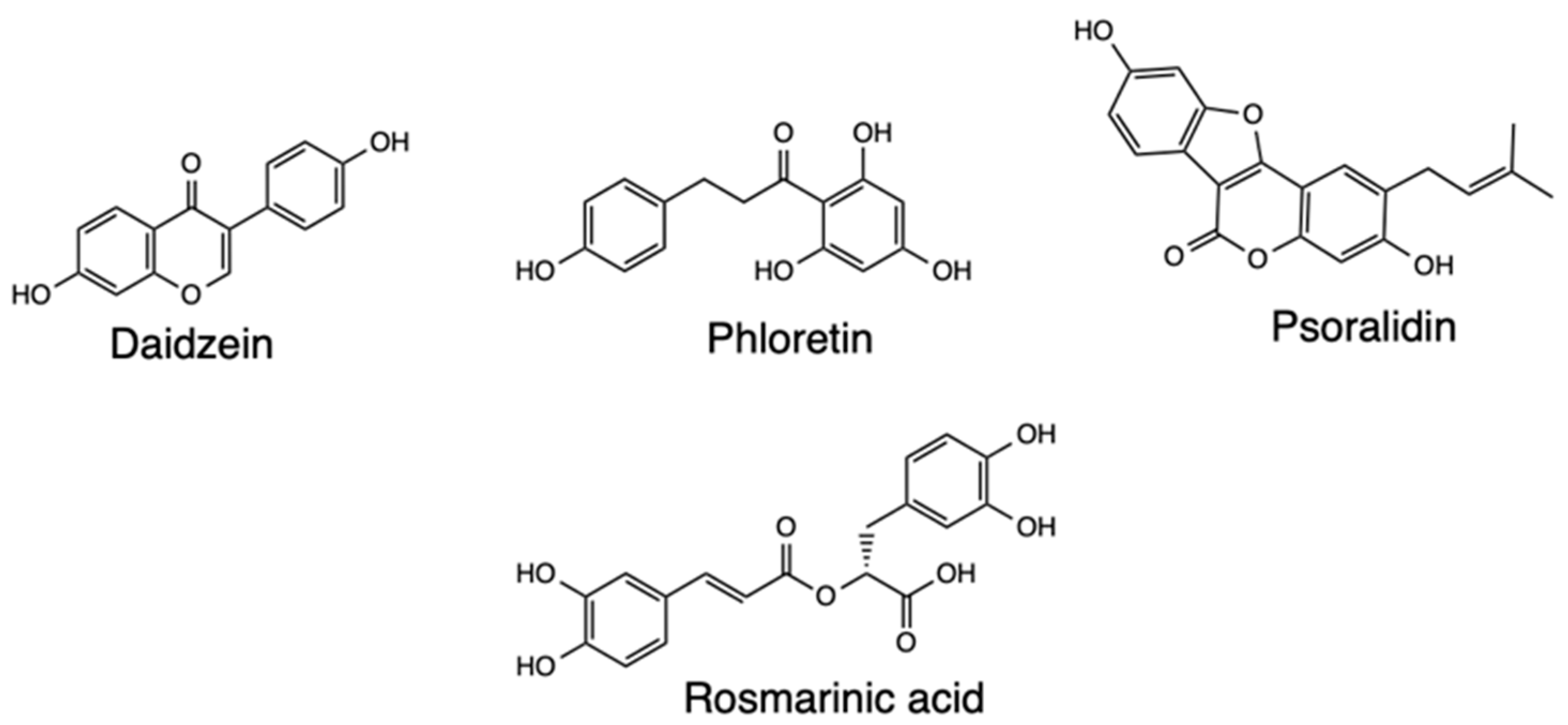
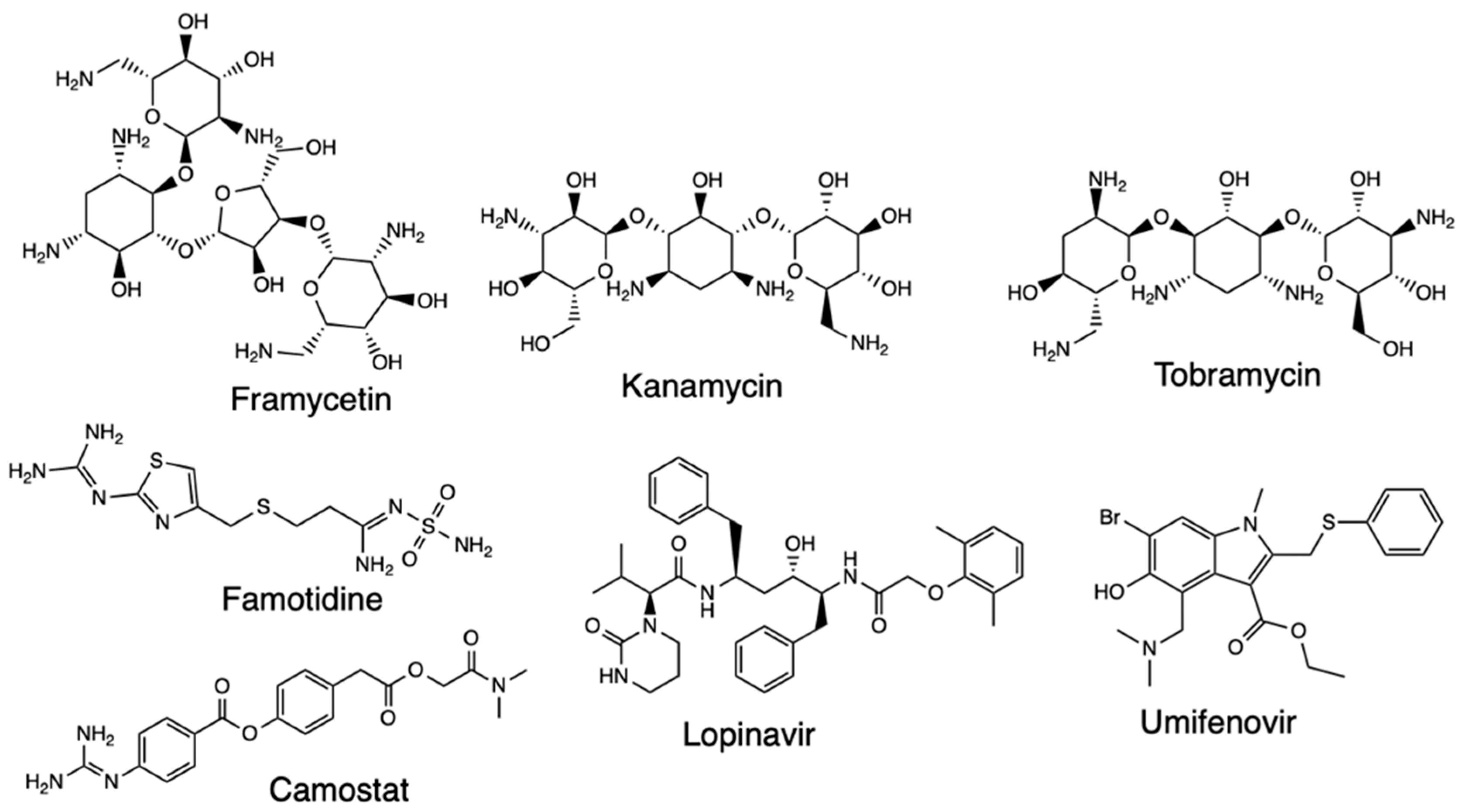
2.5. Homology Modeling
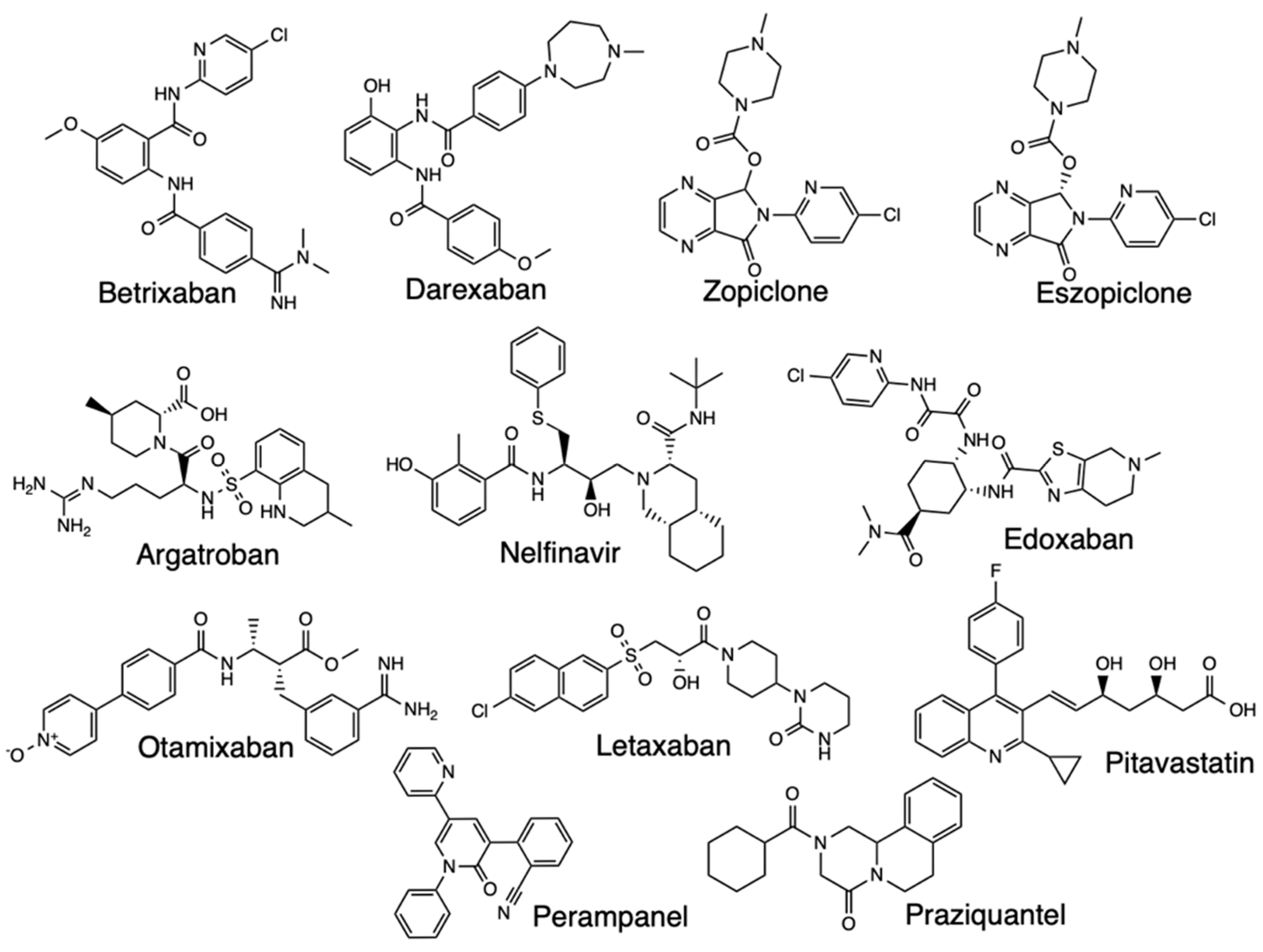
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EUA Emergency Use Authorization |
| CADD Computer-aided Drug Design |
| WHO World Health Organization |
| FDA Food and Drug Administration |
| NIAID National Institute for Allergy and Infectious Diseases |
| RdRp RNA-dependent RNA polymerase |
| 3D Three-dimensional |
| PDB Protein Data Bank |
| SBDD Structure-based drug design |
| LBDD Ligand-based drug design |
| Mpro Main protease |
| QSAR Quantitative structure-activity relationships |
| VS Virtual screening |
| NRP-1 Neuropilin-1 |
| ADT Auto Dock Tools |
| nB0 Non-H bond |
| SNar Narumi simple topological index |
| nHAcc Number of hydrogen bond acceptors |
| SMILES Simplified molecular-input line-entry system |
| 3CLpro 3C-like protease |
| RMSD Root Mean Square Deviation |
| MD Molecular Dynamic |
| ACE2 Angiotensin-converting enzyme 2 |
| hACE2 Human angiotensin-converting enzyme 2 |
References
- Kalita, P., Tripathi, T. and Padhi, A.K., 2023. Computational Protein Design for COVID-19 Research and Emerging Therapeutics. ACS Central Science, 9(4), pp.602-613. [CrossRef]
- Cascella, M., Rajnik, M., Aleem, A., Dulebohn, S. and Di Napoli, R., 2023. Features, evaluation, and treatment of coronavirus (COVID-19). StatPearls.
- Niknam, Z., Jafari, A., Golchin, A., Danesh Pouya, F., Nemati, M., Rezaei-Tavirani, M. and Rasmi, Y., 2022. Potential therapeutic options for COVID-19: an update on current evidence. European Journal of Medical Research, 27, pp.1-15. [CrossRef]
- Chera, A. and Tanca, A., 2022. Remdesivir: the first FDA-approved anti-COVID-19 Treatment for Young Children. Discoveries, 10(2). [CrossRef]
- Beigel, J.H., Tomashek, K.M., Dodd, L.E., Mehta, A.K., Zingman, B.S., Kalil, A.C., Hohmann, E., Chu, H.Y., Luetkemeyer, A., Kline, S. and Lopez de Castilla, D., 2020. Remdesivir for the treatment of Covid-19—Final Report. New England Journal of Medicine, 383(19), pp.1813-1826.
- Rezagholizadeh, A., Khiali, S., Sarbakhsh, P. and Entezari-Maleki, T., 2021. Remdesivir for treatment of COVID-19; An updated systematic review and meta-analysis. European Journal of Pharmacology, 897, p.173926. [CrossRef]
- Abd-Elsalam, S., Salama, M., Soliman, S., Naguib, A.M., Ibrahim, I.S., Torky, M., Abd El Ghafar, M.S., Abdul-Baki, E.A.R.M. and Elhendawy, M., 2022. Remdesivir efficacy in COVID-19 treatment: A randomized controlled trial. The American Journal of Tropical Medicine and Hygiene, 106(3), p.886. [CrossRef]
- Gupte, V., Hegde, R., Sawant, S., Kalathingal, K., Jadhav, S., Malabade, R. and Gogtay, J., 2022. Safety and clinical outcomes of remdesivir in hospitalised COVID-19 patients: a retrospective analysis of active surveillance database. BMC Infectious Diseases, 22(1), p.1. [CrossRef]
- WHO Solidarity Trial Consortium, 2022. Remdesivir and three other drugs for hospitalised patients with COVID-19: Final results of the WHO Solidarity randomised trial and updated meta-analyses. The Lancet, 399(10339), pp.1941-1953. [CrossRef]
- Balakrishnan, V. and Lakshminarayanan, K., 2021. Screening of FDA approved drugs against SARS-CoV-2 main protease: coronavirus disease. International journal of peptide research and therapeutics, 27(1), pp.651-658.
- US Food and Drug Administration (FDA), 2023, June 28. Know your treatment options for Covid-19. https://www.fda.gov/consumers/consumer-updates/know-your-treatment-options-covid-19.
- Toussi, S.S., Hammond, J.L., Gerstenberger, B.S. and Anderson, A.S., 2023. Therapeutics for COVID-19. Nature Microbiology, 8(5), pp.771-786.
- Muratov, E.N., Amaro, R., Andrade, C.H., Brown, N., Ekins, S., Fourches, D., Isayev, O., Kozakov, D., Medina-Franco, J.L., Merz, K.M. and Oprea, T.I., 2021. A critical overview of computational approaches employed for COVID-19 drug discovery. Chemical Society Reviews, 50(16), pp.9121-9151.
- Saloni, Kumari, D., Ranjan, P. and Chakraborty, T., 2022. A computational study of potential therapeutics for COVID-19 invoking conceptual density functional theory. Structural Chemistry, 33(6), pp.2195-2204. [CrossRef]
- Napolitano, F., Xu, X. and Gao, X., 2022. Impact of computational approaches in the fight against COVID-19: An AI guided review of 17 000 studies. Briefings in bioinformatics, 23(1), p.bbab456. [CrossRef]
- Ghosh, D., Ghosh Dastidar, D., Roy, K., Ghosh, A., Mukhopadhyay, D., Sikdar, N., Biswas, N.K., Chakrabarti, G. and Das, A., 2022. Computational prediction of the molecular mechanism of statin group of drugs against SARS-CoV-2 pathogenesis. Scientific Reports, 12(1), p.6241.
- Singh, P.K., Pathania, S. and Rawal, R.K., 2020. Exploring RdRp–remdesivir interactions to screen RdRp inhibitors for the management of novel coronavirus 2019-nCoV. SAR and QSAR in Environmental Research, 31(11), pp.857-867. [CrossRef]
- Chang, C.C., Hsu, H.J., Wu, T.Y. and Liou, J.W., 2022. Computer-aided discovery, design, and investigation of COVID-19 therapeutics. Tzu-Chi Medical Journal, 34(3), p.276. [CrossRef]
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X. and Zheng, M., 2020. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10(5), pp.766-788.
- Dong, J., Varbanov, M., Philippot, S., Vreken, F., Zeng, W.B. and Blay, V., 2023. Ligand-based discovery of coronavirus main protease inhibitors using MACAW molecular embeddings. Journal of Enzyme Inhibition and Medicinal Chemistry, 38(1), pp.24-35. [CrossRef]
- Sharma, V., Wakode, S. and Kumar, H., 2021. Structure-and ligand-based drug design: Concepts, approaches, and challenges. Chemoinformatics and bioinformatics in the pharmaceutical sciences, pp.27-53.
- Alméciga-Díaz, C.J., Pimentel-Vera, L.N., Caro, A., Mosquera, A., Castellanos Moreno, C.A., Manosalva Rojas, J.P. and Díaz-Tribaldos, D.C., 2020. Virtual screening of potential inhibitors for SARS-CoV-2 main protease. Preprints.org, . [CrossRef]
- Charoute, H., Elkarhat, Z., Elkhattabi, L., El Fahime, E., Oukkache, N., Rouba, H. and Barakat, A., 2022. Computational screening of potential drugs against COVID-19 disease: The Neuropilin-1 receptor as molecular target. VirusDisease, 33(1), pp.23-31. [CrossRef]
- Marinho, E.M., de Andrade Neto, J.B., Silva, J., da Silva, C.R., Cavalcanti, B.C., Marinho, E.S. and Júnior, H.V.N., 2020. Virtual screening based on molecular docking of possible inhibitors of Covid-19 main protease. Microbial Pathogenesis, 148, p.104365.
- Eweas, A.F., Alhossary, A.A. and Abdel-Moneim, A.S., 2021. Molecular docking reveals ivermectin and remdesivir as potential repurposed drugs against SARS-CoV-2. Frontiers in Microbiology, 11, p.3602.
- Gholivand, K., Barzegari, A., Mohammadpanah, F., Yaghoubi, R., Roohzadeh, R. and Valmoozi, A.A.E., 2022. Synthesis, characterized, QSAR studies and molecular docking of some phosphonates as COVID-19 inhibitors. Polyhedron, 221, p.115824. [CrossRef]
- Costa, A.S., Martins, J.P.A. and de Melo, E.B., 2022. SMILES-based 2D-QSAR and similarity search for identification of potential new scaffolds for development of SARS-CoV-2 MPRO inhibitors. Structural Chemistry, 33(5), pp.1691-1706.
- Qiao, J., Li, Y.S., Zeng, R., Liu, F.L., Luo, R.H., Huang, C., Wang, Y.F., Zhang, J., Quan, B., Shen, C. and Mao, X., 2021. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science, 371(6536), pp.1374-1378.
- Butt, A.A. and Kanwal, F., 2012. Boceprevir and telaprevir in the management of hepatitis C virus–infected patients. Clinical infectious diseases, 54(1), pp.96-104. [CrossRef]
- Li, L. and Huang, S., 2021. Newly synthesized Mpro inhibitors as potential oral anti-SARS-CoV-2 agents. Signal Transduction and Targeted Therapy, 6(1), p.138.
- Fischer, C. and Feys, J.R., 2023. SARS-CoV-2 Mpro inhibitors: Achieved diversity, developing resistance and future strategies. Future Pharmacology, 3(1), pp.80-107.
- La Monica, G., Bono, A., Lauria, A. and Martorana, A., 2022. Targeting SARS-CoV-2 main protease for treatment of COVID-19: Covalent inhibitors structure–activity relationship insights and evolution perspectives. Journal of medicinal chemistry, 65(19), pp.12500-12534.
- Alves, V.M., Bobrowski, T., Melo-Filho, C.C., Korn, D., Auerbach, S., Schmitt, C., Muratov, E.N. and Tropsha, A., 2021. QSAR modeling of SARS-CoV Mpro inhibitors identifies sufugolix, cenicriviroc, proglumetacin, and other drugs as candidates for repurposing against SARS-CoV-2. Molecular informatics, 40(1), p.2000113.
- Ničkčović, V.P., Nikolić, G.R., Nedeljković, B.M., Mitić, N., Danić, S.F., Mitić, J., Marčetić, Z., Sokolović, D. and Veselinović, A.M., 2022. In silico approach for the development of novel antiviral compounds based on SARS-COV-2 protease inhibition. Chemical Papers, 76(7), pp.4393-4404. [CrossRef]
- Sharma S., Khan Y., Saini K., Bhatia V., 2022). Role of QSAR in filling in the gaps of CoVID-19 Therapeutics. Chem Informatics, 8 (5).
- Oubahmane, M., Hdoufane, I., Delaite, C., Sayede, A., Cherqaoui, D. and El Allali, A., 2023. Design of Potent Inhibitors Targeting the Main Protease of SARS-CoV-2 Using QSAR Modeling, Molecular Docking, and Molecular Dynamics Simulations. Pharmaceuticals, 16(4), p.608.
- Arun, K.G., Sharanya, C.S., Abhithaj, J., Francis, D. and Sadasivan, C., 2021. Drug repurposing against SARS-CoV-2 using E-pharmacophore based virtual screening, molecular docking and molecular dynamics with main protease as the target. Journal of Biomolecular Structure and Dynamics, 39(13), pp.4647-4658.
- Maiorov, V.N. and Crippen, G.M., 1994. Significance of root-mean-square deviation in comparing three-dimensional structures of globular proteins. Journal of molecular biology, 235(2), pp.625-634. [CrossRef]
- Cao, J.F., Gong, Y., Wu, M., Yang, X., Xiong, L., Chen, S., Xiao, Z., Li, Y., Zhang, L., Zan, W. and Zhang, X., 2022. Exploring the mechanism of action of licorice in the treatment of COVID-19 through bioinformatics analysis and molecular dynamics simulation. Frontiers in pharmacology, 13, p.1003310. [CrossRef]
- Kumar, S., Sharma, P.P., Shankar, U., Kumar, D., Joshi, S.K., Pena, L., Durvasula, R., Kumar, A., Kempaiah, P., Poonam and Rathi, B., 2020. Discovery of new hydroxyethylamine analogs against 3CLpro protein target of SARS-CoV-2: Molecular docking, molecular dynamics simulation, and structure–activity relationship studies. Journal of Chemical Information and Modeling, 60(12), pp.5754-5770.
- Rudrapal, M., Celik, I., Khan, J., Ansari, M.A., Alomary, M.N., Alatawi, F.A., Yadav, R., Sharma, T., Tallei, T.E., Pasala, P.K. and Sahoo, R.K., 2022. Identification of bioactive molecules from Triphala (Ayurvedic herbal formulation) as potential inhibitors of SARS-CoV-2 main protease (Mpro) through computational investigations. Journal of King Saud University-Science, 34(3), p.101826.
- Nutho, B., Mahalapbutr, P., Hengphasatporn, K., Pattaranggoon, N.C., Simanon, N., Shigeta, Y., Hannongbua, S. and Rungrotmongkol, T., 2020. Why are lopinavir and ritonavir effective against the newly emerged coronavirus 2019? Atomistic insights into the inhibitory mechanisms. Biochemistry, 59(18), pp.1769-1779. [CrossRef]
- Padhi, A.K., Seal, A., Khan, J.M., Ahamed, M. and Tripathi, T., 2021. Unraveling the mechanism of arbidol binding and inhibition of SARS-CoV-2: Insights from atomistic simulations. European journal of pharmacology, 894, p.173836.
- Vankadari, N., 2020. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. International Journal of Antimicrobial Agents, 56(2), p.105998.
- Deganutti, G., Prischi, F. and Reynolds, C.A., 2021. Supervised molecular dynamics for exploring the druggability of the SARS-CoV-2 spike protein. Journal of computer-aided molecular design, 35, pp.195-207.
- Razizadeh, M., Nikfar, M. and Liu, Y., 2021. Small molecule therapeutics to destabilize the ACE2-RBD complex: A molecular dynamics study. Biophysical Journal, 120(14), pp.2793-2804. [CrossRef]
- Kumar, V., Liu, H. and Wu, C., 2021. Drug repurposing against SARS-CoV-2 receptor binding domain using ensemble-based virtual screening and molecular dynamics simulations. Computers in Biology and Medicine, 135, p.104634.
- Pirolli, D., Righino, B., Camponeschi, C., Ria, F., Di Sante, G. and De Rosa, M.C., 2023. Virtual screening and molecular dynamics simulations provide insight into repurposing drugs against SARS-CoV-2 variants Spike protein/ACE2 interface. Scientific Reports, 13(1), p.1494.
- Al-Karmalawy, A.A., Dahab, M.A., Metwaly, A.M., Elhady, S.S., Elkaeed, E.B., Eissa, I.H. and Darwish, K.M., 2021. Molecular docking and dynamics simulation revealed the potential inhibitory activity of ACEIs against SARS-CoV-2 targeting the h ACE2 receptor. Frontiers in Chemistry, 9, p.661230.
- Saeed, M., Saeed, A., Alam, M.J. and Alreshidi, M., 2021. Receptor-based pharmacophore modeling in the search for natural products for COVID-19 Mpro. Molecules, 26(6), p.1549. [CrossRef]
- Rampogu, S. and Lee, K.W., 2021. Pharmacophore modelling-based drug repurposing approaches for SARS-CoV-2 therapeutics. Frontiers in Chemistry, 9, p.636362.π.
- Aldahham, B.J., Al-Khafaji, K., Saleh, M.Y., Abdelhakem, A.M., Alanazi, A.M. and Islam, M.A., 2022. Identification of naphthyridine and quinoline derivatives as potential Nsp16-Nsp10 inhibitors: a pharmacoinformatics study. Journal of Biomolecular Structure and Dynamics, 40(9), pp.3899-3906. [CrossRef]
- Das, S., Sarmah, S., Lyndem, S. and Singha Roy, A., 2021. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 39(9), pp.3347-3357.
- Pandit, N.K., Mann, S.S., Mohanty, A. and Meena, S.S., 2023. e-Pharmacophore modeling and in silico study of CD147 receptor against SARS-CoV-2 drugs. Genomics & Informatics, 21(2).
- Halimi, M. and Bararpour, P., 2022. Natural inhibitors of SARS-CoV-2 main protease: structure based pharmacophore modeling, molecular docking and molecular dynamic simulation studies. Journal of Molecular Modeling, 28(9), p.279.
- Rensi, S., Keys, A., Lo, Y.C., Derry, A., McInnes, G., Liu, T. and Altman, R., 2020. Homology modeling of TMPRSS2 yields candidate drugs that may inhibit entry of SARS-CoV-2 into human cells. ChemRxiv.
- Xu, Z., Peng, C., Shi, Y., Zhu, Z., Mu, K., Wang, X. and Zhu, W., 2020. Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. BioRxiv, pp.2020-01.
- Yang, H. and Yang, J., 2021. A review of the latest research on M pro targeting SARS-COV inhibitors. RSC Medicinal Chemistry, 12(7), pp.1026-1036.
- Sargolzaei, M., 2021. Effect of nelfinavir stereoisomers on coronavirus main protease: Molecular docking, molecular dynamics simulation and MM/GBSA study. Journal of Molecular Graphics and Modelling, 103, p.107803. [CrossRef]
- Mengist, H.M., Dilnessa, T. and Jin, T., 2021. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Frontiers in Chemistry, 9, p.622898.
- Schrödinger Release 2023-3: Maestro, Schrödinger, LLC, New York, NY, 2023.
- Schrödinger Release 2023-3: LigPrep, Schrödinger, LLC, New York, NY, 2023.
- Friesner, R. A.; Murphy, R. B.; Repasky, M. P.; Frye, L. L.; Greenwood, J. R.; Halgren,T. A.; Sanschagrin, P. C.; Mainz, D. T., "Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes," J. Med. Chem., 2006, 49, 6177–6196 . [CrossRef]
- Schrödinger Release 2023-3: Glide, Schrödinger, LLC, New York, NY, 2023.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




