1. Introduction
Colon polyps are mucosal protrusions within the colonic lumen. Based on their size, they are categorized as minuscule (5 mm or less), small (6 - 9 mm), and large (1 cm or greater in diameter) [
1]. Despite being predominantly asymptomatic, these polyps are commonly identified during screening colonoscopies. Predominantly originating from the mucosa, colon polyps can be benign, adenomatous, or serrated [
2]. The prevalence of adenomas among North American patients aged 50 - 75 was 30.2% [
3]. While a malignant potential is inherent to certain polyps, it is crucial to note that not all polyps undergo malignant transformation. Adenomatous and serrated polyps present malignant potential, whereas hyperplastic, post-inflammatory, and hamartomatous polyps generally do not [
4]. Given the potential for malignancy in certain polyps, standard procedure dictates their resection, followed by a detailed pathological analysis to ascertain their histological profile. This protocol is often underscored because over 95% of colorectal cancers are derived from adenomatous polyps [
1]. Most notably, colonic adenomas serve as the precursor to a significant proportion of colon cancers, exhibiting progression in size and degrees of dysplasia [
5]. Timely identification and intervention concerning polyps are pivotal, as they directly impact colon cancer mortality rates because colon cancer is the second most common cause of cancer death in the United States [
6].
According to the current guidelines, people at an average risk of colon cancer in the general population are advised to start colon screening at the age of 45 [
7]. There are also screening guidelines for people who have relatives affected by colon cancer. These recommendations suggest that individuals should start their screenings at the age of 40 or ten years before a first-degree relative was diagnosed with colon cancer, whichever comes first. Additionally, if there are two first-degree relatives who had been diagnosed with colon cancer at any age, it is advised to begin screenings without delay [
8]. Colon cancer screening is usually individualized after the age of 75, considering the patient’s screening history and health [
9].
Colonoscopy is a standard diagnostic and therapeutic tool to detect any colon polyps that could potentially be malignant. However, a meta-analysis of 43 publications with more than 15,000 tandem colonoscopies shows a miss rate of 26% for adenomas, 9% for advanced adenomas and 27% for serrated polyps [
10]. Based on previous literature, factors that may contribute to missed polyps include inadequate bowel preparation, suboptimal procedure techniques, and incomplete removal of polyps [
11,
12]. Methods to ensure thorough polyp removal and reduce the incidence of interval colon cancer include cecal intubation time, satisfactory bowel preparation, and a colonoscopic withdrawal period lasting at least 6 minutes or more [
11]. However, colon polyps might still go undetected during routine surveillance.
Machine learning image detection has made significant strides in the medical field, especially in colonic polyp detection. Several studies have explored the application of convolutional neural networks (CNNs) and deep learning techniques to detect polyps in colonoscopy images automatically, illustrating their clinical importance in minimizing undetected polyps and enhancing polyp detection rates [
13,
14,
15]. Research conducted at Xiangya Hospital of Central South University analyzed 681 colonoscopy images, utilizing the DeFrame system, they achieved an impressive 100% recall and 80% sensitivity. This system effectively identifies polyps with diverse morphologies and locations within colonoscopy footage [
12]. In addition, Wan et al. proposed an attentive YOLOv5 model, which achieved a precision of 0.915 and the recall rate of 0.899 on the Kvasir dataset [
15]. Misawa et al. (2019) developed a 3-D convolutional network model, which worked nearly in real-time and achieved a sensitivity of 90% and a specificity of 63.3% and accuracy of 76.5% with a training set of 411 short videos [
16]. These AI-based systems have shown promising results in improving the detection rate of polyps, which is critical for early diagnosis and intervention in colorectal cancer.
Research on machine learning polyp detection models is promising; however, further research and validation on larger and more diverse datasets are essential to establish the generalizability and robustness of these AI-driven solutions. For example, the selection of the most suitable model can be a major challenge. In the current study, this issue will be addressed by comparing three prominent machine learning image identification models: Google Teachable Machine (GTM) , Roboflow 3.0 (RF3), and You Only Live Once Version 8 (YOLOV8). The focus will be on the ability of each model to get trained and tested on the same set of images to accurately differentiate between normal colonic tissue, resected polyps, and polyps. The current study is of paramount importance, given that colon cancer ranks as the second leading cause of cancer-related deaths in the U.S., and colonic polyps can be missed easily due to a multitude of reasons.
2. Methods
2.1. Data Collection and Preprocessing
An open-source online gastrointestinal endoscopy database was used to train the machine-learning models. The database used in this research is titled “HyperKvasir” curated by Borgli et al. (2020) and sourced from Simula Datasets [
17]. The dataset consists of 110,079 images of the GI tract, out of which, 10,662 are labeled and 99,417 are unlabeled images. The labeled image dataset consisted of anatomic landmarks such as the cecum, ileum and retroflex-rectum. The dataset also has pathologic findings such as polyps as well as therapeutic interventions such as dyed-resection margins.
A total of 601 images were used to train the image classification models to train the models on differentiating between normal colonic tissue, polyp and resected polyp. 201 images of the cecum were used as “normal colonic tissue.” The model was trained using a dataset consisting of 200 images of polyps found in various sections of the lower gastrointestinal tract, along with an additional set of 200 images depicting dyed-resected polyp margins. The purpose of this training was to differentiate between polyps that were still intact and those that had been surgically removed. All the images were cropped to ensure a 1:1 aspect ratio (∼650x650 pixels). All the images were also ensured to be the same format of Joint Photographic Experts Group (JPEG). Any images that were obstructed or over/ under exposed were subsequently removed. Three different image classification models were trained: Google Teachable Machine (GTM), Roboflow 3.0 Object Detection (RF3), and You Only Look Once version 8 (YOLOv8).
2.2. Google Teachable Machine Setup and Image Classification
Once the dataset was processed, Google Teachable Machine’s web tool was used to develop an image classification model for polyp detection. GTM is an online platform that allows users to train, test and deploy machine learning image classification models quickly [
18]. To utilize the Image classification model in GTM, all 601 images from the pre-processed dataset were imported into GTM. The images were labeled into 3 separate classes: normal, polyp and resected polyp according to their labels assigned in the dataset. The data was trained for 300 epochs with a batch size of 32 and learning rate of 0.0001. Epoch refers to the number of times each image is fed through the training model. Batch size refers to the number of images used in one iteration of training. The learning rate determines how much the model’s parameters are adjusted in each update step during training. The batch size of 16 and learning rate of 0.001 are the default settings from GTM; however, fine adjustment of the program has shown that a batch size of 32 had a higher accuracy without significant differences in training time. In addition, the learning rate is small (0.0001) in order to ensure that the model gradually adjusts its parameters; prior testing with the dataset has shown better accuracy with lowering of the learning rate from 0.001 to 0.0001. GTM also does not provide further documentation on the specific deep learning architecture for their image classification model. Since there are 601 images altogether and 32 images per batch, there are a total of 19 batches. Once all batches go through the dataset, one epoch is complete. GTM also generates its own evaluation train/test split as 85% of the images are split into training samples (170 images per class) and 15% into test samples (30 images per class). This split cannot be changed by the user.
2.3. RoboFlow 3.0 Object Detection Setup and Image Classification
In order to generate an image classification model using RF3, the images first need to be annotated correctly. Computer Vision Annotation Tool (CVAT) was used to annotate the images. CVAT was used to annotate normal colonic tissue, polyps and resected polyps for all 601 images. The annotation data (class and position) of the image was exported as text files from CVAT. The text files were uploaded along with the 601 images onto the RoboFlow website. RoboFlow also generates its own train/validate/test split; however, the end-user can change the split. For the current study, the train/validate/test split was not changed from the default setting, with 70% of the images for training (∼140 images per class), 20% of the images for validation (40 images per class) and 10% for testing (20 images per class). RoboFlow also allows the user to preprocess the data on their own website; however, since the images were already pre-processed, this was not done on RoboFlow. Finally, the images were trained using “RoboFlow 3.0 Object Detection (FAST).” RoboFlow also recommends using a pre-trained benchmark model to give the model prior information to improve performance; Microsoft Common Objects in Context version 7 (MS COCO v7) was used as the pre-trained benchmark. RoboFlow automatically determines the number of epochs necessary for proper training of the model; the current model was trained for 300 epochs.
2.4. You Only Look Once version 8 Object Detection Setup and Image Classification
Prior to using YOLOv8, the images needed to be annotated correctly as with the RF3 model. The same CVAT annotations were used as above for all 601 images. The annotation data of the images was exported as text files from CVAT, and the text files along with the images were uploaded onto the YOLOv8 website. The same 70-20-10 train-validate-test split used for RF3 was exported with the YOLOv8 format. This downloaded file contains images and labels that are split into training, validation and testing groups. YOLOv8 is implemented as a deep learning model in Python, and the model architecture and weights must be generated or downloaded using Python code. Google Colaboratory (Colab) was used to write and execute the code to generate a YOLOV8 model. Before creating and training the YOLOv8 model, it is imperative to import all the images and labels into a specific Google Drive folder. In the current study, there were two overarching folders named “images” and “labels,” and within each folder there was a folder for “train”, “val” and “test,” which contained the images and labels for each split (i.e. images for the training split go into the “images” → “train” folder). Once the folders are configured correctly, a .YAML file will be used to ensure that the correct images train, validate and test the model. Before using the .YAML file, make sure to open the file and rename the path to the proper folder name. After configuring the .YAML file, the YOLOV8 model can be created and trained. The YOLOv8 model used in this study is based on the Ultralytics YOLOv8.0.137 implementation; specifically, the YOLOv8 nano (YOLOV8n) model was used for the current study. The model was set to train for 300 epochs with early stopping patience of 50 epochs, and a batch size of 32 is used. All other settings were left as is. The AdamW optimizer is used for training with a learning rate of 0.001429, momentum of 0.9, and weight decay of 0.0005. The YOLOv8 model consists of 225 layers and has a total of 3,011,433 parameters. The training data contains 421 images; the validation data consists of 120 images. Data augmentation techniques, including blur, median blur, grayscale conversion, and Contrast Limited Adaptive Histogram Equalization (CLAHE) are applied to the training data using the Albumentations library. The rest of the 60 images will be used for testing and generating metrics.
2.5. Metrics from the different models and their meanings
GTM generates its own confusion matrix, accuracy data as well as train-test performance graphs; however, it does not provide extra parameters such as precision, recall and F1 score. The confusion matrix generated by GTM are not comparable to the graphs generated by RF3 and YOLOV8 because the confusion matrix in GTM is made of a random 15% of the dataset, so the same 10% (60 images) generated by RF3 train-validate-test split were used to generate another confusion matrix as well as metrics such as precision, recall and F1 score. RF3 does not generate its own confusion matrix, the images from the test split were fed back into the RF3 model and the TPs, TNs, FPs and FNs were each recorded similar to
Figure 1. The generated confusion matrix was then used to generate different metrics such as precision, recall, F1 score and overall accuracy. YOLOV8 generates its own confusion matrix as well as precision and loss graphs; it also provides information on precision, recall, mAP. In order to ensure that metrics from all three models are comparable to each other, precision, recall, F1 score are calculated from a confusion matrix. Precision, recall, and F1 score are used to analyze the performance of the machine learning model because they are commonly used evaluation metrics in the field. Accuracy of the whole model can also be calculated from values generated in a confusion matrix, and this provides some more information on the models.
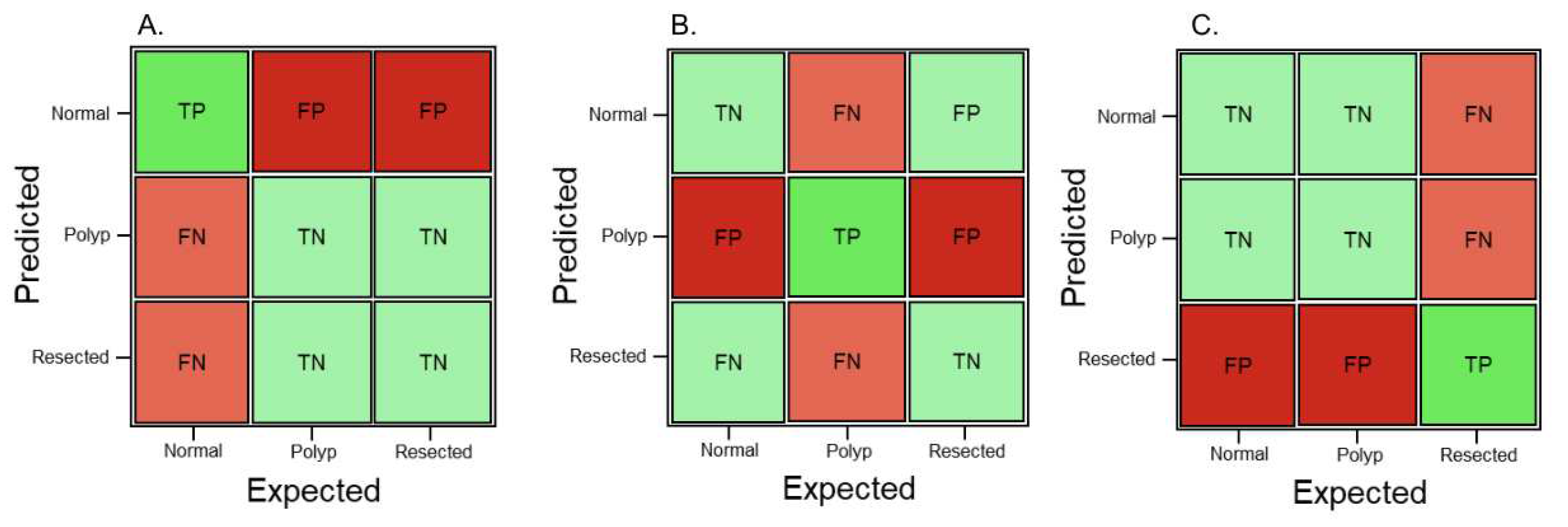
Figure 1 shows three confusion matrices with the corresponding true positive (TP), true negative (TN), false positive (FP) and false negative (FN) values based on which class will be predicted using the machine learning model.
Precision refers to the percentage of instances the classifier labels as positive with respect to the total predictive positive instances, i.e., the TP divided by TP + FP (Equation
1).
A high precision score indicates that the model is making fewer false positive predictions. Precision is also known as positive predictive value (PPV). Recall refers to the proportion of events that actually was of a certain class that was classified as that class. It is derived by dividing the true positives to all positives, so it is derived by dividing the true positive by predicted results (Equation
2).
High recall score indicates that the model is identifying a larger number of actual positive examples. Recall is also known as sensitivity. The F-1 score is a combination of precision and recall; it is the weighted average of the precision and recall scores (Equation
3).
It ranges from 0 to 1. A value of 1 indicates perfect precision and recall, while a value of 0 indicates poor precision or recall. Overall accuracy refers to the sum of correctly classified values divided by the total number of values in the confusion matrix (Equation
4).
2.6. Assessing external validity of all models
In order to assess external validity of the models, 90 extra images (30 normal, 30 polyp and 30 resected polyp) were extracted from the HyperKvasir database. All three models do not have access to these 90 extra images, and the 90 images are externally fed into all three models. After each image is fed into each model, new confusion matrices are generated and precision, recall, F-1 score and accuracy are calculated as described previously.
3. Results
3.1. GTM Metrics
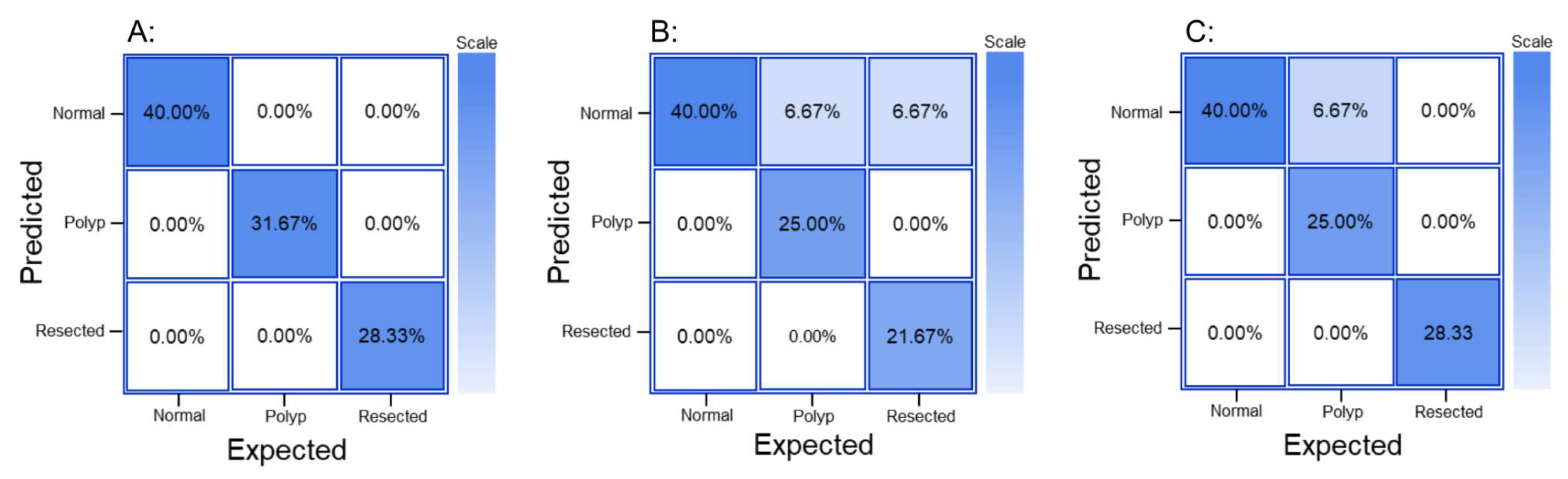
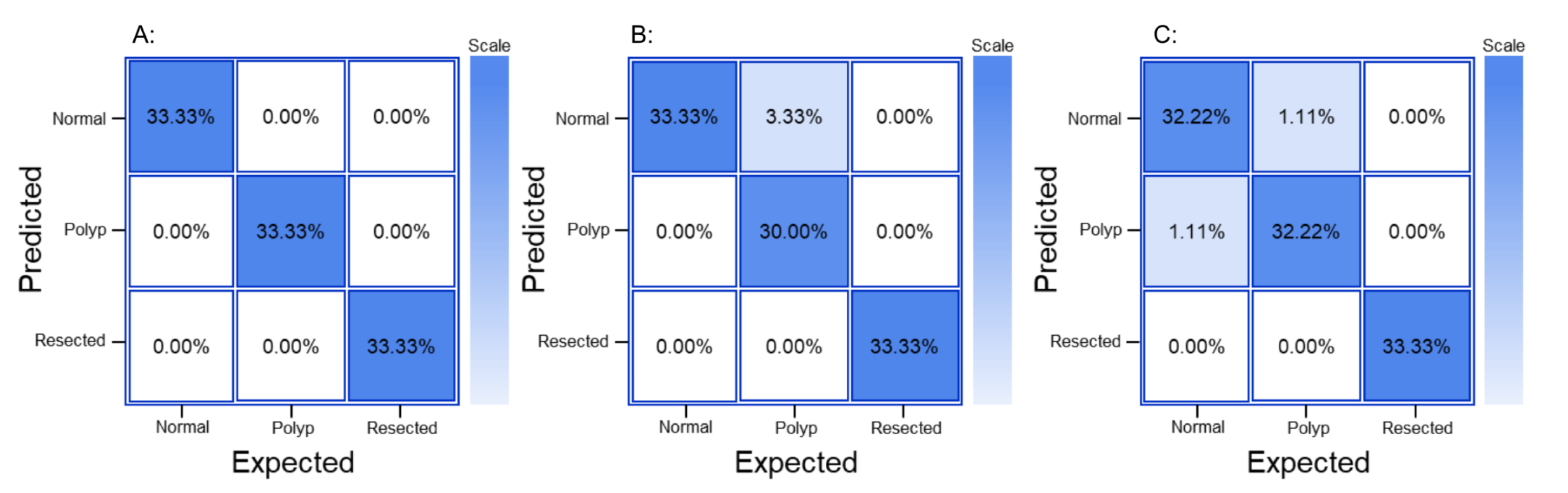
The GTM produced a confusion matrix, which was subsequently normalized to display values as percentages (
Figure 2(A)). GTM does not generate other metrics, so the main metrics were generated from the confusion matrix, including: precision, recall and F-1 score (
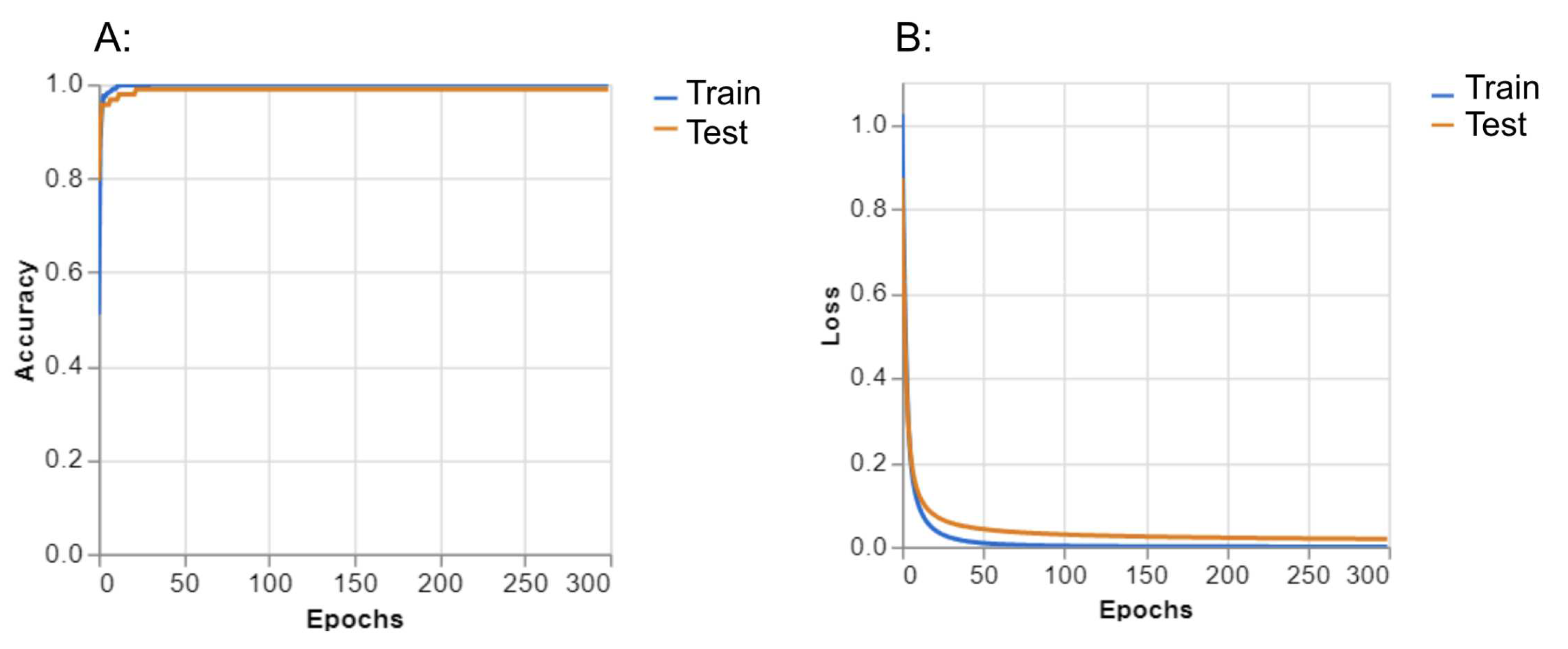
Table 1). For the “normal” class, precision was 1.00, recall was 1.00 and F-1 score was 1.00. For the “polyp” class, precision was 0.97, recall was 1.00 and F-1 score was 0.98. For the “resected polyp” class, precision was 1.00, recall was 0.97, and F-1 score was 0.98. The overall accuracy of the GTM model was 0.99 (Table 4). Performance graphs generated by GTM showed that as the number of epochs increased, the accuracy of the train and test split equally increased (
Figure 3(A)). In addition, as the number of epochs increased, the loss of the train and test split decreased equally (
Figure 3(B)). Loss represents how well the model is performing during training.
3.2. RF3 Metrics
The generated confusion matrix for RF3 was normalized to display values as percentages (
Figure 2(B)). Further metrics such as precision, recall and F-1 score were generated from the confusion matrix (
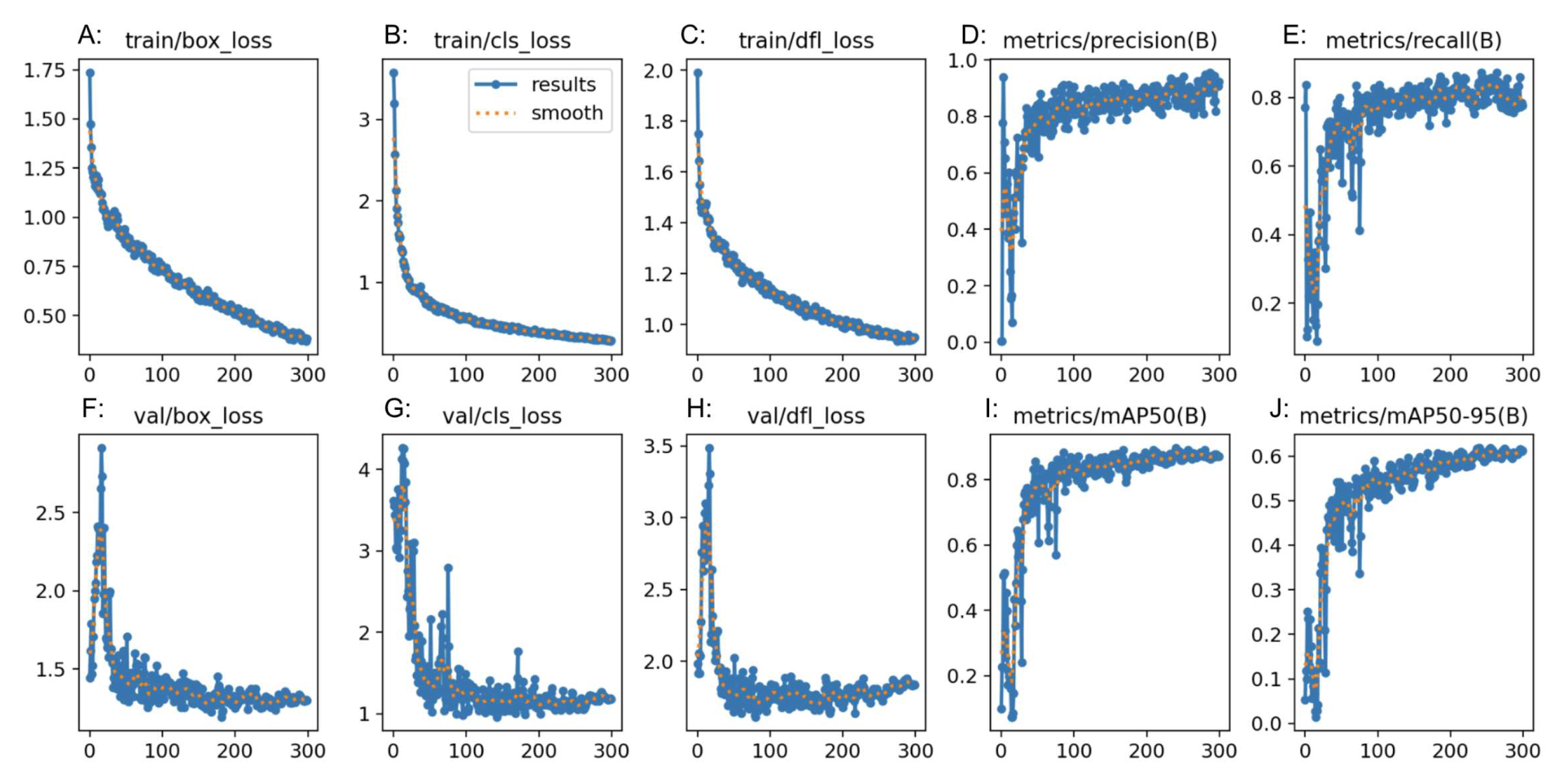
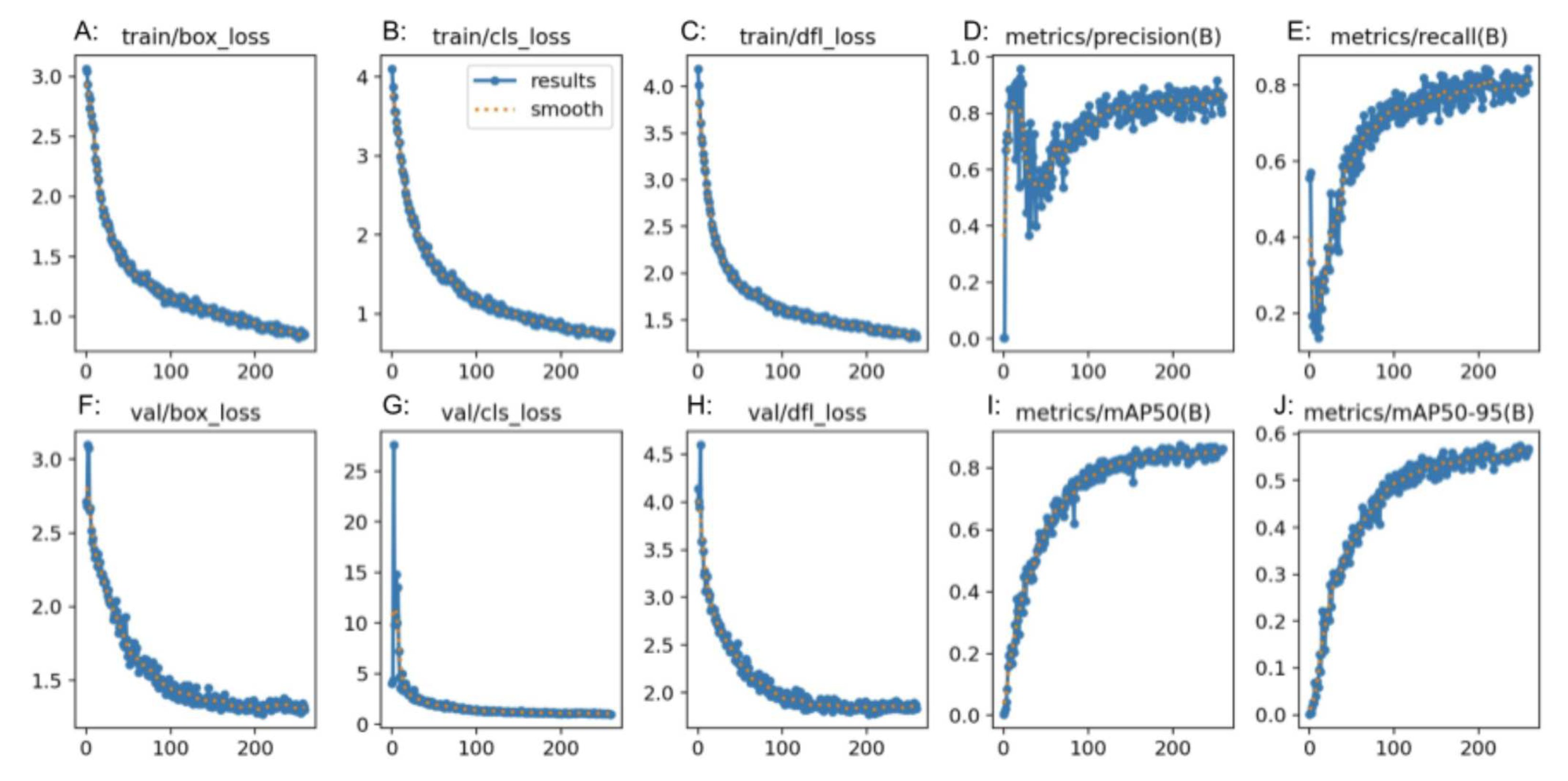
Table 2). For the “normal” class, precision was 0.75, recall was 1.00 and F-1 score was 0.86. For the “polyp” class, precision was 1.00, recall was 0.79 and F-1 score was 0.88. For the “resected polyp” class, precision was 1.00, recall was 0.76, and F-1 score was 0.87. Additional metrics from RoboFlow itself showed an average precision and mAP for all 3 classes as 0.89 with an average recall of 0.82. The overall accuracy of the RF3 model was 0.87 (Table 4). Performance graphs generated by RF3 showed that as the number of epochs increased, the bounding box regression loss (box_loss) (
Figure 4(A and F)), classification loss (cls_loss) (
Figure 4(B and G)), and deformable convolution layer loss (dfl_loss) (
Figure 4(C and H)) decreased. In addition, as the number of epochs increased, the precision, mAP (
Figure 4(D, I and J)) and recall (
Figure 4(E)) increased. “Box_loss” measures the error in predicting bounding box coordinates and dimension, so a lower “box_loss” indicates higher accuracy in detecting the location of polyps. “Cls_loss” refers to the error in the predicted class probabilities for each object in the image, so a lower “cls_loss” indicates higher accuracy of the model in predicting the class that an image belongs to (normal, polyp or resected polyp in the current study). “Dfl_loss” measures the error in the deformable convolution layers, which allow the model to detect objects in images of different scales and aspect ratios, so a lower “dfl_loss” indicates that the model is better at handling images with different variations and aspect ratios.
3.3. YOLOv8 Metrics
The generated confusion matrix for YOLOV8 was normalized to display values as percentages (
Figure 2(C)). Further metrics such as precision, recall and F-1 score were generated from the confusion matrix (
Table 3). For the “normal” class, precision was 0.96, recall was 1.00 and F-1 score was 0.98. For the “polyp” class, precision was 0.86, recall was 0.75 and F-1 score was 0.80. For the “resected polyp” class, precision was 0.90, recall was 0.95, and F-1 score was 0.92. Additional metrics from YOLOv8 itself showed an average precision for all 3 classes as 0.90, mAP of 0.95 with an average recall of 0.90. The overall accuracy of the RF3 model was 0.84 (
Table 4). The performance graphs generated by RF3 showed that as the number of epochs increased, the bounding box regression loss (box_loss) (
Figure 5(A and F)), classification loss (cls_loss) (
Figure 5(B and G)), and deformable convolution layer loss (dfl_loss) (
Figure 5(C and H)) decreased in both the training and validation plots. In addition, as the number of epochs increased, the precision, mAP (
Figure 5(D, I and J)) and recall (
Figure 5(E)) increased.
3.4. External Validity Assessment
In assessing external validity of the GTM model, the “normal”, “polyp” and “resected polyp” classes had a precision, recall and F1 score of 1.00 (
Table 6). The average confidence in the prediction of the “normal” class in GTM was 99.70, in the “polyp” class was 98.13, in the “resected polyp” class was 99.10. The overall accuracy of the GTM model was 1.00 (
Table 5).
For the RF3 model, the “normal” class had a precision of 0.91, recall of 1.00 and F1 score of 0.95 and a confidence of 93.26 (
Table 7). The “polyp” class had a precision of 1.00, recall of 0.90 and F1 score of 0.95 and a confidence of 80.60. The “resected polyp” class had a precision of 1.00, recall of 1.00 and F1 score of 1.00 and a confidence of 89.97. The overall accuracy of the RF3 model is 0.97 (
Table 5).
For the YOLOv8 model, the “normal” class had a precision of 0.97, recall of 0.97 and F1 score of 0.97 and a confidence of 85.5 (
Table 8). The “polyp” class had a precision of 0.97, recall of 0.97 and F1 score of 0.97 and a confidence of 79.87. The “resected polyp” class had a precision of 1.00, recall of 1.00 and F1 score of 1.00 and a confidence of 75.73. The overall accuracy of the RF3 model is 0.97 (
Table 5).
4. Discussion
In the current study, the performance of three machine learning image classification models, Google Teachable Machine (GTM), Roboflow3 (RF3), and YOLOv8, was compared in distinguishing between normal colon, polyps, and resected polyps in the colon. Metrics generated by GTM demonstrated excellent performance in classifying images into their respective classes. Analysis generated from the confusion matrix revealed the highest precision, recall, and F1 scores for all classes. The performance graphs generated by GTM indicated that as the number of epochs increased during training, the accuracy and loss of both train and test splits improved simultaneously, suggesting that the model effectively learned to generalize well on both the training and test data, and there are minimal signs of “overfitting” of the data. “Overfitting” in machine learning means that the model is memorizing features of images instead of trying to extrapolate the features that allow it to make generalizations on the image belonging to a particular class [
19].
RF3 also demonstrated strong performance in the classification task, however with slightly lower metrics compared to GTM. The confusion matrix analysis showed good precision and recall values for all classes. The F1 score was comparable for all three models, and was satisfactory. The "normal" class had a lower precision than the rest of the classes, and the “polyp” and “resected polyp” classes had a lower recall than the “normal” class. This could be explained by the lower number of samples in the testing split for classes “polyp” (19 images) and “resected polyp” (17 images) as compared to the “normal” (24 images) class. The test split of images created by Roboflow did not evenly split the images between the three classes, so misclassifications can create bigger differences in the different metrics. Therefore, the metrics generated from the external validity test are more comparable between the three models. The plots generated by RF3 also show good performance with minimal signs of overfitting as the loss decreased equally for the training (
Figure 4(A, B, C)) and validation (
Figure 4(F, G, H)) plots as the number of epochs increased; however, the validation plots had some spikes in loss.
YOLOv8 also demonstrated competitive performance in the classification task, with metrics falling between those of GTM and RF3. The confusion matrix analysis showed good precision and recall values for all classes. The F1 score for all three classes was also better than the RF3 model but not as high as GTM. The "normal" class had a lower precision than the rest of the classes, and the “polyp” and “resected polyp” classes had a lower recall than the “normal” class. This is very similar to the trends found in the RF3 model, and this could be explained by the same reasoning of unevenly split images between the three classes. Therefore, the metrics generated from the external validity test are more comparable between the three models. The plots generated by YOLOv8 also show good performance with lower signs of overfitting than the RF3 model as the loss decreased equally for the training (Figure
Figure 4(A, B, C)) and validation (
Figure 4(F, G, H)) plots without any sudden spikes.
In the external validity assessment, all three models demonstrated robust generalization to new, unseen data. GTM achieved perfect precision, recall, and F1 scores for all classes, along with a remarkable overall accuracy of 1.00. RF3 and YOLOV8 also performed well, with precision, recall, and F1 scores close to their original evaluation on the test set. Overall, the study revealed that GTM, RF3, and YOLOV8 are all capable of effectively classifying colon images into the categories of "normal," "polyp," and "resected polyp." GTM exhibited outstanding performance with high precision, recall, and F1 scores, as well as excellent external validity. RF3 and YOLOv8 also performed well, with competitive metrics and strong generalization to new data. The choice of the most suitable model may depend on specific use cases, computational resources, and the required level of precision and recall for the application.
Comparing all three models, GTM demonstrated exceptional performance in the classification task, achieving perfect precision, recall, and F1 scores for all classes on both the original test set and the external validation set. GTM’s key advantages include its ease of use and accessibility, making it an excellent option for researchers or practitioners without extensive machine learning expertise. RF3 performed well, with competitive metrics on both the original test set and the external validation set. One of the significant advantages of RF3 is its ability to handle more complex tasks, thanks to its capability for bounding box regression and object detection using deformable convolution layers. YOLOv8 also displayed competitive performance, with metrics falling between those of GTM and RF3. One of the primary advantages of YOLOv8 lies in its efficiency and speed, making it an excellent choice for real-time or near-real-time image classification applications.
Despite their strengths, all three models exhibited certain limitations that merit consideration. GTM’s main limitation is its relatively simple architecture, which might not be suitable for more complex image classification tasks. In addition, GTM is a classification model, whereas RF3 and YOLOv8 models used are image detection programs that detect the polyp rather than simple classification. Image classification refers to the process of categorizing images into specific classes or categories based on their visual content. This task is essential for applications such as object recognition, scene understanding, and image retrieval. On the other hand, image detection focuses on localizing and identifying specific objects or regions within an image [
20]. Compared to image classification, which only requires determining the category or class of an entire image, image detection involves identifying and localizing objects within an image.
RF3 and YOLOv8 rely on more complex architectures that demand substantial computational resources and longer training times. Additionally, these models may require more fine-tuning and parameter adjustments to achieve optimal performance, making them less user-friendly for those without extensive machine learning expertise. However, the outcome of RF3 and YOLOv8 are more applicable for clinical practice as it can be used to ensure that colonic polyps are not missed. Comparing the RF3 and YOLOv8 models, the YOLOv8 model can be done remotely or in a private Google Drive. For RF3, for a free user, all the images that will be used for training, validation and testing must be uploaded onto the RoboFlow website, and they are not private. In addition, free users also only get a limited amount of credits to generate RF3 models. However, RoboFlow is extremely user friendly and requires no understanding of coding to implement via the RoboFlow website; whereas, YOLOV8 requires some understanding of coding. Future research could focus on optimizing and fine-tuning the hyperparameters of RF3 and YOLOv8 to enhance their performance while considering the computational cost. Furthermore, exploring ensemble methods that combine the strengths of different models could potentially lead to even better classification results.
5. Conclusion
In conclusion, the comparison of Google Teachable Machine (GTM), Roboflow3 (RF3), and YOLOv8 revealed that each model possesses unique advantages and limitations. GTM is exceptionally user-friendly and effective for quick prototyping, while RF3 excels in more complex tasks with precise object detection. YOLOv8 offers computational efficiency and real-time capabilities, making it suitable for time-sensitive applications. The choice of the most appropriate model will depend on the specific requirements of the image classification task, available resources, and the desired level of accuracy and efficiency.
Author Contributions
Conceptualization, A.A., R.J., J.A., J.J., T.J., A.K., H.A., P.P., J.S., and M.T.; methodology, A.A., R.J., J.A., J.J., T.J., A.K., H.A., P.P., J.S., and M.T.; software, A.A., R.J., J.A., J.J., T.J., A.K., H.A., P.P., J.S., and M.T.; validation, A.A., R.J., J.A., J.J., T.J., A.K., H.A., P.P., J.S., and M.T.; formal analysis, A.A., R.J., J.A., J.J., T.J., A.K., H.A., P.P., J.S., and M.T.; investigation, A.A., R.J., J.A., J.J., T.J., A.K., H.A., P.P., J.S., and M.T.; resources, A.A., R.J., J.A., J.J., T.J., A.K., H.A., P.P., J.S., and M.T.; data curation, A.A., R.J., J.A., J.J., T.J., A.K., H.A., P.P., J.S., and M.T.; writing—original draft preparation, A.A., R.J., J.A., J.J., T.J., A.K., H.A., P.P., J.S., and M.T.; writing—review and editing, A.A., R.J., J.A., J.J., T.J., A.K., H.A., P.P., J.S., and M.T.; visualization, A.A., R.J., J.A., J.J., T.J., A.K., H.A., P.P., J.S., and M.T.; supervision, M.T.; project administration, M.T.; funding acquisition, M.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| GI |
Gastrointestinal |
| GTM |
Google Teachable Machine |
| RF3 |
Roboflow 3.0 Object Detection |
| YOLOv8 |
You Only Look Once version 8 |
| CVAT |
Computer Vision Annotation Tool |
| MS COCO v7 |
Microsoft Common Objects in Context version 7 |
| Colab |
Google Colaboratory |
| .YAML |
YAML Ain’t Markup Language |
| CLAHE |
Contrast Limited Adaptive Histogram Equalization |
| AI |
Artificial Intelligence |
| JPEG |
Joint Photographic Experts Group |
| YOLOV8n |
YOLOv8 nano |
| AdamW |
Adaptive Moment Estimation version W optimizer |
| PPV |
Positive Predictive Value |
| mAP |
Mean Average Precision |
| box_loss |
Bounding Box Regression Loss |
| cls_loss |
Classification Loss |
| dfl_loss |
Deformable Convolution Layer Loss |
References
- Meseeha, M.; Attia, M. Colon Polyps; StatPearls Publishing: StatPearls [Internet], 2023. [Google Scholar]
- Summers, R.M. Polyp Size Measurement at CT Colonography: What Do We Know and What Do We Need to Know? Radiology 2010, 255, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Heitman, S.J.; Ronksley, P.E.; Hilsden, R.J.; Manns, B.J.; Rostom, A.; Hemmelgarn, B.R. Prevalence of Adenomas and Colorectal Cancer in Average Risk Individuals: A Systematic Review and Meta-analysis. Clinical Gastroenterology and Hepatology 2009, 7, 1272–1278. [Google Scholar] [CrossRef] [PubMed]
- Bonnington, S.N. Surveillance of colonic polyps: Are we getting it right? World Journal of Gastroenterology 2016, 22, 1925. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Fearon, E.R.; Hamilton, S.R.; Kern, S.E.; Preisinger, A.C.; Leppert, M.; Smits, A.M.; Bos, J.L. Genetic Alterations during Colorectal-Tumor Development. New England Journal of Medicine 1988, 319, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA: A Cancer Journal for Clinicians 2023, 73, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Jayasinghe, M.; Prathiraja, O.; Caldera, D.; Jena, R.; Coffie-Pierre, J.A.; Silva, M.S.; Siddiqui, O.S. Colon Cancer Screening Methods: 2023 Update. Cureus 2023. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, B.A.; Marshall, J.L. Colon Cancer in Young Adults: Trends and Their Implications. Current Oncology Reports 2019, 21. [Google Scholar] [CrossRef] [PubMed]
- Gornick, D.; Kadakuntla, A.; Trovato, A.; Stetzer, R.; Tadros, M. Practical considerations for colorectal cancer screening in older adults. World Journal of Gastrointestinal Oncology 2022, 14, 1086–1102. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, S.; Pan, P.; Xia, T.; Chang, X.; Yang, X.; Guo, L.; Meng, Q.; Yang, F.; Qian, W.; Xu, Z.; Wang, Y.; Wang, Z.; Gu, L.; Wang, R.; Jia, F.; Yao, J.; Li, Z.; Bai, Y. Magnitude, Risk Factors, and Factors Associated With Adenoma Miss Rate of Tandem Colonoscopy: A Systematic Review and Meta-analysis. Gastroenterology 2019, 156, 1661–1674.e11. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Jung, Y.S.; Jeong, W.S.; Yang, H.J.; Park, S.K.; Choi, K.; Park, D.I. Miss rate of colorectal neoplastic polyps and risk factors for missed polyps in consecutive colonoscopies. Intestinal Research 2017, 15, 411. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Lu, S.; Tang, Y.; Wang, D.; Sun, X.; Yi, J.; Liu, B.; Cao, Y.; Chen, Y.; Liu, X. A Machine Learning-Based System for Real-Time Polyp Detection (DeFrame): A Retrospective Study. Frontiers in Medicine 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- ei Kudo, S.; Mori, Y.; Misawa, M.; Takeda, K.; Kudo, T.; Itoh, H.; Oda, M.; Mori, K. Artificial intelligence and colonoscopy: Current status and future perspectives. Digestive Endoscopy 2019, 31, 363–371. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, Y.; Liu, M.; Lai, Y.; Liu, P.; Wang, Z.; Xing, T.; Huang, Y.; Li, Y.; Li, A.; Wang, Y.; Luo, X.; Liu, S.; Han, Z. Artificial Intelligence-Assisted Colonoscopy for Detection of Colon Polyps: a Prospective, Randomized Cohort Study. Journal of Gastrointestinal Surgery 2020, 25, 2011–2018. [Google Scholar] [CrossRef]
- Wan, J.; Chen, B.; Yu, Y. Polyp Detection from Colorectum Images by Using Attentive YOLOv5. Diagnostics 2021, 11, 2264. [Google Scholar] [CrossRef] [PubMed]
- Misawa, M.; ei Kudo, S.; Mori, Y.; Cho, T.; Kataoka, S.; Yamauchi, A.; Ogawa, Y.; Maeda, Y.; Takeda, K.; Ichimasa, K.; Nakamura, H.; Yagawa, Y.; Toyoshima, N.; Ogata, N.; Kudo, T.; Hisayuki, T.; Hayashi, T.; Wakamura, K.; Baba, T.; Ishida, F.; Itoh, H.; Roth, H.; Oda, M.; Mori, K. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology 2018, 154, 2027–2029.e3. [Google Scholar] [CrossRef] [PubMed]
- Borgli, H.; Thambawita, V.; Smedsrud, P.H.; Hicks, S.; Jha, D.; Eskeland, S.L.; Randel, K.R.; Pogorelov, K.; Lux, M.; Nguyen, D.T.D.; Johansen, D.; Griwodz, C.; Stensland, H.K.; Garcia-Ceja, E.; Schmidt, P.T.; Hammer, H.L.; Riegler, M.A.; Halvorsen, P.; de Lange, T. HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Scientific Data 2020, 7. [Google Scholar] [CrossRef] [PubMed]
- Carney, M.; Webster, B.; Alvarado, I.; Phillips, K.; Howell, N.; Griffith, J.; Jongejan, J.; Pitaru, A.; Chen, A. Teachable Machine: Approachable Web-Based Tool for Exploring Machine Learning Classification. Extended Abstracts of the 2020 CHI Conference on Human Factors in Computing Systems. ACM, 2020. [CrossRef]
- Ying, X. An Overview of Overfitting and its Solutions. Journal of Physics: Conference Series 2019, 1168, 022022. [Google Scholar] [CrossRef]
- Zhang, Y. A Fine-Grained Image Classification and Detection Method Based on Convolutional Neural Network Fused with Attention Mechanism. Computational Intelligence and Neuroscience 2022, 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Sample confusion matrix showing TP, TN, FP, and FN, if predictions are required for (A) Normal colonic tissue, (B) Polyp, and (C) Resected polyp.
Figure 1.
Sample confusion matrix showing TP, TN, FP, and FN, if predictions are required for (A) Normal colonic tissue, (B) Polyp, and (C) Resected polyp.
Figure 2.
Normalized confusion matrix from A: GTM, B: RF3, and C: YOLOv8.
Figure 2.
Normalized confusion matrix from A: GTM, B: RF3, and C: YOLOv8.
Figure 3.
Accuracy (A) and Loss (B) plots from GTM.
Figure 3.
Accuracy (A) and Loss (B) plots from GTM.
Figure 4.
Precision (D, I, J), recall (E) and loss (A, B, C, F, G, H) plots from RF3.
Figure 4.
Precision (D, I, J), recall (E) and loss (A, B, C, F, G, H) plots from RF3.
Figure 5.
Precision (D, I, J), recall (E) and loss (A, B, C, F, G, H) plots from YOLOv8.
Figure 5.
Precision (D, I, J), recall (E) and loss (A, B, C, F, G, H) plots from YOLOv8.
Figure 6.
Normalized confusion matrix for external validity dataset with A: GTM, B: RF3, C: YOLOv8.
Figure 6.
Normalized confusion matrix for external validity dataset with A: GTM, B: RF3, C: YOLOv8.
Table 1.
Precision, Recall and F-1 score for GTM.
Table 1.
Precision, Recall and F-1 score for GTM.
| Class |
Precision |
Recall |
F-1 Score |
| Normal |
1.00 |
1.00 |
1.00 |
| Polyp |
1.00 |
1.00 |
1.00 |
| Resected Polyp |
1.00 |
1.00 |
1.00 |
Table 2.
Precision, Recall and F-1 score for RF3.
Table 2.
Precision, Recall and F-1 score for RF3.
| Class |
Precision |
Recall |
F-1 Score |
| Normal |
0.75 |
1.00 |
0.86 |
| Polyp |
1.00 |
0.79 |
0.88 |
| Resected Polyp |
1.00 |
0.76 |
0.87 |
Table 3.
Precision, Recall and F-1 score for YOLOv8.
Table 3.
Precision, Recall and F-1 score for YOLOv8.
| Class |
Precision |
Recall |
F-1 Score |
| Normal |
0.86 |
1.00 |
0.92 |
| Polyp |
1.00 |
0.79 |
0.88 |
| Resected Polyp |
1.00 |
1.00 |
1.00 |
Table 4.
Accuracy measurement differences from GTM, RF3 and YOLOv8.
Table 4.
Accuracy measurement differences from GTM, RF3 and YOLOv8.
| Model |
Accuracy |
| GTM |
0.99 |
| RF3 |
0.87 |
| YOLOv8 |
0.84 |
Table 5.
Accuracy measurement differences from GTM, RF3 and YOLOV8 for external validation dataset.
Table 5.
Accuracy measurement differences from GTM, RF3 and YOLOV8 for external validation dataset.
| Model |
Accuracy |
| GTM |
1.00 |
| RF3 |
0.97 |
| YOLOv8 |
0.97 |
Table 6.
Precision, Recall and F1 score for GTM for external validation dataset.
Table 6.
Precision, Recall and F1 score for GTM for external validation dataset.
| Class |
Precision |
Recall |
F-1 Score |
Average Confidence |
| Normal |
1.00 |
1.00 |
1.00 |
99.70 |
| Polyp |
1.00 |
1.00 |
1.00 |
98.13 |
| Resected Polyp |
1.00 |
1.00 |
1.00 |
99.10 |
Table 7.
Precision, Recall and F1 score for RF3 for external validation dataset.
Table 7.
Precision, Recall and F1 score for RF3 for external validation dataset.
| Class |
Precision |
Recall |
F-1 Score |
Average Confidence |
| Normal |
0.91 |
1.00 |
0.95 |
93.26 |
| Polyp |
1.00 |
0.90 |
0.95 |
80.60 |
| Resected Polyp |
1.00 |
1.00 |
1.00 |
89.97 |
Table 8.
Precision, Recall and F1 score for YOLOv8 for external validation dataset.
Table 8.
Precision, Recall and F1 score for YOLOv8 for external validation dataset.
| Class |
Precision |
Recall |
F-1 Score |
Average Confidence |
| Normal |
0.97 |
0.97 |
0.98 |
85.50 |
| Polyp |
0.97 |
0.97 |
0.98 |
79.78 |
| Resected Polyp |
1.00 |
1.00 |
1.00 |
75.73 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).