Submitted:
05 August 2023
Posted:
08 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Enzymatic cleavages (cryptides)
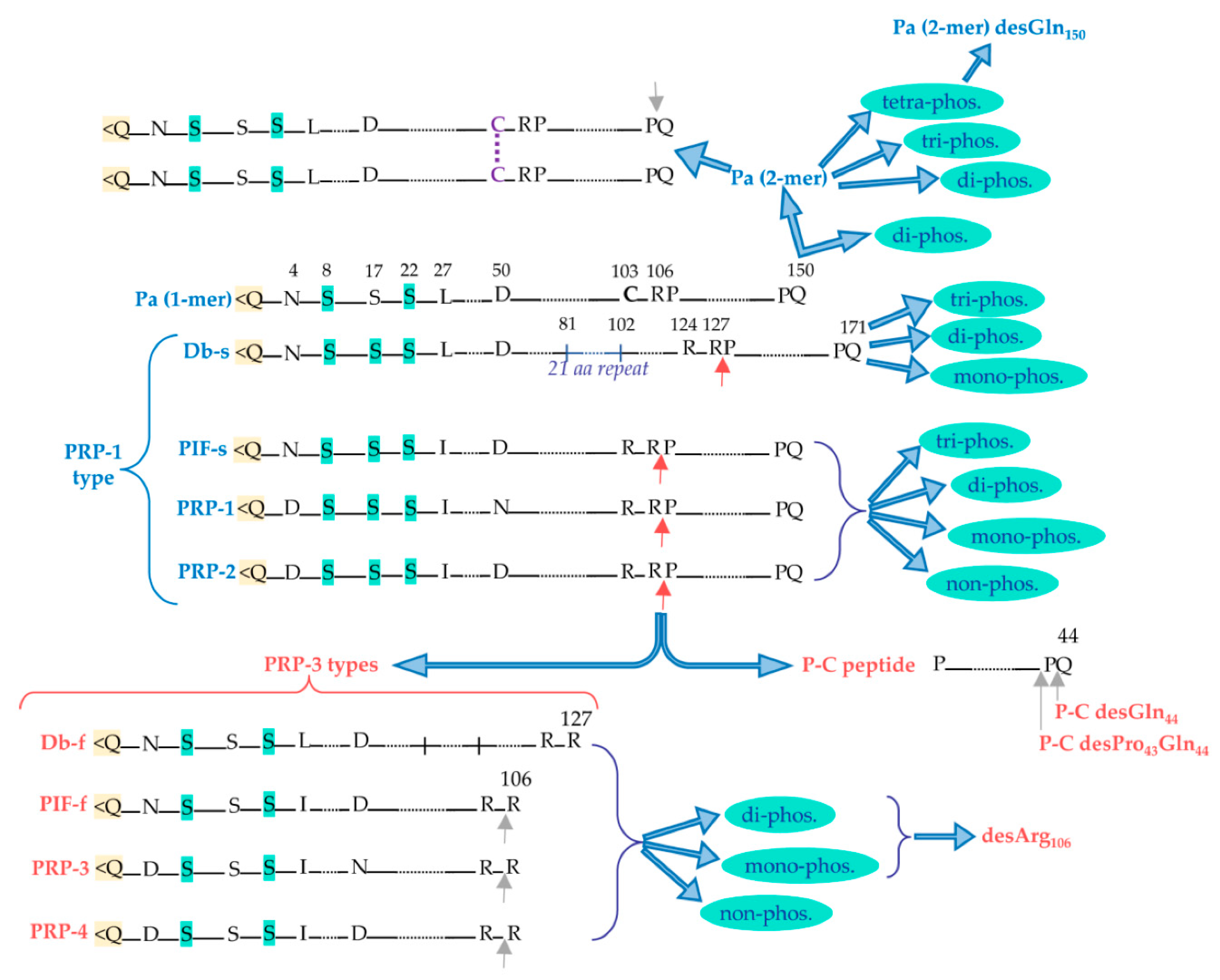
2.1. Pre-secretory cleavages
2.1.1. Proline-rich proteins (PRPs)
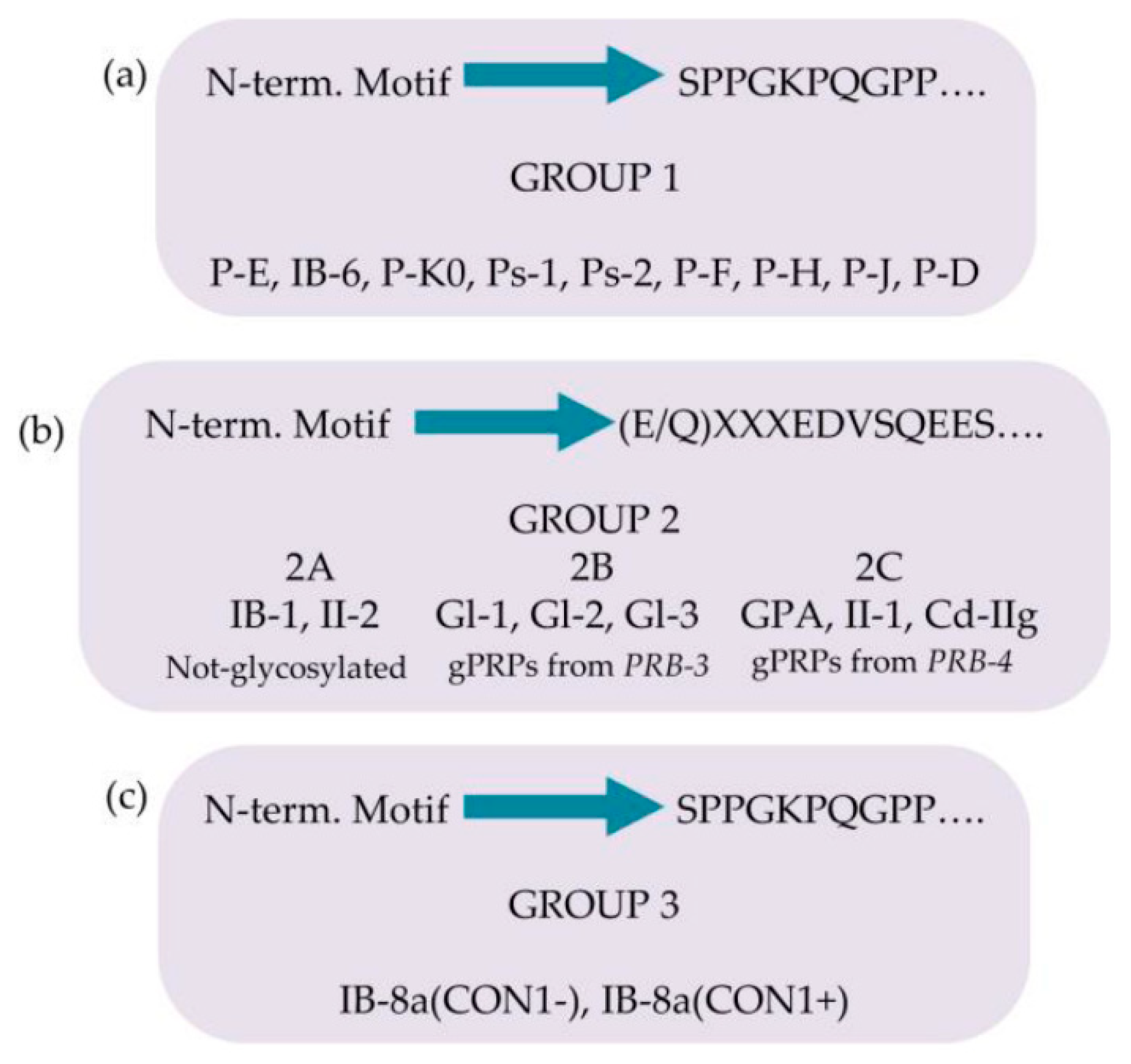
2.1.1.1. Acidic proline-rich-proteins (aPRPs)
2.1.1.2. Basic-proline-rich-proteins (bPRPs)
- Products of Locus PRB-1:
- Products of Locus PRB-2:
- Products of Locus PRB-4:
2.1.2. Further pre-secretory cleavages of PRPs
2.1.3. Role of PRPs
2.1.4. Statherin and P-B peptides
2.1.5. Histatins (Hst)
2.1.6. Cystatins
2.2. Post-secretory cleavages
2.3. Proteolytic cleavages: variations related to age and pathologies
3. Phosphorylation
3.3. Variation of phosphorylation as a function of age and for the diagnosis of different diseases
4. Sulfation
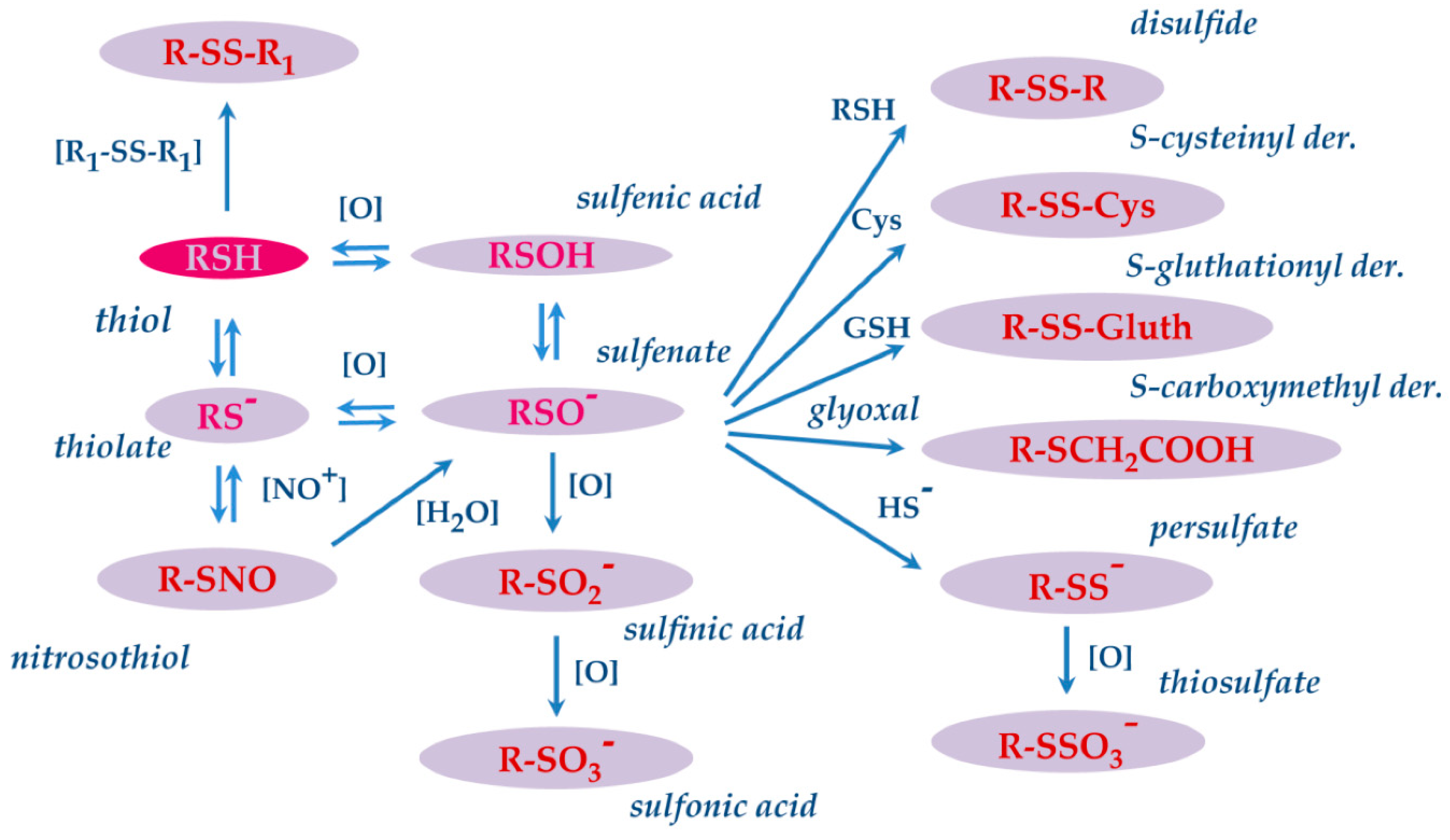
5. S-modifications
5.1. S-modifications of salivary proteins in pathologies
6. Transglutamination
7. Glycosylation
7.1. Non enzymatic glycosylation (glycation)
7.2. Enzymatic glycosylation
8. Citrullination
9. N-terminal modifications
9.1. Excision of initiatory methionine and Nt-acetylation
9.2. Pyroglutamic acid modification
10. C-terminal modifications
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Boroumand, M.; Olianas, A.; Cabras, T.; Manconi, B.; Fanni, D.; Faa, G.; Desiderio, C.; Messana, I.; Castagnola, M. Saliva, abodily fluid with recognized and potential diagnostic applications. J. Sep. Sci. 2021, 44, 3677–3690. [Google Scholar] [CrossRef] [PubMed]
- Helmerhorst, E.J.; Oppenheim, F.G. Saliva: a dynamic proteome. J. Dent. Res. 2007, 86, 680–693. [Google Scholar] [CrossRef] [PubMed]
- Lau, W.W.; Hardt, M.; Zhang, Y.H.; Freire, M.; Ruhl, S. The Human Salivary Proteome Wiki: A Community-Driven Research Platform. J. Dent. Res. 2021, 100, 1510–1519. [Google Scholar] [CrossRef] [PubMed]
- Iavarone, F.; Desiderio, C.; Vitali, A.; Messana, I.; Martelli, C.; Castagnola, M.; Cabras, T. Cryptides: latent peptides eve- rywhere. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 246–263. [Google Scholar] [CrossRef]
- Castle, D.; Castle, A. Intracellular transport and secretion of salivary proteins. Crit. Rev. Oral Biol. Med. 1998, 9, 4–22. [Google Scholar] [CrossRef]
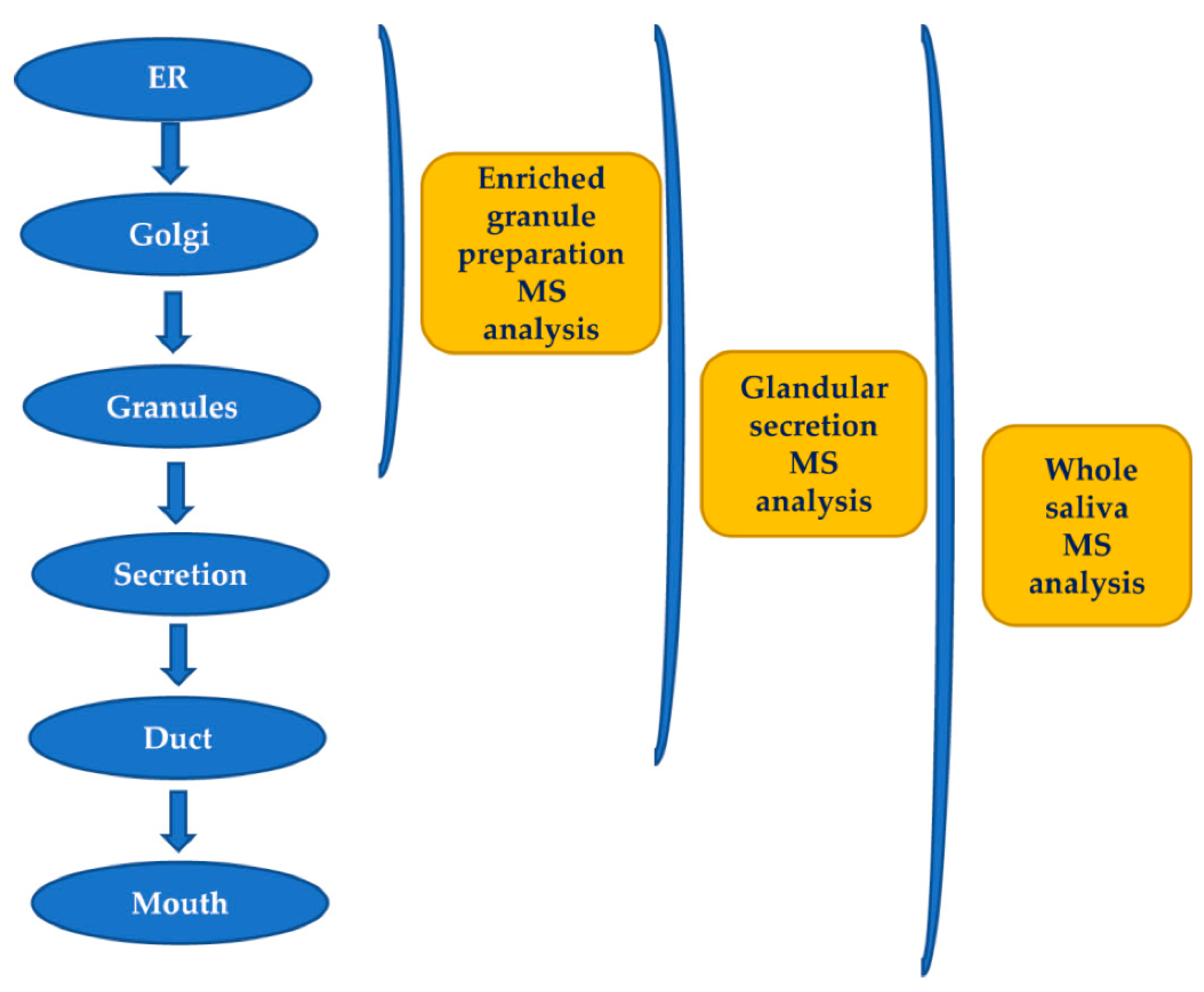
- Messana, I.; Cabras, T.; Pisano, E.; Sanna, M. T.; Olianas, A.; Manconi, B.; Pellegrini, M.; Paludetti, G.; Scarano, E.; Fiorita, A.; et al. Trafficking and postsecretory events responsible for the formation of secreted human salivary peptides: a proteomics ap- proach. Mol. Cell. Proteomics 2008, 7, 911–926. [Google Scholar] [CrossRef]
- Chan, M.; Bennick, A. Proteolytic processing of a human salivary proline-rich protein precursor by proprotein convertases. Eur. J. Biochem. 2001, 268, 3423–3431. [Google Scholar] [CrossRef]
- Cai, K.; Bennick, A. Processing of acidic proline-rich proprotein by human salivary gland convertase. Arch. Oral Biol. 2004, 49, 871–879. [Google Scholar] [CrossRef]
- Inzitari, R.; Cabras, T.; Onnis, G.; Olmi, C.; Mastinu, A.; Sanna, M.T.; Pellegrini, M.G.; Castagnola, M.; Messana, I. Different isoforms and post-translational modifications of human salivary acidic proline-rich proteins. Proteomics 2005, 5, 805–815. [Google Scholar] [CrossRef]
- Kauffman, D. L.; Keller, P.J. The basic proline-rich proteins in human parotid saliva from a single subject. Arch. Oral Biol. 1979, 24, 249–256. [Google Scholar] [CrossRef]
- Kauffman, D.; Wong, R.; Bennick, A.; Keller, P. Basic proline-rich proteins from human parotid saliva: complete covalent struc- ture of protein IB-9 and partial structure of protein IB-6, members of a polymorphic pair. Biochemistry 1982, 21, 6558–6562. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, D.; Hofmann, T.; Bennick, A.; Keller, P. Basic proline-rich proteins from human parotid saliva: complete covalent structures of proteins IB-1 and IB-6. Biochemistry 1986, 25, 2387–2392. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, D. L.; Bennick, A.; Blum, M.; Keller, P. J. Basic proline-rich proteins from human parotid saliva: relationships of the covalent structures of ten proteins from a single individual. Biochemistry 1991, 30, 3351–3356. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, E.; Isemura, S.; Sanada, K. Complete amino acid sequence of a basic proline-rich peptide, P-D, from human parotid saliva. J. Biochem. 1983, 93, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, E.; Isemura, S.; Sanada, K. Complete amino acid sequence of a basic proline-rich peptide, P-F, from human parotid saliva. J. Biochem. 1983, 93, 883–888. [Google Scholar] [CrossRef]
- Saitoh, E.; Isemura, S.; Sanada, K. Further fractionation of basic proline-rich peptides from human parotid saliva and complete amino acid sequence of basic proline-rich peptide P-H. J. Biochem. 1999. [Google Scholar] [CrossRef]
- Isemura, S.; Saitoh, E.; Sanada, K. Isolation and amino acid sequences of proline-rich peptides of human whole saliva. J. Biochem. 1979, 86, 79–86. [Google Scholar]
- Isemura, S.; Saitoh, E.; Sanada, K. Fractionation and characterization of basic proline-rich peptides of human parotid saliva and the amino acid sequence of proline-rich peptide P-E. J. Biochem. 1982, 91, 2067–2075. [Google Scholar] [CrossRef]
- Messana, I.; Cabras, T.; Inzitari, R.; Lupi, A.; Zuppi, C.; Olmi, C.; Fadda, M. B.; Cordaro, M.; Giardina, B.; Castagnola, M. Char- acterization of the human salivary basic proline-rich protein complex by a proteomic approach. J. Proteome Res. 2004, 3, 792–800. [Google Scholar] [CrossRef]
- Cabras, T.; Pisano, E.; Boi, R.; Olianas, A.; Manconi, B.; Inzitari, R.; Fanali, C.; Giardina, B.; Castagnola, M.; Messana, I. Age- dependent modifications of the human salivary secretory protein complex. J. Proteome Res. 4134. [Google Scholar] [CrossRef]
- Maeda, N.; Kim, H.S.; Azen, E.A.; Smithies, O. Differential RNA splicing and post-translational cleavages in the human salivary proline-rich protein gene system. J. Biol. Chem. 1985, 260, 11123–11130. [Google Scholar] [CrossRef]
- Messana, I.; Inzitari, R.; Fanali, C.; Cabras, T.; Castagnola, M. Facts and artifacts in proteomics of body fluids. What proteomics of saliva is telling us? J. Sep. Sci. 2008, 31, 1948–1963. [Google Scholar] [CrossRef]
- Padiglia, A.; Orrù, R.; Boroumand, M.; Olianas, A.; Manconi, B.; Sanna, M.T.; Desiderio, C.; Iavarone, F.; Liori, B.; Messana, I.; et al. Extensive Characterization of the Human Salivary Basic Proline-Rich Protein Family by Top-Down Mass Spectrometry. J. Proteome Res. 2018, 17, 3292–3307. [Google Scholar] [CrossRef] [PubMed]
- Manconi, B.; Castagnola, M.; Cabras, T.; Olianas, A.; Vitali, A.; Desiderio, C.; Sanna, M.T.; Messana, I. The intriguing heteroge- neity of human salivary proline-rich proteins: Short title: Salivary proline-rich protein species. J. Proteomics 2016, 134, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Messana, I.; Cabras, T.; Iavarone, F.; Manconi, B.; Huang, L.; Martelli, C.; Olianas, A.; Sanna, M.T.; Pisano, E.; Sanna, M.; et al. Chrono-proteomics of human saliva: variations of the salivary proteome during human development. J. Proteome Res. 2015, 14, 1666–1677. [Google Scholar] [CrossRef] [PubMed]
- Manconi, B.; Cabras, T.; Pisano, E.; Sanna, M.T.; Olianas, A.; Fanos, V.; Faa, G.; Nemolato, S.; Iavarone, F.; Castagnola, M.; Messana, I. Modifications of the acidic soluble salivary proteome in human children from birth to the age of 48 months investi- gated by a top-down HPLC-ESI-MS platform. J. Proteomics 2013, 91, 536–543. [Google Scholar] [CrossRef]
- Lu, Y.; Bennick, A. Interaction of tannin with human salivary proline-rich proteins. Arch. Oral Biol. 1998, 43, 717–728. [Google Scholar] [CrossRef]
- Cannon, R.D.; Chaffin, W.L. Oral colonization by Candida albicans. Crit. Rev. Oral Biol Med. 1999, 359–383. [Google Scholar] [CrossRef]
- Ruhl, S.; Sandberg, A.L.; Cisar, J.O. Salivary receptors for the proline-rich protein-binding and lectin-like adhesins of oral acti- nomyces and streptococci. J. Dent. Res. 2004, 83, 505–510. [Google Scholar] [CrossRef]
- Cabras, T.; Melis, M.; Castagnola, M.; Padiglia, A.; Tepper, B.J.; Messana, I.; Tomassini Barbarossa, I. Responsiveness to 6-n- propylthiouracil (PROP) is associated with salivary levels of two specific basic proline-rich proteins in humans. PLoS One 2012, 7, e30962. [Google Scholar] [CrossRef]
- Vitorino, R.; Calheiros-Lobo, M.J.; Williams, J.; Ferrer-Correia, A.J.; Tomer, K.B.; Duarte, J.A.; Domingues, P.M.; Amado, F.M. (2007). Peptidomic analysis of human acquired enamel pellicle. Biomed. Chromatogr. 2007, 21, 1107–1117. [Google Scholar] [CrossRef]
- Palmerini, C.A.; Mazzoni, M.; Radicioni, G.; Marzano, V.; Granieri, L.; Iavarone, F.; Longhi, R.; Messana, I.; Cabras, T. , Sanna, M.T.; et al. Antagonistic Effect of a Salivary Proline-Rich Peptide on the Cytosolic Ca2+ Mobilization Induced by Progesterone in Oral Squamous Cancer Cells. PLoS One, 0147. [Google Scholar] [CrossRef]
- Macias, M. J.; Wiesner, S.; Sudol, M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002, 513, 30–37. [Google Scholar] [CrossRef]
- Righino, B.; Pirolli, D.; Radicioni, G.; Marzano, V.; Longhi, R.; Arcovito, A.; Sanna, M.T.; De Rosa, M.C.; Paoluzi, S.; Cesareni, G.; et al. Structural studies and SH3 domain binding properties of a human antiviral salivary proline-rich peptide. Biopolymers 2016, 106, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Azen, E.A. Genetics of salivary protein polymorphisms. Crit. Rev. Oral Biol. Med. [CrossRef]
- Sabatini, L.M.; Carlock, L.R.; Johnson, G.W.; Azen, E.A. cDNA cloning and chromosomal localization (4q11-13) of a gene for statherin, a regulator of calcium in saliva. Am. J. Hum. Genet. 1987, 41, 1048–1060. [Google Scholar] [PubMed]
- Raj, P.A.; Johnsson, M.; Levine, M.J.; Nancollas, G.H. Salivary statherin. Dependence on sequence, charge, hydrogen bonding potency, and helical conformation for adsorption to hydroxyapatite and inhibition of mineralization. J. Biol. Chem. 1992, 267, 5968–5976. [Google Scholar] [CrossRef] [PubMed]
- Niemi, L.D.; Johansson, I. Salivary statherin peptide-binding epitopes of commensal and potentially infectious Actinomyces spp. delineated by a hybrid peptide construct. Infect. Immun. 2004, 72, 782–787. [Google Scholar] [CrossRef]
- Gibbons, R.J.; Hay, D.I. Human salivary acidic proline-rich proteins and statherin promote the attachment of Actinomyces vis- cosus LY7 to apatitic surfaces. Infect. Immun. 1988, 56, 439–445. [Google Scholar] [CrossRef]
- Isemura, S. Nucleotide sequence of gene PBII encoding salivary proline-rich protein P-B. J. Biochem. 2000, 127, 393–398. [Google Scholar] [CrossRef]
- Inzitari, R.; Cabras, T.; Rossetti, D.V.; Fanali, C.; Vitali, A.; Pellegrini, M.; Paludetti, G.; Manni, A.; Giardina, B.; Messana, I.; et al. Detection in human saliva of different statherin and P-B fragments and derivatives. Proteomics 2006, 6, 6370–6379. [Google Scholar] [CrossRef]
- Strawich, E.; Glimcher, M.J. Tooth ‘enamelins’ identified mainly as serum proteins. Major ‘enamelin’ is albumin. Eur. J. Biochem. 1990, 191, 47–56. [Google Scholar] [CrossRef]
- Ayad, M.; Van Wuyckhuyse, B.C.; Minaguchi, K.; Raubertas, R.F.; Bedi, G.S.; Billings, R.J.; Bowen, W.H.; Tabak, L.A. The asso ciation of basic proline-rich peptides from human parotid gland secretions with caries experience. J. Dent. Res. 2000, 79, 976–982. [Google Scholar] [CrossRef]
- Jensen, J.L.; Lamkin, M.S.; Troxler, R.F.; Oppenheim, F.G. Multiple forms of statherin in human salivary secretions. Arch. Oral Biol. 1991, 36, 529–534. [Google Scholar] [CrossRef]
- Sabatini, L.M.; He, Y.Z.; Azen, E.A. Structure and sequence determination of the gene encoding human salivary statherin. Gene 1990, 89, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, R.; Lobo, M.J.; Duarte, J.R.; Ferrer-Correia, A.J.; Domingues, P.M.; Amado, F.M. The role of salivary peptides in dental caries. Biomed. Chromatogr. 2005, 19, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, L.M.; Azen, E.A. Histatins, a family of salivary histidine-rich proteins, are encoded by at least two loci (HIS1 and HIS2). Biochem. Biophys. Res. Commun. 1989, 160, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, F.G.; Xu, T.; McMillian, F.M.; Levitz, S.M.; Diamond, R.D.; Offner, G.D.; Troxler, R.F. Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Can dida albicans. J. Biol. Chem. 1988, 263, 7472–7477. [Google Scholar] [CrossRef] [PubMed]
- Castagnola, M.; Inzitari, R.; Rossetti, D.V.; Olmi, C.; Cabras, T.; Piras, V.; Nicolussi, P.; Sanna, M.T.; Pellegrini, M.; Giardina, B.; et al. A cascade of 24 histatins (histatin 3 fragments) in human saliva. Suggestions for a pre-secretory sequential cleavage path way. J. Biol. Chem. 2004, 279, 41436–41443. [Google Scholar] [CrossRef]
- Torres, P.; Díaz, J.; Arce, M.; Silva, P.; Mendoza, P.; Lois, P.; Molina-Berríos, A.; Owen, G.I.; Palma, V.; Torres, V.A. The salivary peptide histatin-1 promotes endothelial cell adhesion, migration, and angiogenesis. FASEB J. 2017, 31, 4946–4958. [Google Scholar] [CrossRef]
- Oudhoff, M.J.; Bolscher, J.G.; Nazmi, K.; Kalay, H.; van ‘t Hof, W.; Amerongen, A.V.; Veerman, E.C. Histatins are the major 1317 wound-closure stimulating factors in human saliva as identified in a cell culture assay. FASEB J. 2008, 22, 3805–3812. [Google Scholar] [CrossRef]
- Amado, F.; Lobo, M.J.; Domingues, P.; Duarte, J.A.; Vitorino, R. Salivary peptidomics. Expert. Rev. Proteomics 2010, 7, 709–721. [Google Scholar] [CrossRef]
- Castagnola, M.; Scarano, E.; Passali, G.C.; Messana, I.; Cabras, T.; Iavarone, F.; Di Cintio, G.; Fiorita, A.; De Corso, E.; Paludetti, G. Salivary biomarkers and proteomics: future diagnostic and clinical utilities. Acta Otorhinolaryngol. Ital. 2017, 37, 94–101. [Google Scholar] [CrossRef]
- Kos, J.; Lah, T.T. Cysteine proteases and their endogenous inhibitors: target proteins for prognosis, diagnosis and therapy in cancer (review). Oncol. Rep. 1998, 5, 1349–13461. [Google Scholar] [CrossRef]
- Henskens, Y.M.; Veerman, E.C.; Nieuw Amerongen, A.V. Cystatins in health and disease. Biol. Chem. Hoppe Seyler 1996, 377, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Calkins, C.C.; Sloane, B.F. Mammalian cysteine protease inhibitors: biochemical properties and possible roles in tumor progres sion. Biol. Chem. Hoppe Seyler 1995, 376, 71–80. [Google Scholar] [PubMed]
- Manconi, B.; Liori, B.; Cabras, T.; Vincenzoni, F.; Iavarone, F.; Castagnola, M.; Messana, I.; Olianas, A. Salivary cystatins: explor ing new post-translational modifications and polymorphisms by top-down mass spectrometry. J. Proteome Res. 2017, 16, 4196–4207. [Google Scholar] [CrossRef]
- Lupi, A.; Messana, I.; Denotti, G.; Schininà, M.E.; Gambarini, G.; Fadda, M.B.; Vitali, A.; Cabras, T.; Piras, V.; Patamia, M.; et al. 1334 Identification of the human salivary cystatin complex by the coupling of high-performance liquid chromatography and ion trap mass spectrometry. Proteomics 2003, 3, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Ryan, C.M.; Souda, P.; Halgand, F.; Wong, D.T.; Loo, J.A.; Faull, K.F.; Whitelegge, J.P. Confident assignment of intact mass tags to human salivary cystatins using top-down Fourier-transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom. 2010, 21, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Iavarone, F.; Cabras, T.; Pisano, E.; Sanna, M.T.; Nemolato, S.; Vento, G.; Tirone, C.; Romagnoli, C.; Cordaro, M.; Fanos, V.; et al. Top-down HPLC-ESI-MS detection of S-glutathionylated and S-cysteinylated derivatives of cystatin B and its 1-53 and 54 98 fragments in whole saliva of human preterm newborns. J. Proteome Res. 2013, 12, 917–926. [Google Scholar] [CrossRef]
- Requena, T.; Velasco, M. The human microbiome in sickness and in health. Rev. Clin. Esp. (Barc). 2021, 221, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Chimenos-Küstner, E.; Giovannoni, M.L.; Schemel-Suárez, M. Dysbiosis as a determinant factor of systemic and oral pathology: importance of microbiome. Med. Clin. (Barc). 2017, 149, 305–309. [Google Scholar] [CrossRef]
- Helmerhorst, E.J.; Sun, X.; Salih, E.; Oppenheim, F.G. Identification of Lys-Pro-Gln as a novel cleavage site specificity of saliva associated proteases. J. Biol. Chem. 2008, 283, 19957–19966. [Google Scholar] [CrossRef]
- Zamakhchari, M.; Wei, G.; Dewhirst, F.; Lee, J.; Schuppan, D.; Oppenheim, F.G.; Helmerhorst, E.J. Identification of Rothia bac teria as gluten-degrading natural colonizers of the upper gastro-intestinal tract. PloS One 2011, 6, e24455. [Google Scholar] [CrossRef]
- Asano, M.; Komiyama, K. Polymeric immunoglobulin receptor. J. Oral Sci. 2011, 53, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Kaetzel, C.S. The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal sur faces. Immunol. Rev. 2005, 206, 83–99. [Google Scholar] [CrossRef]
- Ramachandran, P.; Boontheung, P.; Xie, Y.; Sondej, M.; Wong, D.T.; Loo, J.A. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 2006, 5, 1493–1503. [Google Scholar] [CrossRef] [PubMed]
- Cabras, T.; Sanna, M.; Manconi, B.; Fanni, D.; Demelia, L.; Sorbello, O.; Iavarone, F.; Castagnola, M.; Faa, G.; Messana, I. Proteo mic investigation of whole saliva in Wilson’s disease. J. Proteomics 2015, 128, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Boroumand, M.; Iavarone, F.; Manconi, B.; Pieroni, L.; Greco, V.; Vento, G.; Tirone, C.; Desiderio, C.; Fiorita, A.; Faa, G.; et al. HPLC-ESI-MS top-down analysis of salivary peptides of preterm newborns evidenced high activity of some exopeptidases and convertases during late fetal development. Talanta 2021, 222, 121429. [Google Scholar] [CrossRef] [PubMed]
- Cabras, T.; Pisano, E.; Mastinu, A.; Denotti, G.; Pusceddu, P.P.; Inzitari, R.; Fanali, C.; Nemolato, S.; Castagnola, M.; Messana, I. 1362 Alterations of the salivary secretory peptidome profile in children affected by type 1 diabetes. Mol. Cell. Proteomics 2010, 9, 2099–2108. [Google Scholar] [CrossRef]
- Hardt, M.; Thomas, L.R.; Dixon, S.E.; Newport, G.; Agabian, N.; Prakobphol, A.; Hall, S.C.; Witkowska, H.E.; Fisher, S.J. Toward defining the human parotid gland salivary proteome and peptidome: identification and characterization using 2D SDS-PAGE, ultrafiltration, HPLC, and mass spectrometry. Biochemistry 2005, 44, 2885–2899. [Google Scholar] [CrossRef]
- Huq, N.L.; Cross, K.J.; Ung, M.; Myroforidis, H.; Veith, P.D.; Chen, D.; Stanton, D.; Huiling, H.; Ward, B.R.; Reynolds, E.C. A Review of the Salivary Proteome and Peptidome and Saliva-derived Peptide Therapeutics. Int. J. Pept. Res. Ther. [CrossRef]
- Manconi, B.; Liori, B.; Cabras, T.; Vincenzoni, F.; Iavarone, F.; Lorefice, L.; Cocco, E.; Castagnola, M.; Messana, I.; Olianas, A. Top-down proteomic profiling of human saliva in multiple sclerosis patients. J. Proteomics. [CrossRef]
- Serrao, S.; Firinu, D.; Olianas, A.; Deidda, M.; Contini, C.; Iavarone, F.; Sanna, M.T.; Boroumand, M.; Amado, F.; Castagnola, M. 1374 Top-Down Proteomics of Human Saliva Discloses Significant Variations of the Protein Profile in Patients with Mastocytosis. J. 1375 Proteome. Res. 2020, 19, 3238–3253. [Google Scholar] [CrossRef]
- Contini, C. , Serrao, S., Manconi, B., Olianas, A., Iavarone, F., Bizzarro, A., Masullo, C., Castagnola, M., Messana, I., Diaz, G., et al. Salivary Proteomics Reveals Significant Changes in Relation to Alzheimer’s Disease and Aging. J. Alzheimers Dis. 2022, 89, 605–622. [Google Scholar] [CrossRef]
- Olianas, A.; Guadalupi, G.; Cabras, T.; Contini, C.; Serrao, S.; Iavarone, F.; Castagnola, M.; Messana, I.; Onali, S.; Chessa, L.; et al. Top-Down Proteomics Detection of Potential Salivary Biomarkers for Autoimmune Liver Diseases Classification. Int. J. Mol. Sci. 2023, 24, 959. [Google Scholar] [CrossRef]
- Contini, C.; Olianas, A.; Serrao, S.; Deriu, C.; Iavarone, F.; Boroumand, M.; Bizzarro, A.; Lauria, A.; Faa, G.; Castagnola, M.; et al. Top-Down Proteomics of Human Saliva Highlights Anti-inflammatory, Antioxidant, and Antimicrobial Defense Responses in Alzheimer Disease. Front. Neurosci. 2021, 15, 668852. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L.J.; Linley, A.; Hammond, D.E.; Hood, F.E.; Coulson, J.M.; MacEwan, D.J.; Ross, S.J.; Slupsky, J.R.; Smith, P.D.; Eyers, P.A. New Perspectives, Opportunities, and Challenges in Exploring the Human Protein Kinome. Cancer Res. 2018, 78, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Salih, E.; Siqueira, W.L.; Helmerhorst, E.J.; Oppenheim, F.G. Large-scale phosphoproteome of human whole saliva using disul fidethiol interchange covalent chromatography and mass spectrometry. Anal. Biochem. [CrossRef]
- Stone, M.D. .; Chen, X.; McGowan, T.; Bandhakavi, S.; Cheng, B.; Rhodus, N.L.; Griffin, T.J. Large-scale phosphoproteomics analysis of whole saliva reveals a distinct phosphorylation pattern. J. Proteome Res. Tagliabracci, V.S.; Pinna L.A.; Dixon, J.E. Secreted protein kinases. Trends Biochem. Sci. , , -130, doi:10.1016/j.tibs.2012.11.008. 2011, 10, 1728–1736. [Google Scholar] [CrossRef] [PubMed]
- Tagliabracci, V.S.; Wiley, S.E.; Guo, X.; Kinch, L.N.; Durrant, E.; Wen, J.; Xiao, J.; Cui, J.; Nguyen, K.B.; Engel, J.L.; et al. A Single Kinase Generates the Majority of the Secreted Phosphoproteome. Cell 2015, 161, 1619–1632. [Google Scholar] [CrossRef]
- Isemura, S.; Saitoh, E.; Sanada, K.; Minakata, K. Identification of full-sized forms of salivary (S-type) cystatins (cystatin SN, cystatin SA, cystatin S, and two phosphorylated forms of cystatin S) in human whole saliva and determination of phosphory lation sites of cystatin S. J Biochem. 1991, 110, 648–654. [Google Scholar] [CrossRef]
- Madapallimattam, G.; Bennick, A. Phosphorylation of salivary proteins by salivary gland protein kinase. J. Dent. Res. 1986, 65, 405–411. [Google Scholar] [CrossRef]
- Iavarone, F.; D’Alessandro, A.; Tian, N.; Cabras, T.; Messana, I.; Helmerhorst, E.J.; Oppenheim, F.G.; Castagnola, M. Castagnola, M. High resolution high-performance liquid chromatography with electrospray ionization mass spectrometry and tandem mass spec trometry characterization of a new isoform of human salivary acidic proline-rich proteins named Roma-Boston Ser22 (Phos). 1405. [Google Scholar]
- Phe variant. J Sep Sci. 2014, 37, 1896–1902. [CrossRef]
- Halgand, F.; Zabrouskov, V.; Bassilian, S.; Souda, P.; Loo, J.A.; Faull, K.F.; Wong, D.T.; &, *!!! REPLACE !!!*; Whitelegge, J.P. ; & Whitelegge, J.P. Defining intact protein primary structures from saliva: a step toward the human proteome project. Anal. Chem. [CrossRef]
- Castagnola, M.; Inzitari, R.; Fanali, C.; Iavarone, F.; Vitali, A.; Desiderio, C.; Vento, G.; Tirone. C.; Romagnoli, C.; Cabras T.; et al. The surprising composition of the salivary proteome of preterm human newborn. Mol. Cell Proteomics 2011, 10, M110–003467. [Google Scholar] [CrossRef]
- Wahid, A.; Sohail, A.; Wang, H.; Guo, M.; Zhang, L.; Ji, Y.; Wang, P.; Xiao, H. Titanium(IV) immobilized affinity chromatog raphy facilitated phosphoproteomics analysis of salivary extracellular vesicles for lung cancer. Anal. Bioanal. Chem. 2022, 414, 3697–3708. [Google Scholar] [CrossRef]
- Sun, J.; Wang, X.; Ding, Y.; Xiao, B.; Wang, X.; Ali, M.M.; Ma, L.; Xie, Z.; Gu, Z.; Chen, G.; et al. Proteomic and phosphoproteomic landscape of salivary extracellular vesicles to assess OSCC therapeutical outcomes. Proteomics 2023, 23, e2200319. [Google Scholar] [CrossRef]
- Inzitari, R.; Vento, G.; Capoluongo, E.; Boccacci, S.; Fanali, C.; Cabras, T.; Romagnoli, C.; et al. Proteomic analysis of salivary acidic proline-rich proteins in human pre-term and at-term new-borns. J. Proteome Res. 2007, 6, 1371–1377. [Google Scholar] [CrossRef] [PubMed]
- Messana, I.; Cabras, T.; Iavarone, F.; Manconi, B.; Huang, L.; Martelli, C.; Olianas, A.; Sanna, M.T.; Pisano, E.; Sanna, M.; et al. Chrono-proteomics of human saliva: variations of the salivary proteome during human development. J. Proteome Res. 2015, 14, 1666–1677. [Google Scholar] [CrossRef] [PubMed]
- Castagnola, M.; Messana, I.; Inzitari, R.; Fanali, C.; Cabras, T.; Morelli, A.; Pecoraro, A.M.; Neri, G.; Torrioli, M.G. , Gurrieri, F. Hypo-phosphorylation of salivary peptidome as a clue to the molecular pathogenesis of autism spectrum disorders. J. Proteome Res. 2008, 7, 5327–5332. [Google Scholar] [CrossRef]
- Gamage, N.; Barnett, A.; Hempel, N.; Duggleby, R.G.; Windmill, K.F.; Martin, J.L.; McManus, M.E. Human sulfotransferases and their role in chemical metabolism. Toxicol. Sci. 2006, 90, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Leyh, T.S. The physical biochemistry and molecular genetics of sulfate activation. Crit. Rev. Biochem. Mol. Biol. 1993, 28, 515–542. [Google Scholar] [CrossRef] [PubMed]
- Venkatachalam, K.V. Human 3’-phosphoadenosine 5’-phosphosulfate (PAPS) synthase: biochemistry, molecular biology and genetic deficiency. IUBMB Life 2003, 55, 1–11. [Google Scholar] [CrossRef]
- Schröder, E.; Gebel, L.; Eremeev, A.A.; Morgner, J.; Grum, D.; Knauer, S.K.; Bayer, P.; Mueller, J.W. Human PAPS synthase 1433 isoforms are dynamically regulated enzymes with access to nucleus and cytoplasm PLoS One. 7, 2955; e9. [Google Scholar] [CrossRef]
- Gamage, N.; Barnett, A.; Hempel, N.; Duggleby, R.G.; Windmill, K.F.; Martin, J.L.; McManus, M.E. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006, 90, 5–22. [Google Scholar] [CrossRef]
- Negishi, M.; Pedersen, L.G.; Petrotchenko, E.; Shevtsov, S.; Gorokhov, A.; Kakuta, Y.; Pedersen, L.C. Structure and function of sulfotransferases. Arch. Biochem. Biophys. 2001, 390, 149–157. [Google Scholar] [CrossRef]
- Falany, C.N. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997, 11, 206–216. [Google Scholar] [CrossRef]
- Pedersen, L.C.; Yi, M.; Pedersen, L.G.; Kaminski, A.M. From steroid and drug metabolism to glycobiology, using sulfotransfer ase structures to understand and tailor function. Drug Metab Dispos. 2022, 50, 1027–1041. [Google Scholar] [CrossRef]
- Kurogi, K.; Rasool, M.I.; Alherz, F.A.; El Daibani, A.A.; Bairam, A.F.; Abunnaja, M.S.; Yasuda, S.; Wilson, L.J.; Hui, Y.; Liu, M.C. SULT genetic polymorphisms: physiological, pharmacological and clinical implications. Expert Opin. Drug Metab. Toxicol. 2021, 17, 767–784. [Google Scholar] [CrossRef] [PubMed]
- Riches, Z.; Stanley, E.L.; Bloomer, J.C.; Coughtrie, M.W. Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: the SULT “pie”. Drug Metab. Dispos. [CrossRef]
- Cabras, T.; Fanali, C.; Monteiro, J.A.; Amado, F.; Inzitari, R.; Desiderio, C.; Giardina, B.; Castagnola, M.; Messana, I. Tyrosine polysulfation of human salivary histatin 1. A post-translational modification specific of the submandibular gland. J. Proteome Res. 2007, 6, 2472–2480. [Google Scholar] [CrossRef] [PubMed]
- Poole, L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015, 80, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C.; Hampton, M.B. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008, 45, 549–561. [Google Scholar] [CrossRef]
- Cabras, T.; Manconi, B.; Iavarone, F.; Fanali, C.; Nemolato, S.; Fiorita, A.; Scarano, E.; Passali, G.C.; Manni, A.; Cordaro, M.; et al. RP-HPLC-ESI-MS evidenced that salivary cystatin B is detectable in adult human saliva mostly as S-modified derivatives: S glutathionyl, S-cysteinyl and S-S 2-mer. J. Proteomics 2012, 75, 908–913. [Google Scholar] [CrossRef] [PubMed]
- Pol, E.; Björk, I. Contributions of individual residues in the N-terminal region of cystatin B (stefin B) to inhibition of cysteine proteinases. Biochim. Biophys. Acta 2003, 1645, 105–112. [Google Scholar] [CrossRef]
- Grek, C.L.; Zhang, J.; Manevich, Y.; Townsend, D.M.; Tew, K.D. Causes and consequences of cysteine S-glutathionylation. J. Biol. Chem. 2013, 288, 26497–26504. [Google Scholar] [CrossRef]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef]
- Leclerc, E.; Fritz, G.; Vetter, S.W.; Heizmann, C.W. Binding of S100 proteins to RAGE: an update. Biochim. Biophys. Acta 2009, 1793, 993–1007. [Google Scholar] [CrossRef]
- Zackular, J.P.; Chazin, W.J.; Skaar, E.P. Nutritional Immunity: S100 Proteins at the Host-Pathogen Interface. J. Biol. Chem. 2015, 290, 18991–18998. [Google Scholar] [CrossRef]
- Lim, S.Y.; Raftery, M.J.; Goyette, J.; Hsu, K.; Geczy, C.L. Oxidative modifications of S100 proteins: functional regulation by redox. J. Leukoc. Biol. 2009, 86, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.Y.; Raftery, M.J.; Goyette, J.; Geczy, C.L. S-glutathionylation regulates inflammatory activities of S100A9. J. Biol. Chem. 2010, 285, 14377–14388. [Google Scholar] [CrossRef] [PubMed]
- Boroumand, M.; Manconi, B.; Serrao, S.; Iavarone, F.; Olianas, A.; Cabras, T.; Contini, C.; Pieroni, L.; Sanna, M.T.; Vento, G.; et al. Investigation by top-down high-performance liquid chromatography–mass spectrometry of glutathionylation and cysteinyl ation of salivary S100A9 and cystatin B in preterm newborns. Sep. Sci. plus 2022, 5, 17–27. [Google Scholar] [CrossRef]
- Iavarone, F.; Melis, M.; Platania, G.; Cabras, T.; Manconi, B.; Petruzzelli, R.; Cordaro, M.; Siracusano, A.; Faa, G.; Messana, I.; et al. Characterization of salivary proteins of schizophrenic and bipolar disorder patients by top-down proteomics. J. Proteomics 2014, 103, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Contini, C.; Firinu, D.; Serrao, S.; Manconi, B.; Olianas, A.; Cinetto, F.; Cossu, F.; Castagnola, M.; Messana, I.; Del Giacco, S.; et al. RP-HPLC-ESI-IT Mass Spectrometry Reveals Significant Variations of the Human Salivary Protein Profile Associated with Predominantly Antibody Deficiencies. J. Clin. Immunol. 2020, 40, 329–339. [Google Scholar] [CrossRef]
- Hoskin, T.S.; Crowther, J.M.; Cheung, J.; Epton, M.J.; Sly, P.D.; Elder, P.A.; Dobson, R.C.J.; Kettle, A.J.; Dickerhof, N. Oxidative cross-linking of calprotectin occurs in vivo, altering its structure and susceptibility to proteolysis. Redox Biol. 2019, 24, 101202. [Google Scholar] [CrossRef]
- Gomes, L.H.; Raftery, M.J.; Yan, W.X.; Goyette, J.D.; Thomas, P.S.; Geczy, C.L. S100A8 and S100A9-oxidant scavengers in in flammation. Free Radic. Biol. Med. 2013, 58, 170–186. [Google Scholar] [CrossRef]
- Smith, T.S.; Bennett, J.P.Jr. Mitochondrial toxins in models of neurodegenerative diseases. I: In vivo brain hydroxyl radical production during systemic MPTP treatment or following microdialysis infusion of methylpyridinium or azide ions. Brain Res. 1997, 765, 183–188. [Google Scholar] [CrossRef]
- Karlík, M.; Valkovič, P.; Hančinová, V.; Krížová, L.; Tóthová, L.; Celec, P. Markers of oxidative stress in plasma and saliva in patients with multiple sclerosis. Clin. Biochem. 2015, 48, 24–28. [Google Scholar] [CrossRef]
- Schipper, H.M.; Arnold, D.; GrandʼMaison, F.; Melmed, C.; Moore, F.; Levental, M. Constantin Su, M., Stril, J.L. Godin J., Tol erability and safety of combined glatiramer acetate and N-acetylcysteine in relapsing-remitting multiple sclerosis. Clin. Neuro pharmacol, 38. [CrossRef]
- Yao, Y.; Berg, E.A.; Costello, C.E.; Troxler, R.F.; Oppenheim, F.G. Identification of protein components in human acquired enamel pellicle and whole saliva using novel proteomics approaches. J. Biol. Chem. [CrossRef]
- Schüpbach, P.; Oppenheim, F.G.; Lendenmann, U.; Lamkin, M.S.; Yao, Y.; Guggenheim, B. Electron-microscopic demonstration of proline-rich proteins, statherin, and histatins in acquired enamel pellicles in vitro. Eur J Oral Sci. 2001, 109, 60–68. [Google Scholar] [CrossRef]
- Hannig, C.; Hannig, M.; Kensche, A.; Carpenter, G. The mucosal pellicle - An underestimated factor in oral physiology. Arch. Oral Biol. 2017, 80, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Presland, R.B.; Dale, B.A. Epithelial structural proteins of the skin and oral cavity: function in health and disease. Crit. Rev. Oral Biol. Med. 2000, 11, 383–408. [Google Scholar] [CrossRef] [PubMed]
- Bradway, S. D.; Bergey, E.J.; Jones, P.C.; Levine, M.J. Oral mucosal pellicle. Adsorption and transpeptidation of salivary com ponents to buccal epithelial cells. Biochem. J. 1989, 261, 887–896. [Google Scholar] [CrossRef]
- Cabiddu, G.; Maes, P.; Hyvrier, F.; Olianas, A.; Manconi, B.; Brignot, H.; Canon, F.; Cabras, T.; Morzel, M. Proteomic character ization of the mucosal pellicle formed in vitro on a cellular model of oral epithelium. J. Proteomics 2020, 222, 103797. [Google Scholar] [CrossRef]
- Neyraud, E.; Morzel, M. Biological films adhering to the oral soft tissues: Structure, composition, and potential impact on taste perception. J. Texture Stud. 2019, 50, 19–26. [Google Scholar] [CrossRef]
- Perez Alea, M.; Thomas, V.; Martin, G.; El Alaoui, S. Identification of human salivary transglutaminases. Amino Acids 2013, 44, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Griffin, M. TG2, a novel extracellular protein with multiple functions. Amino Acids 2012, 42, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Boroumand, M.; Olianas, A.; Manconi, B.; Serrao, S.; Iavarone, F.; Desiderio, C.; Pieroni, L.; Faa, G.; Messana, I.; Castagnola, M.; et al. Mapping of transglutaminase 2 sites of human salivary small basic proline-rich proteins by HPLC high resolution ESI MS/MS. J. Proteome Res. 2020, 19, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Cabras, T.; Inzitari, R.; Fanali, C.; Scarano, E.; Patamia, M. Sanna, M.T.; Pisano, E.; Giardina, B.; Castagnola, M.; Messana, I. HPLC-MS characterization of cyclo-statherin Q-37, a specific cyclization product of human salivary statherin generated by transglutaminase 2. J. Sep. Sci. 2006, 29, 2600–2608. [Google Scholar] [CrossRef]
- Hannig, C.; Hannig, M.; Attin, T. Enzymes in the acquired enamel pellicle. Eur. J. Oral Sci. 2005, 113, 2–13. [Google Scholar] [CrossRef]
- Siqueira, W.L.; Custodio, W.; McDonald, E.E. New insights into the composition and functions of the acquired enamel pellicle. J. Dent. Res. 2012, 91, 1110–1118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ames, J.M.; Smith, R.D.; Baynes, J.W.; Metz, T.O. A Perspective on the Maillard Reaction and the Analysis of protein Glycation by Mass Spectrometry: Probing the Pathogenesis of Chronic Disease. J. Proteome Res. 2009, 8, 754–769. [Google Scholar] [CrossRef] [PubMed]
- Rahbar, S.; Blumenfe, O.; Ranney, H.M. Studies of an unusual hemoglobin in patients with diabetes mellitus. Biochem. Biophys. Re.s Commun. 1969, 36, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, I.; Morimoto, K.; Takeshita, T.; Toda, M. Correlation between Saliva Glycated and Blood Glycated Proteins. Environ. Health Prev. Med. 2003, 8, 95–99. [Google Scholar] [CrossRef]
- Goldstein, D.E.; Little, R.R.; Lorenz, R.A.; Malone, J.I.; Nathan, D.; Peterson, C.M. Tests of glycemia in diabetes. Diabetes Care 1995, 18, 896–909. [Google Scholar] [CrossRef]
- Ramasamy. R.; Vannucci, S.J.; Yan, S.S.; Herold, K.; Yan, S.F.; Schmidt, A.M. Advanced glycation end products and RAGE: a common thread in aging, diabetes, neurodegeneration, and inflammation. Glycobiology 2005, 15, 16R–28R. [Google Scholar] [CrossRef]
- Corica, D.; Pepe, G.; Currò, M.; Aversa, T.; Tropeano, A.; Ientile, R.; Wasniewska, M. Methods to investigate advanced glycation end-product and their application in clinical practice. Methods. 2022, 203, 90–102. [Google Scholar] [CrossRef]
- McCarthy, A.D.; Cortizo, A.M.; Giménez Segura, G.; Bruzzone, L.; Etcheverry, S.B. Non-enzymatic glycosylation of alkaline phosphatase alters its biological properties. Mol. Cell. Biochem. 1998, 181, 63–69. [Google Scholar] [CrossRef]
- Muraoka, M.Y.; Justino, A.B; Caixeta, D.C.; Queiroz, J.S.; Sabino-Silva, R.; Espindola, F.S. Fructose and methylglyoxal-induced glycation alters structural and functional properties of salivary proteins, albumin and lysozyme. PLoS One 2022, 17, e0262369. [Google Scholar] [CrossRef]
- Tessier, F.J. The Maillard reaction in the human body. The main discoveries and factors that affect glycation. Pathol. Biol. (Paris) 2010, 58, 214–219. [Google Scholar] [CrossRef]
- Shin, A.; Connolly, S.; Kabytaev, K. Protein glycation in diabetes mellitus. Adv. Clin. Chem. 2023, 113, 101–156. [Google Scholar] [CrossRef] [PubMed]
- Li. Y.M. Glycation ligand binding motif in lactoferrin. Implication in diabetic infection. Adv. Exp. Med. Biol. 1998, 443, 57–63. [Google Scholar] [CrossRef]
- Belce, A.; Uslu, E.; Kucur, M.; Umut, M.; Ipbuker, A.; Seymen, H.O. Evaluation of salivary sialic acid level and Cu-Zn superoxide dismutase activity in type 1 diabetes mellitus. Tohoku J. Exp. Med. 2000, 192, 219–225. [Google Scholar] [CrossRef]
- Ilea, A.; Băbţan, A.M.; Boşca, B.A.; Crişan, M.; Petrescu, N.B.; Collino, M.; Sainz, R.M.; Gerlach, J.Q.; Câmpian, R.S. Advanced Glycation end products (AGEs) in oral pathology. Arch. Oral Biol. 2018, 93, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Manig, F.; Hellwig, M.; Pietz, F.; Henle, T. Quantitation of free glycation compounds in saliva. PLoS One 2019, 14, e0220208. [Google Scholar] [CrossRef]
- Landesberg, R.; Woo, V.; Huang, L.; Cozin, M.; Lu, Y.; Bailey, C.; Qu, W.; Pulse, C.; Schmidt, A.M. The expression of the receptor for glycation endproducts (RAGE) in oral squamous cell carcinomas. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2008, 105, 617–624. [Google Scholar] [CrossRef]
- Maciejczyk, M.; Nesterowicz, M.; Szulimowska, J.; Zalewska, A. Oxidation, Glycation, and Carbamylation of Salivary Biomol ecules in Healthy Children, Adults, and Elderly: Can Saliva Be Used in the assessment of Aging? J. Inflam. Res. 2022, 15, 2051–2073. [Google Scholar] [CrossRef]
- Goyette, J.; Geczy, C.L. Inflammation-associated S100 proteins: new mechanisms that regulate function. Amino Acids 2011, 41, 821–842. [Google Scholar] [CrossRef]
- Agho, E.T.; Owotade, F.J.; Kolawole, B.A.; Oyetola, E.O.; Adedeji, T.A. ; Salivary inflammatory biomarkers and glycated hae moglobin among patients with type 2 diabetic mellitus. BMC Oral Health 2021, 21, 101. [Google Scholar] [CrossRef]
- Helenius, A.; Aebi, M. Intracellular functions of N-linked glycans. Science 2001, 291, 2364–2369. [Google Scholar] [CrossRef]
- Xu, C.; Ng, D.T.W. Glycosylation-directed quality control of protein folding. Nat. Rev. Mol. Cell. Biol. 2015, 16, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Minakata, S.; Manabe, S.; Inai, Y.; Ikezaki, M.; Nishitsuji, K.; Ito, Y.; Ihara, Y. Protein C-Mannosylation and C-Mannosyl Tryp- tophan in Chemical Biology and Medicine. Molecules 2021, 26, 5258. [Google Scholar] [CrossRef] [PubMed]
- Franc, V.; Yang, Y.; Heck, A.J. Proteoform profile mapping of the human serum complement component C9 revealing unex- pected new features of N-, O-, and C-glycosylation. Anal. Chem. 2017, 89, 3483–3491. [Google Scholar] [CrossRef] [PubMed]
- Siew, J.J.; Chern, Y. Microglial Lectins in Health and Neurological Diseases. Front. Mol. Neurosci. 2018, 11, 158. [Google Scholar] [CrossRef] [PubMed]
- Duca, M.; Malagolini, N.; Dall’Olio, F. The Mutual Relationship between Glycosylation and Non-Coding RNAs in Cancer and Other Physio-Pathological Conditions. Int. J. Mol. Sci. 2022, 23, 15804. [Google Scholar] [CrossRef] [PubMed]
- Borga, C.; Meeran, S.M.; Fassan, M. Non-coding RNAs, a real Next-Gen Class of Biomarkers? Noncoding RNA Res. 2019, 4, 80–81. [Google Scholar] [CrossRef]
- Flynn, R.A.; Pedram, K.; Malaker, S.A.; Batista, P.J.; Smith, B.A.H.; Johnson, A.G.; George, B.M. ; Majzoub. K.; Villalta, P.W.; Carette, J.E.; et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell, 3109. [Google Scholar] [CrossRef]
- Morelle, W.; Canis, K.; Chirat, F.; Faid, V.; Michalski, J.C. The use of mass spectrometry for the proteomic analysis of glycosyl- ation. Proteomics 2006, 6, 3993–4015. [Google Scholar] [CrossRef]
- Sondej, M.; Denny, P.A.; Xie, Y.; Ramachandran, P.; Si, Y.; Takashima, J.; Shi, W.; Wong, D.T.; Loo, J.A.; Denny, P.C. Glycop ro- filing of the Human Salivary Proteome. Clin. Proteomics. 2009, 5, 52–68. [Google Scholar] [CrossRef]
- Cabras, T.; Boi, R.; Pisano, E.; Iavarone, F.; Fanali, C.; Nemolato, S.; Faa, G.; Castagnola, M.; Messana, I. HPLC-ESI-MS and MS/MS structural characterization of multifucosylated N-glycoforms of the basic proline-rich protein IB-8a CON1(+) in human saliva. J. Sep. Sci. 2012, 35, 1079–1086. [Google Scholar] [CrossRef]
- Manconi, B.; Cabras, T.; Sanna, M.; Piras, V.; Liori, B.; Pisano, E.; Iavarone, F.; Vincenzoni, F.; Cordaro, M.; Faa, G.; et al. N- and O-linked glycosylation site profiling of the human basic salivary proline-rich protein 3M. J. Sep. Sci. 2016, 39, 1987–1997. [Google Scholar] [CrossRef]
- Liu, S.; Wang, H.; Jiang, X.; Ji, Y.; Wang, Z.; Zhang, Y.; Wang, P.; Xiao, H. Integrated N-glycoproteomics Analysis of Human Saliva for Lung Cancer. J. Proteome Res. 2022, 21, 1589–1602. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Kobeissy, F.; Mondello, S.; Barsa, C.; Mechref, Y. MS-based glycomics: An analytical tool to assess nervous system diseases. Front. Neurosci. 2022, 16, 1000179. [Google Scholar] [CrossRef] [PubMed]
- Robertson, L.A.; Moya, K.L.; Breen, K.C. The potential role of tau protein O-glycosylation in Alzheimer’s disease. J. Alzheimers Dis. 2004, 6, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer. 2015, 15, 540–555. [Google Scholar] [CrossRef]
- György, B.; Tóth, E.; Tarcsa, E.; Falus, A.; Buzás, E.I. Citrullination: a posttranslational modification in health and disease. Int. J. Biochem. Cell. Biol. 2006, 38, 1662–1677. [Google Scholar] [CrossRef]
- Anzilotti, C.; Pratesi, F.; Tommasi, C.; Migliorini, P. Peptidylarginine deiminase 4 and citrullination in health and disease. Au- toimmun. Rev. 9,. [CrossRef]
- Alghamdi, M.; Alasmari, D.; Assiri, A.; Mattar, E.; Aljaddawi, A.A.; Alattas, S.G.; Redwan, E.M. An Overview of the Intrinsic Role of Citrullination in Autoimmune Disorders. J. Immunol. Res. 2019, 2019, 7592851. [Google Scholar] [CrossRef]
- van Venrooij, W.J.; Pruijn, G.J. Citrullination: a small change for a protein with great consequences for rheumatoid arthritis. Arthritis Res. 2000, 2, 249–251. [Google Scholar] [CrossRef]
- Ciesielski, O.; Biesiekierska, M.; Panthu, B.; Soszyński, M.; Pirola, L.; Balcerczyk, A. Citrullination in the pathology of inflam matory and autoimmune disorders: recent advances and future perspectives. Cell. Mol. Life Sci. 2022, 79, 94. [Google Scholar] [CrossRef]
- Bay-Jensen, A.C.; Sand, J.M.B.; Genovese, F.; Siebuhr, A.S.; Nielsen, M.J.; Leeming, D.J.; Manon-Jensen, T.; Karsdal, M.A. Chap ter 31 - Structural Biomarkers. In Biochemistry of Collagens, Laminins and Elastin, 1st ed.; Karsdal, M., Ed.; Elsevier: Radarweg 29, 1043 NX Amsterdam, The Netherlands, 2016; pp. 203–233. [Google Scholar] [CrossRef]
- Yasuda, T.; Tahara, K.; Sawada, T. Detection of salivary citrullinated cytokeratin 13 in healthy individuals and patients with rheumatoid arthritis by proteomics analysis. PLoS One. 2022, 17, e0265687. [Google Scholar] [CrossRef]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Anzilotti, C.; Pratesi, F.; Tommasi, C.; Migliorini, P. Peptidylarginine deiminase 4 and citrullination in health and disease. Au toimmun. Rev. 2010, 9, 158–160. [Google Scholar] [CrossRef] [PubMed]
- Tar, I.; Csősz, É.; Végh, E.; Lundberg, K.; Kharlamova, N.; Soós, B.; Szekanecz, Z.; Márton, I. Salivary citrullinated proteins in rheumatoid arthritis and associated periodontal disease. Sci. Rep. 2021, 11, 13525. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, W.; To, M.; Yamamoto, Y.; Inaba, K.; Yakeishi, M.; Saruta, J.; Fuchida, S.; Hamada, N.; Tsukinoki, K. Detection of anti-citrullinated protein antibody (ACPA) in saliva for rheumatoid arthritis using DBA mice infected with Porphyromonas gingivalis. Arch. Oral. Biol. 2019, 108, 104510. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Esparza, R; Rodríguez-Rodríguez, M. ; Pérez-Pérez, M.E.; Badillo-Soto, M.A.; Torres-Del-Muro, F.; Bollain-Y-Goytia, J.J.; Pacheco-Tovar, D.; Avalos-Díaz, E. Posttranslational Protein Modification in the Salivary Glands of Sjögren’s Syndrome Patients. Autoimmune Dis. 2013, 2013, 548064. [Google Scholar] [CrossRef]
- Giglione, C.; Fieulaine, S.; Meinnel, T. N-terminal protein modifications: bringing back into play the ribosome. Biochimie 2015, 114, 134–146. [Google Scholar] [CrossRef]
- Deng, S.; Marmorstein, R. Protein N-Terminal Acetylation: Structural Basis, Mechanism, Versatility, and Regulation. Trends Biochem. Sci. 2021, 46, 15–27. [Google Scholar] [CrossRef]
- Vijayan, D.K.; Zhang, K.Y.J. Human glutaminyl cyclase: Structure, function, inhibitors and involvement in Alzheimer’s disease. Pharmacol. Res. 2019, 147, 104342. [Google Scholar] [CrossRef]
- Castagnola, M.; Cabras, T.; Iavarone, F.; Vincenzoni, F.; Vitali, A.; Pisano, E.; Nemolato, S.; Scarano, E.; Fiorita, A.; Vento, G.; et al. Top-down platform for deciphering the human salivary proteome. J. Matern. Fetal Neonatal Med. 2012, 25, 27–43. [Google Scholar] [CrossRef]
- Manconi, B.; Cabras, T.; Pisano, E.; Nemolato, S.; Inzitari, R.; Iavarone, F.; Fanali, C.; Sanna, M.T.; Tirone, C.; Vento, G.; et al. 1637 Characterization of two isoforms of SPRR3 from preterm human newborn saliva, autoptic fetal oral mucosa, parotid and sub mandibular gland samples. Biochem. Biophys, Res. Commun. 2010, 398, 477–481. [Google Scholar] [CrossRef]
- Cabras, T.; Iavarone, F.; Manconi, B.; Olianas, A.; Sanna, M.T.; Castagnola, M.; Messana, I. Top-down analytical platforms for the characterization of the human salivary proteome. Bioanalysis 2014, 6, 563–581. [Google Scholar] [CrossRef]
- Van Damme, P.; Hole, K.; Pimenta-Marques, A.; Helsens, K.; Vandekerckhove, J.; Martinho, R.G.; Gevaert, K.; Arnesen, T. NatF Contributes to an Evolutionary Shift in Protein N-Terminal acetylation and is important for normal chromosome segrega tion. PLoS Genet. 2011, 7, e1002169. [Google Scholar] [CrossRef] [PubMed]
- Varland, S.; Osberg, C.; Arnesen, T. N-terminal modifications of cellular proteins: the enzymes involved, their substrate speci ficity and biological effects. Proteomics 2015, 15, 2385–2401. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, R.; Alves, R.; Barro, A.; Caseiro, A.; Ferreira, R.; Lobo, M.C.; Bastos, A.; Duarte, J.; Carvalho, D.; Santos, L.L. Finding new translational modifications in salivary proline-rich proteins. Proteomics, 3742. [Google Scholar] [CrossRef]
- Schilling, S.; Wasternack, C.; Demuth, H.U. Glutaminyl cyclase from animals and plants: a case of functionally convergent protein evolution. Biol. Chem. 2008, 389, 983–991. [Google Scholar] [CrossRef]
- Schlenzig, D.; Manhart, S.; Cinar, Y.; Kleinschmidt, M.; Hause, G.; Willbold, D.; Funke, S.A.; Schilling, S.; Demuth, H.U. Py roglutamate formation influences solubility and amyloidogenicity of amyloid peptides. Biochemistry 2009, 48, 7072–7078. [Google Scholar] [CrossRef] [PubMed]
- Wirths, O.; Breyhan, H.; Cynis, H.; Schilling, S.; Demuth, H.U.; Bayer, T.A. Intraneuronal pyroglutamate-Abeta 3-42 triggers neurodegeneration and lethal neurological deficits in a transgenic mouse model. Acta Neuropathol. 2009, 118, 487–496. [Google Scholar] [CrossRef]
- Bersin, L.M.; Patel, S.M.; Topp, E.M. Effect of ‘pH’ on the rate of pyroglutamate formation in solution and lyophilized sol ids. Mol. Pharm. 2021, 18, 3116–3124. [Google Scholar] [CrossRef]
- Castagnola, M.; Cabras, T.; Vitali, A.; Sanna, M.T.; Messana, I. Biotechnological implication of the salivary proteome. Trends Biotechnol. 2011, 29, 409–418. [Google Scholar] [CrossRef]
- Schulte, S.J.; Huang, J.; Pierce, N.A. ; Hybridization Chain Reaction Lateral Flow Assays for Amplified Instrument-Free At Home SARS-CoV-2 Testing. ACS Infect. Dis. 2023, 9, 450–458. [Google Scholar] [CrossRef]
- Yu, H.; Lee, H.; Cheong, J.; Woo, S.W.; Oh, J.; Oh, H.K.; Lee, J.H.; Zheng, H.; Castro, C.M.; Yoo, Y.E.; et al. A rapid assay provides on-site quantification of tetrahydrocannabinol in oral fluid. Sci. Transl. Med. 2021, 13, eabe2352. [Google Scholar] [CrossRef]

| Protein and Peptides | C-term. cleavage |
|---|---|
| aPRP Pa-dimer | …..QSP↓Q |
| PRP3/PRP4/PIF-f | …..RPP↓R |
| P-C (or IB-8b) | ….QS↓P↓Q |
| bPRP II-2 | …..RSP↓R |
| bPRP IB-1 | .….RSP↓R |
| bPRP PF (or IB-8c) precursor | .…RSA↓R |
| bPRP PE (or IB-9) | ….RSP↓R |
| Protein/peptide | Partial sequence | Consensus | Ref.s |
|---|---|---|---|
| Statherin (Ser2, Ser3) | DSSEEKF… | SXE | [41] |
| Histatin 1 (Ser2) | DSHEKR… | SXE | [103] |
| Cystatin S (Ser3) | SSSKEENR… | SXE | [57,58,59,83] |
| Cystatin S1 (Ser1) | SSS(Phos)KEENR… | SXS(Phos) | “ |
| aPRP (Types 1 and 3) (Ser8) | …DEDVSQEDV…. | SXE | [9] |
| aPRP (Types 1 and 3) (Ser22) | …GGDSEQFIDEER… | SX(3/4)(E/D/S(phos))3 | “ |
| aPRP (Types 1 and 3) (Ser17) | …LVISDGG DS(phos)EQFI… | SX(3/4)(E/D/S(phos))3 | “ |
| bPRP II-2 (Ser8) | …NEDVSQEESPS… | SXE | [23] |
| bPRP IB-1 (Ser8) | …NEDVSQEESPS… | SXE | “ |
| gPRP Gl-2 or PRP-3M (Ser8) | …NEDVSQEESPS… | SXE | “ |
| gPRP Gl-1 or PRP-3L (Ser8)1 | …NEDVSQEESPS… | SXE | “ |
| gPRP Gl-3 or PRP-3S (Ser8)1 | …NEDVSQEESPS… | SXE | “ |
| gPRP Glycosyl. Protein A (Ser8)1 | …SEDVSQEESLFL… | SXE | “ |
| gPRP II-1 (Ser8)1 | …SEDVSQEESLFL… | SXE | “ |
| gPRP Cd-IIg (Ser8)1 | …SEDVSQEESLFL… | SXE | “ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





