Submitted:
03 August 2023
Posted:
04 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample collection, RNA extraction, and quantification
2.2. Next-generation sequencing and analysis
2.3. Sequence comparison and phylogenetic analysis
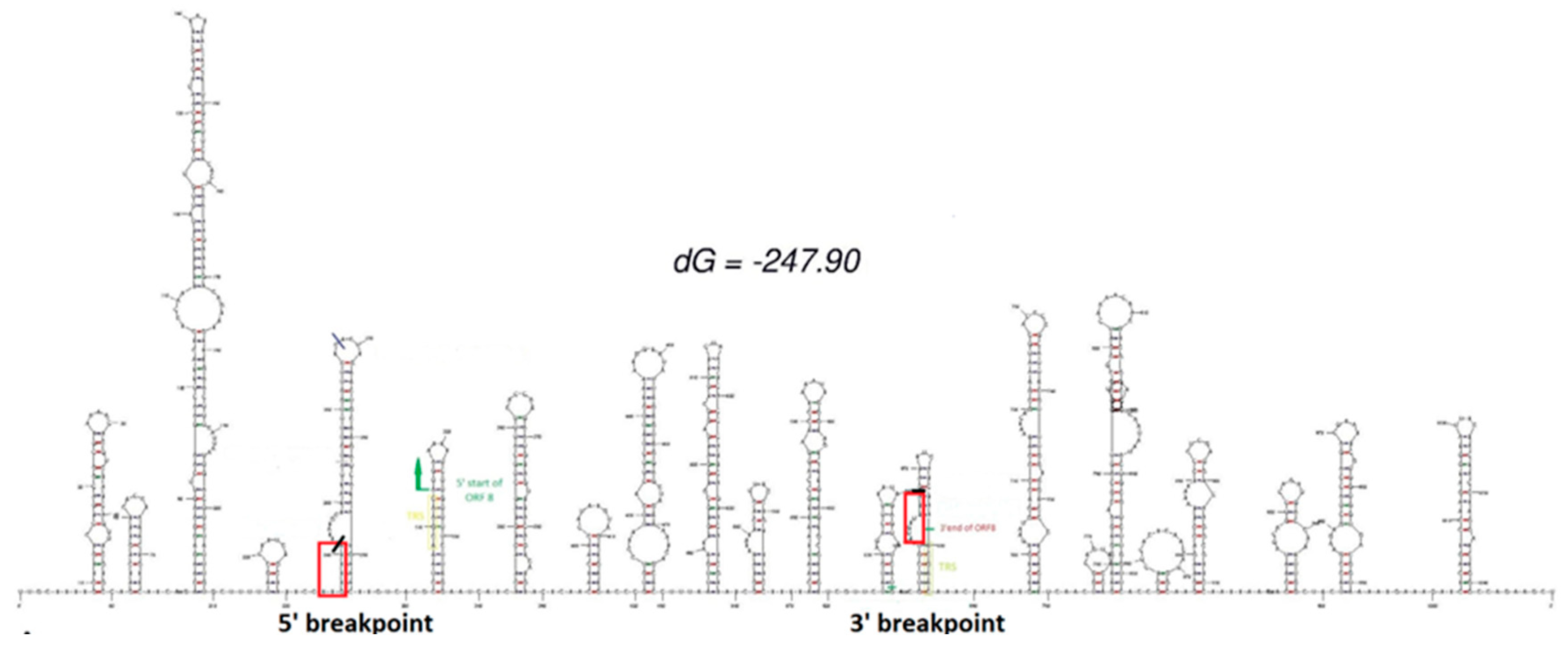
2.4. RNA secondary structure prediction
2.5. RNA retro-transcription, amplification and gel electrophoresis
2.6. PCR clean-up and Sanger sequencing
3. Results
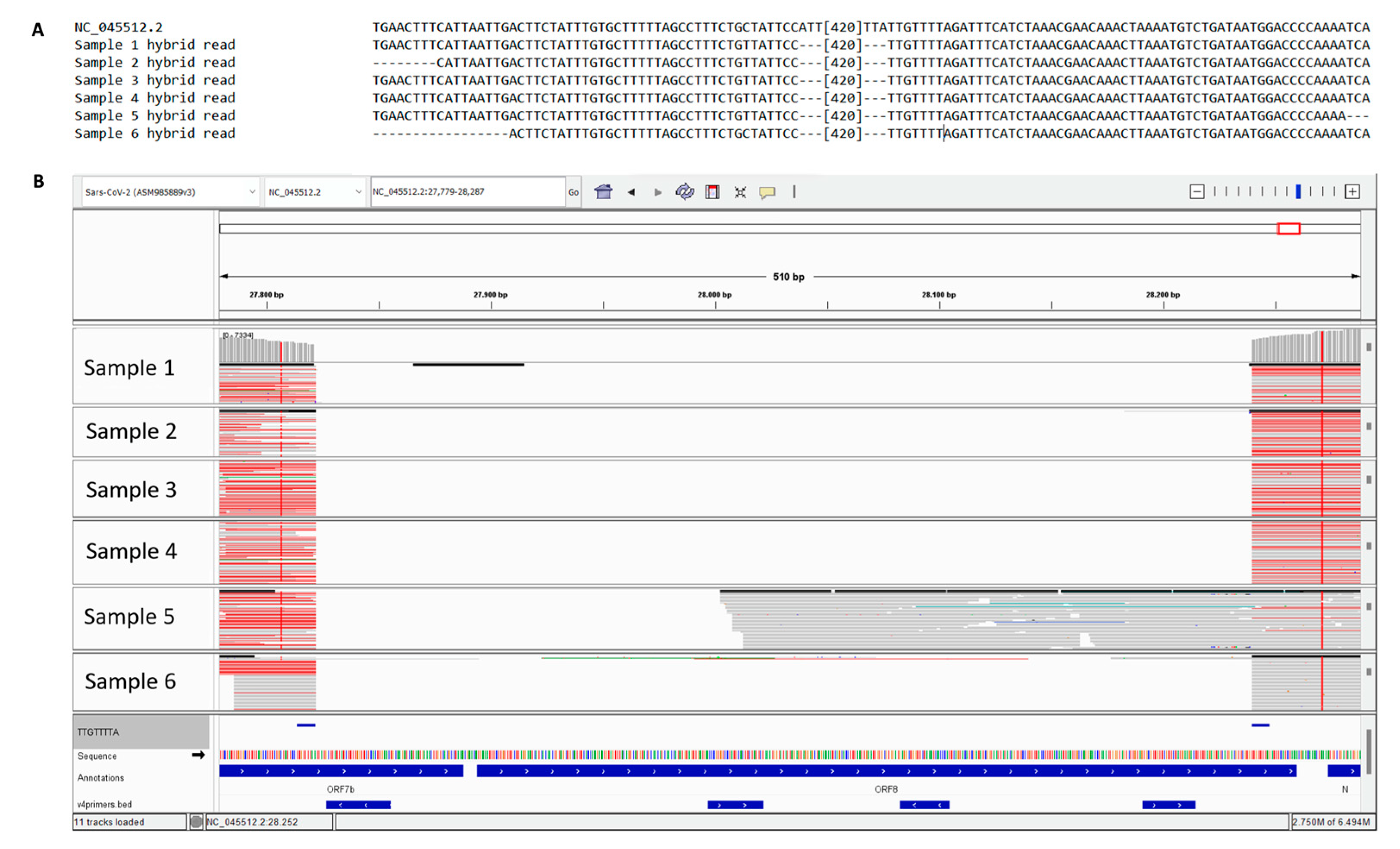
3.1. Determination of 6 cases of SARS-CoV-2 with 426 nt deletions in the ORF7b and ORF8 regions
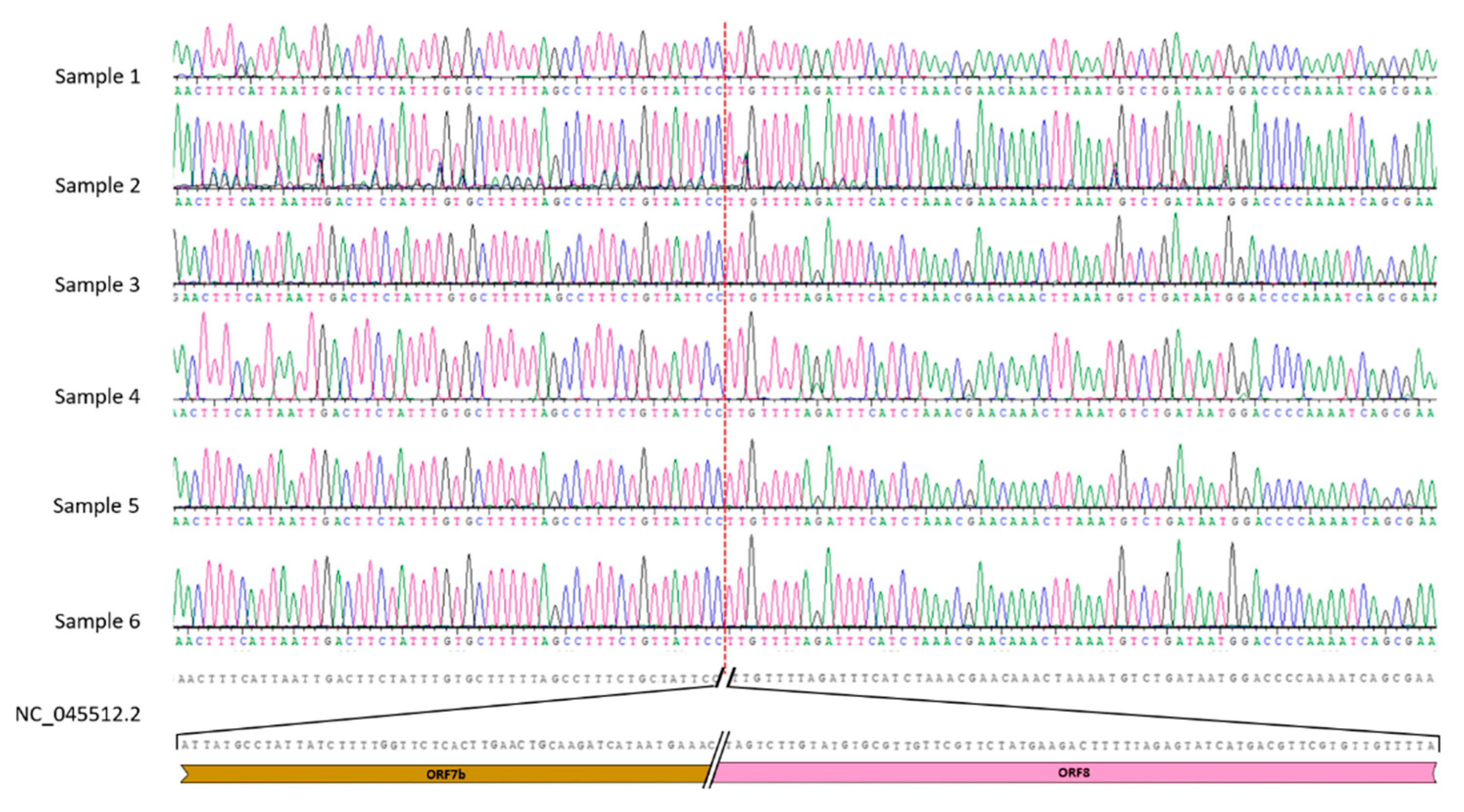
3.2. Sanger analysis confirms the deletion and its consistence
3.3. Phylogenetic analysis of SARS-CoV-2 strains with 426 nt deletions
3.4. Clinical context of patients carrying the Δ426 mutation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.M.; Wang, W.; Song, Z.-G.G.; Hu, Y.; Tao, Z.-W.W.; Tian, J.-H.H.; Pei, Y.-Y.Y.; et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. The Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M. Coronavirus Genomic Nsp14-ExoN, Structure, Role, Mechanism, and Potential Application as a Drug Target. J Med Virol 2021, 93, 4258–4264. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827. [Google Scholar] [CrossRef]
- Focosi, D.; Maggi, F. Recombination in Coronaviruses, with a Focus on SARS-CoV-2. Viruses 2022, 14, 1239. [Google Scholar] [CrossRef]
- Kleynhans, J.; Tempia, S.; Wolter, N.; von Gottberg, A.; Bhiman, J.N.; Buys, A.; Moyes, J.; McMorrow, M.L.; Kahn, K.; Gómez-Olivé, F.X.; et al. SARS-CoV-2 Seroprevalence in a Rural and Urban Household Cohort during First and Second Waves of Infections, South Africa, July 2020-March 2021. Emerg Infect Dis 2021, 27, 3020–3029. [Google Scholar] [CrossRef]
- Arora, P.; Kempf, A.; Nehlmeier, I.; Schulz, S.R.; Jäck, H.M.; Pöhlmann, S.; Hoffmann, M. Omicron Sublineage BQ.1.1 Resistance to Monoclonal Antibodies. Lancet Infect Dis 2023, 23, 22–23. [Google Scholar] [CrossRef] [PubMed]
- Rössler, A.; Riepler, L.; Bante, D.; von Laer, D.; Kimpel, J. SARS-CoV-2 Omicron Variant Neutralization in Serum from Vaccinated and Convalescent Persons. New England Journal of Medicine 2022, 386, 698–700. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; Hill, V.; O’Toole, Á.; McCrone, J.; Ruis, C.; Plessis, L. du; Pybus, O.G. A Dynamic Nomenclature Proposal for SARS-CoV-2 to Assist Genomic Epidemiology. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Scialo, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Castaldo, G.; Bianco, A. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung 2020, 198, 867–877. [Google Scholar] [CrossRef]
- Rahman, M.S.; Islam, M.R.; Alam, A.S.M.R.U.; Islam, I.; Hoque, M.N.; Akter, S.; Rahaman, M.M.; Sultana, M.; Hossain, M.A. Evolutionary Dynamics of SARS-CoV-2 Nucleocapsid Protein and Its Consequences. J Med Virol 2021, 93, 2177–2195. [Google Scholar] [CrossRef]
- Syed, A.M.; Ciling, A.; Taha, T.Y.; Chen, I.P.; Khalid, M.M.; Sreekumar, B.; Chen, P.Y.; Renuka Kumar, G.; Suryawanshi, R.; Silva, I.; et al. Omicron Mutations Enhance Infectivity and Reduce Antibody Neutralization of SARS-CoV-2 Virus-like Particles. Proc Natl Acad Sci U S A 2022, 119, e2200592119. [Google Scholar] [CrossRef]
- SARS-CoV-2 Evolution, Post-Omicron - SARS-CoV-2 Coronavirus / SARS-CoV-2 Molecular Evolution - Virological. Available online: https://virological.org/t/sars-cov-2-evolution-post-omicron/911 (accessed on 17 February 2023).
- Rockett, R.J.; Draper, J.; Gall, M.; Sim, E.M.; Arnott, A.; Agius, J.E.; Johnson-Mackinnon, J.; Fong, W.; Martinez, E.; Drew, A.P.; et al. Co-Infection with SARS-CoV-2 Omicron and Delta Variants Revealed by Genomic Surveillance. Nature communications 2022, 13, 2745. [Google Scholar] [CrossRef]
- Truong, T.T.; Ryutov, A.; Pandey, U.; Yee, R.; Goldberg, L.; Bhojwani, D.; Aguayo-Hiraldo, P.; Pinsky, B.A.; Pekosz, A.; Shen, L.; et al. Increased Viral Variants in Children and Young Adults with Impaired Humoral Immunity and Persistent SARS-CoV-2 Infection: A Consecutive Case Series. EBioMedicine 2021, 67. [Google Scholar] [CrossRef]
- Evolutionary Insight into the Emergence of SARS-CoV-2 Variants of Concern. Nature Medicine 2022, 28, 1357–1358. [CrossRef]
- Jackson, B.; Boni, M.F.; Bull, M.J.; Colleran, A.; Colquhoun, R.M.; Darby, A.C.; Haldenby, S.; Hill, V.; Lucaci, A.; McCrone, J.T.; et al. Generation and Transmission of Interlineage Recombinants in the SARS-CoV-2 Pandemic. Cell 2021, 184, 5179–5188.e8. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, J.; Msomi, N.; et al. Emergence of a SARS-CoV-2 Variant of Concern with Mutations in Spike Glycoprotein. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Xie, X.; Liu, Y.; Liu, J. ✉ Neutralization of SARS-CoV-2 Spike 69/70 Deletion, E484K and N501Y Variants by BNT162b2 Vaccine-Elicited Sera. Nat Med 2021, 27, 620–621. [Google Scholar] [CrossRef]
- Su, Y.C.F.; Anderson, D.E.; Young, B.E.; Linster, M.; Zhu, F.; Jayakumar, J.; Zhuang, Y.; Kalimuddin, S.; Low, J.G.H.; Tan, C.W.; et al. Discovery and Genomic Characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2. mBio 2020, 11. [Google Scholar] [CrossRef]
- Hassan, Sk.S.; Kodakandla, V.; Redwan, E.M.; Lundstrom, K.; Pal Choudhury, P.; Abd El-Aziz, T.M.; Takayama, K.; Kandimalla, R.; Lal, A.; Serrano-Aroca, Á.; et al. An Issue of Concern: Unique Truncated ORF8 Protein Variants of SARS-CoV-2. PeerJ 2022, 10, e13136. [Google Scholar] [CrossRef]
- Vinjamuri, S.; Li, L.; Bouvier, M. SARS-CoV-2 ORF8: One Protein, Seemingly One Structure, and Many Functions. Front Immunol 2022, 13. [Google Scholar] [CrossRef]
- Tan, Y.; Schneider, T.; Leong, M.; Aravind, L.; Zhang, D. Novel Immunoglobulin Domain Proteins Provide Insights into Evolution and Pathogenesis of SARS-CoV-2-Related Viruses. mBio 2020, 11. [Google Scholar] [CrossRef]
- Rashid, F.; Dzakah, E.E.; Wang, H.; Tang, S. The ORF8 Protein of SARS-CoV-2 Induced Endoplasmic Reticulum Stress and Mediated Immune Evasion by Antagonizing Production of Interferon Beta. Virus Res 2021, 296, 198350. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xia, T.; Shin, W.-J.; Yu, K.-M.; Jung, W.; Herrmann, A.; Foo, S.-S.; Chen, W.; Zhang, P.; Lee, J.-S.; et al. Viral Mimicry of Interleukin-17A by SARS-CoV-2 ORF8. mBio 2022, 13. [Google Scholar] [CrossRef]
- Kee, J.; Thudium, S.; Renner, D.M.; Glastad, K.; Palozola, K.; Zhang, Z.; Li, Y.; Lan, Y.; Cesare, J.; Poleshko, A.; et al. SARS-CoV-2 Disrupts Host Epigenetic Regulation via Histone Mimicry. Nature 2022, 610, 381–388. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Li, Y.; Huang, F.; Luo, B.; Yuan, Y.; Xia, B.; Ma, X.; Yang, T.; Yu, F.; et al. The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through down-Regulating MHC-Ι. Proceedings of the National Academy of Sciences 2021, 118. [Google Scholar] [CrossRef]
- Stukalov, A.; Girault, V.; Grass, V.; Karayel, O.; Bergant, V.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multilevel Proteomics Reveals Host Perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021, 594, 246–252. [Google Scholar] [CrossRef]
- Redondo, N.; Zaldívar-López, S.; Garrido, J.J.; Montoya, M. SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns. Front Immunol 2021, 12, 2698. [Google Scholar] [CrossRef]
- Shemesh, M.; Aktepe, T.E.; Deerain, J.M.; McAuley, J.L.; Audsley, M.D.; David, C.T.; Purcell, D.F.J.; Urin, V.; Hartmann, R.; Moseley, G.W.; et al. SARS-CoV-2 Suppresses IFNβ Production Mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but Does Not Suppress the Effects of Added Interferon. PLoS Pathog 2021, 17, e1009800. [Google Scholar] [CrossRef]
- Lisberg Toft-Bertelsen, T.; Gravers Jeppesen, M.; Tzortzini, E.; Xue, K.; Giller, K.; Becker, S.; Mujezinovic, A.; Bentzen, B.H.; Andreas, L.B.; Kolocouris, A.; et al. Amantadine Inhibits Known and Novel Ion Channels Encoded by SARS-CoV-2 in Vitro. Commun Biol 2021, 4, 1347. [Google Scholar] [CrossRef]
- Lopera, J.G.; Falendysz, E.A.; Rocke, T.E.; Osorio, J.E. Attenuation of Monkeypox Virus by Deletion of Genomic Regions. Virology 2015, 475, 129–138. [Google Scholar] [CrossRef]
- Vignuzzi, M.; López, C.B. Defective Viral Genomes Are Key Drivers of the Virus–Host Interaction. Nat Microbiol 2019, 4, 1075–1087. [Google Scholar] [CrossRef]
- Bonfield, J.K.; Marshall, J.; Danecek, P.; Li, H.; Ohan, V.; Whitwham, A.; Keane, T.; Davies, R.M. HTSlib: C Library for Reading/Writing High-Throughput Sequencing Data. Gigascience 2021, 10, giab007. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol 2021. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat Biotechnol 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and Sequence Analysis Tools Services from EMBL-EBI in 2022. Nucleic Acids Res 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Turakhia, Y.; Thornlow, B.; Hinrichs, A.S.; De Maio, N.; Gozashti, L.; Lanfear, R.; Haussler, D.; Corbett-Detig, R. Ultrafast Sample Placement on Existing TRees (UShER) Enables Real-Time Phylogenetics for the SARS-CoV-2 Pandemic. Nat Genet 2021, 53, 809–816. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol Biol Evol 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Zuker, M. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucleic Acids Res 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Young, B.E.; Fong, S.W.; Chan, Y.H.; Mak, T.M.; Ang, L.W.; Anderson, D.E.; Lee, C.Y.P.; Amrun, S.N.; Lee, B.; Goh, Y.S.; et al. Effects of a Major Deletion in the SARS-CoV-2 Genome on the Severity of Infection and the Inflammatory Response: An Observational Cohort Study. The Lancet 2020, 396, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New Capabilities and Interfaces. Nucleic Acids Res 2012, 40, e115–e115. [Google Scholar] [CrossRef]
- Greenberg, J.A.; Hohmann, S.F.; Hall, J.B.; Kress, J.P.; David, M.Z. Validation of a Method to Identify Immunocompromised Patients with Severe Sepsis in Administrative Databases. Ann Am Thorac Soc 2016, 13, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, J.; Gao, K.; Hozumi, Y.; Yin, C.; Wei, G.W. Analysis of SARS-CoV-2 Mutations in the United States Suggests Presence of Four Substrains and Novel Variants. Commun Biol 2021, 4. [Google Scholar] [CrossRef]
- Harper, H.; Burridge, A.; Winfield, M.; Finn, A.; Davidson, A.; Matthews, D.; Hutchings, S.; Vipond, B.; Jain, N.; Edwards, K.; et al. Detecting SARS-CoV-2 Variants with SNP Genotyping. PLoS One 2021, 16, e0243185. [Google Scholar] [CrossRef]
- Yuan, F.; Wang, L.; Fang, Y.; Wang, L. Global SNP Analysis of 11,183 SARS-CoV-2 Strains Reveals High Genetic Diversity. Transbound Emerg Dis 2021, 68, 3288–3304. [Google Scholar] [CrossRef]
- Brandt, D.; Simunovic, M.; Busche, T.; Haak, M.; Belmann, P.; Jünemann, S.; Schulz, T.; Klages, L.J.; Vinke, S.; Beckstette, M.; et al. Multiple Occurrences of a 168-Nucleotide Deletion in SARS-CoV-2 ORF8, Unnoticed by Standard Amplicon Sequencing and Variant Calling Pipelines. Viruses 2021, 13, 1870. [Google Scholar] [CrossRef]
- Chen, S.; Zheng, X.; Zhu, J.; Ding, R.; Jin, Y.; Zhang, W.; Yang, H.Y.; Zheng, Y.; Li, X.; Duan, G. Extended ORF8 Gene Region Is Valuable in the Epidemiological Investigation of Severe Acute Respiratory Syndrome–Similar Coronavirus. J Infect Dis 2020, 222, 223–233. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.L. Origin and Evolution of Pathogenic Coronaviruses. Nat Rev Microbiol 2019, 17, 181. [Google Scholar] [CrossRef]
- Rashid, F.; Xie, Z.; Suleman, M.; Shah, A.; Khan, S.; Luo, S. Roles and Functions of SARS-CoV-2 Proteins in Host Immune Evasion. Front Immunol 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Fogeron, M.-L.; Montserret #, R.; Zehnder, J.; Nguyen, M.-H.; Dujardin, M.; Brigandat, L.; Cole, L.; Ninot-Pedrosa, M.; Lecoq, L.; Böckmann, A. SARS-CoV-2 ORF7b: Is a Bat Virus Protein Homologue a Major Cause of COVID-19 Symptoms? BiorXiV 2021. [Google Scholar] [CrossRef]
- Pereira, F. Evolutionary Dynamics of the SARS-CoV-2 ORF8 Accessory Gene. Infection, Genetics and Evolution 2020, 85, 104525. [Google Scholar] [CrossRef] [PubMed]
- Flower, T.G.; Buffalo, C.Z.; Hooy, R.M.; Allaire, M.; Ren, X.; Hurley, J.H. Structure of SARS-CoV-2 ORF8, a Rapidly Evolving Immune Evasion Protein. Proceedings of the National Academy of Sciences 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, X.; Sun, Y.; Zhao, H.; Cheng, F.; Wang, J.; Yang, F.; Hu, J.; Zhang, H.; Wang, C. chen; et al. SARS-CoV-2 ORF8 Reshapes the ER through Forming Mixed Disulfides with ER Oxidoreductases. Redox Biol 2022, 54. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Feng, Y.; Chen, H.; Luk, H.K.H.; Yang, W.-H.; Li, K.S.M.; Zhang, Y.-Z.; Huang, Y.; Song, Z.-Z.; Chow, W.-N.; et al. Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination. J Virol 2015, 89, 10532–10547. [Google Scholar] [CrossRef]
- DeRonde, S.; Deuling, H.; Parker, J.; Chen, J. Identification of a Novel SARS-CoV-2 Variant with a Truncated Protein in ORF8 Gene by next Generation Sequencing. Sci Rep 2022, 12. [Google Scholar] [CrossRef]
- Molecular Evolution of the SARS Coronavirus During the Course of the SARS Epidemic in China. Science (1979) 2004, 303, 1666–1669. [CrossRef]
- Velazquez-Salinas, L.; Zarate, S.; Eberl, S.; Gladue, D.P.; Novella, I.; Borca, M. V.; Tramontano, E.; Negroni, M.; Foley, B.T.; Velazquez-Salinas, L.; et al. Positive Selection of ORF1ab, ORF3a, and ORF8 Genes Drives the Early Evolutionary Trends of SARS-CoV-2 During the 2020 COVID-19 Pandemic. Frontiers in microbiology 2020, 11, 550674. [Google Scholar] [CrossRef]
- Brandt, D.; Simunovic, M.; Busche, T.; Haak, M.; Belmann, P.; Jünemann, S.; Schulz, T.; Klages, L.J.; Vinke, S.; Beckstette, M.; et al. Multiple Occurrences of a 168-Nucleotide Deletion in SARS-CoV-2 ORF8, Unnoticed by Standard Amplicon Sequencing and Variant Calling Pipelines. Viruses 2021, 13, 1870. [Google Scholar] [CrossRef]
- Mazur-Panasiuk, N.; Rabalski, L.; Gromowski, T.; Nowicki, G.; Kowalski, M.; Wydmanski, W.; Szulc, P.; Kosinski, M.; Gackowska, K.; Drweska-Matelska, N.; et al. Expansion of a SARS-CoV-2 Delta Variant with an 872 Nt Deletion Encompassing ORF7a, ORF7b and ORF8, Poland, July to August 2021. Eurosurveillance 2021, 26. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Omasits, U.; Ahrens, C.H.; Müller, S.; Wollscheid, B. Protter: Interactive Protein Feature Visualization and Integration with Experimental Proteomic Data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Zinzula, L. Lost in Deletion: The Enigmatic ORF8 Protein of SARS-CoV-2. Biochem Biophys Res Commun 2021, 538, 116. [Google Scholar] [CrossRef]
- Hisner, R.; Gueli, F.; Peacock, T.P. Repeated Loss of ORF8 Expression in Circulating SARS-CoV-2 Lineages. Available online: https://virological.org/t/repeated-loss-of-orf8-expression-in-circulating-sars-cov-2-lineages/931 (accessed on 29 June 2023).

| ID | Age | Sex | Sequencing date |
Relevant pathology | COVID-19 treatments |
Other treatments |
|---|---|---|---|---|---|---|
| 1 | 59 | F | October 2022 | RA, PV, RSV coinfection, bacterial pneumonia | Tixagevimab+ Cilgavimab | Methylprednisolone |
| 2 | 64 | F | October 2022 | AKI, peritoneal carcinomatosis | None | Carboplatinum |
| 3 | 52 | F | October 2022 | None | None | None |
| 4 | 74 | M | October 2022 | Non-Hodgkin lymphoma | Tixagevimab+ Cilgavimab | R-COMP |
| 5 | 83 | F | October 2022 | Aspiration-associated pneumonia | Remdesivir | None |
| 6 | 54 | F | August 2022 | None | None | None |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





